Abstract
Despite significant interest and past work to elucidate the phylogeny and photochemistry of species of the Heliobacteriaceae, genomic analyses of heliobacteria to date have been limited to just one published genome, that of the thermophilic species Heliobacterium (Hbt.) modesticaldum str. Ice1T. Here we present an analysis of the complete genome of a second heliobacterium, Heliorestis (Hrs.) convoluta str. HHT, an alkaliphilic, mesophilic, and morphologically distinct heliobacterium isolated from an Egyptian soda lake. The genome of Hrs. convoluta is a single circular chromosome of 3.22 Mb with a GC content of 43.1% and 3263 protein-encoding genes. In addition to culture-based observations and insights gleaned from the Hbt. modesticaldum genome, an analysis of enzyme-encoding genes from key metabolic pathways supports an obligately photoheterotrophic lifestyle for Hrs. convoluta. A complete set of genes encoding enzymes for propionate and butyrate catabolism and the absence of a gene encoding lactate dehydrogenase distinguishes the carbon metabolism of Hrs. convoluta from its close relatives. Comparative analyses of key proteins in Hrs. convoluta, including cytochrome c553 and the Fo alpha subunit of ATP synthase, with those of related species reveal variations in specific amino acid residues that likely contribute to the success of Hrs. convoluta in its highly alkaline environment.
1. Introduction
Heliobacteria comprise a unique group of strictly anaerobic, anoxygenic phototrophs that have been isolated from a wide diversity of soil and aquatic habitats [1,2,3,4]. Unlike all other phototrophic bacteria, heliobacteria use bacteriochlorophyll (Bchl) g as the chief chlorophyll pigment for phototrophic growth [5], but despite their ability to use light as an energy source, heliobacteria are apparently incapable of autotrophic growth and, thus, are obligate heterotrophs [4,6]. Heliobacteria are the only phototrophs of the large bacterial phylum Firmicutes [4,7,8], and although they typically stain Gram-negatively, thin sections of cells of heliobacteria exhibit a Gram-positive cell wall morphology [9,10]. In addition to these distinctive properties, cells of heliobacteria are able to differentiate into heat-resistant endospores [4,11], and some heliobacteria have also demonstrated the ability to reduce toxic metals, such as Hg2+, and therefore may be useful for applications in bioremediation [12,13].
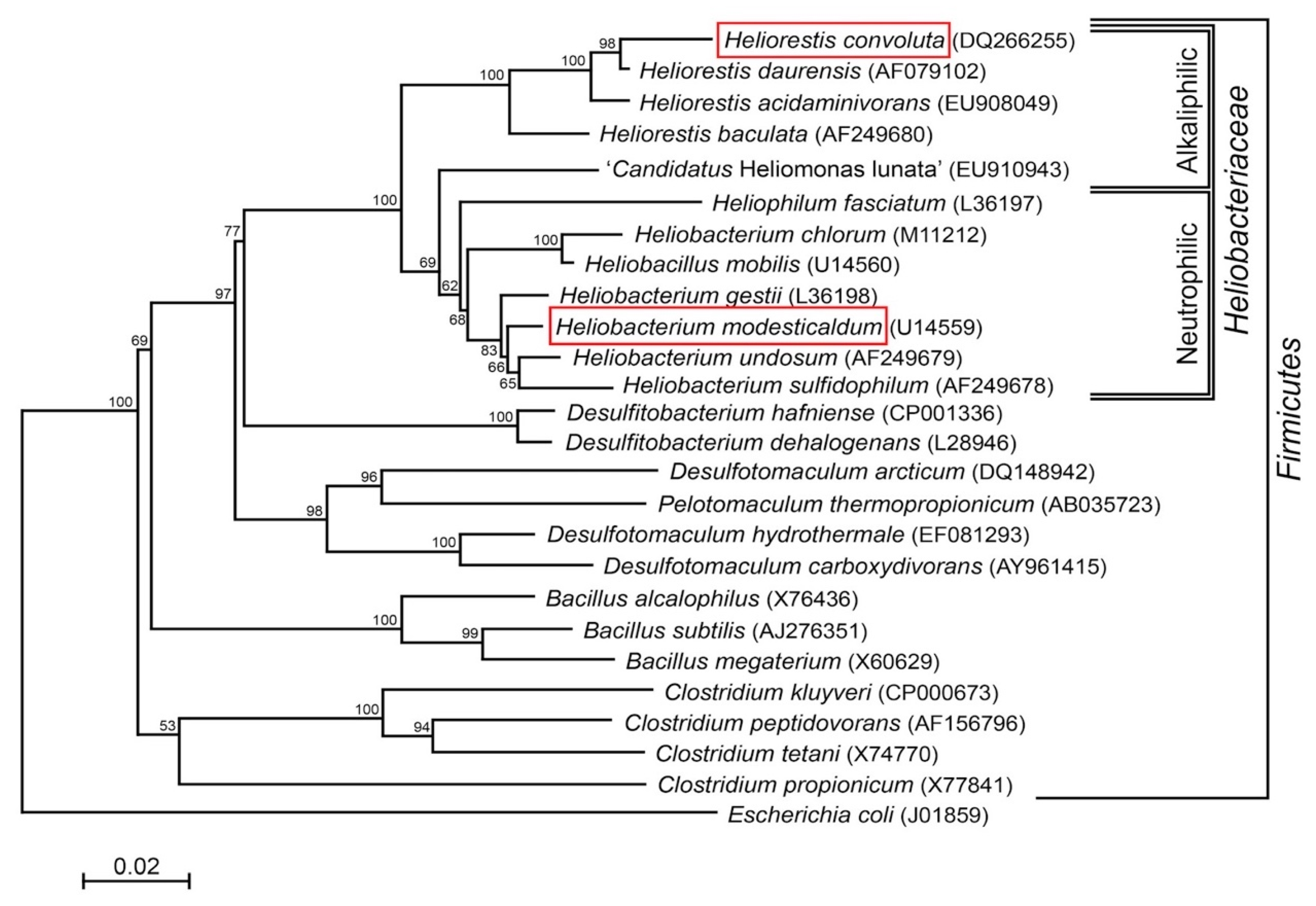
Species of Heliobacteriaceae can be divided into two physiological groups—neutrophiles and alkaliphiles—that track closely with their phylogeny [7] (Figure 1). Included in the neutrophilic clade, the moderate thermophile Hbt. modesticaldum was the first heliobacterium to have its genome sequenced and, with its simple phototrophic machinery consisting of a type I reaction center (RC) and no peripheral antenna photocomplex, has been a model organism for studies of photosynthesis and related photochemistry [6,14]. Like other neutrophilic heliobacteria, Hbt. modesticaldum exhibits both phototrophic growth in the light and chemotrophic growth in the dark [3,15,16,17].
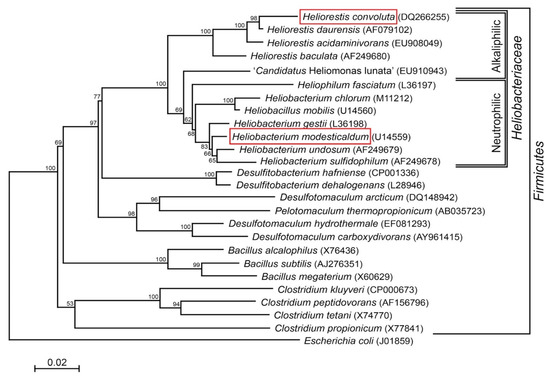
Figure 1.
Phylogenetic (16S rRNA) tree of Heliorestis convoluta and related Firmicutes. Heliobacteria, the only phototrophic Firmicutes, are divided into alkaliphilic and neutrophilic species. Heliobacterium modesticaldum (boxed) is the model organism for physiological and biochemical studies of the heliobacteria; Hrs. convoluta (boxed) is the first alkaliphilic heliobacterium to have a described genome. Note that the branching pattern shown here suggests a possible alkaliphilic origin to the heliobacteria, as previously discussed by Sattley and Swingley [7]. The weighted neighbor-joining method [18] and Jukes-Cantor corrected distance model were used for tree construction. Nodes represent bootstrap values (≥50%) based on 100 replicates, and Escherichia coli was used to root the tree. GenBank accession numbers for each sequence used in the analysis are shown in parentheses, adapted from Sattley and Swingley [7], Adv. Bot. Res. 2013, 66, 67–97, Copyright 2013 Elsevier Ltd.
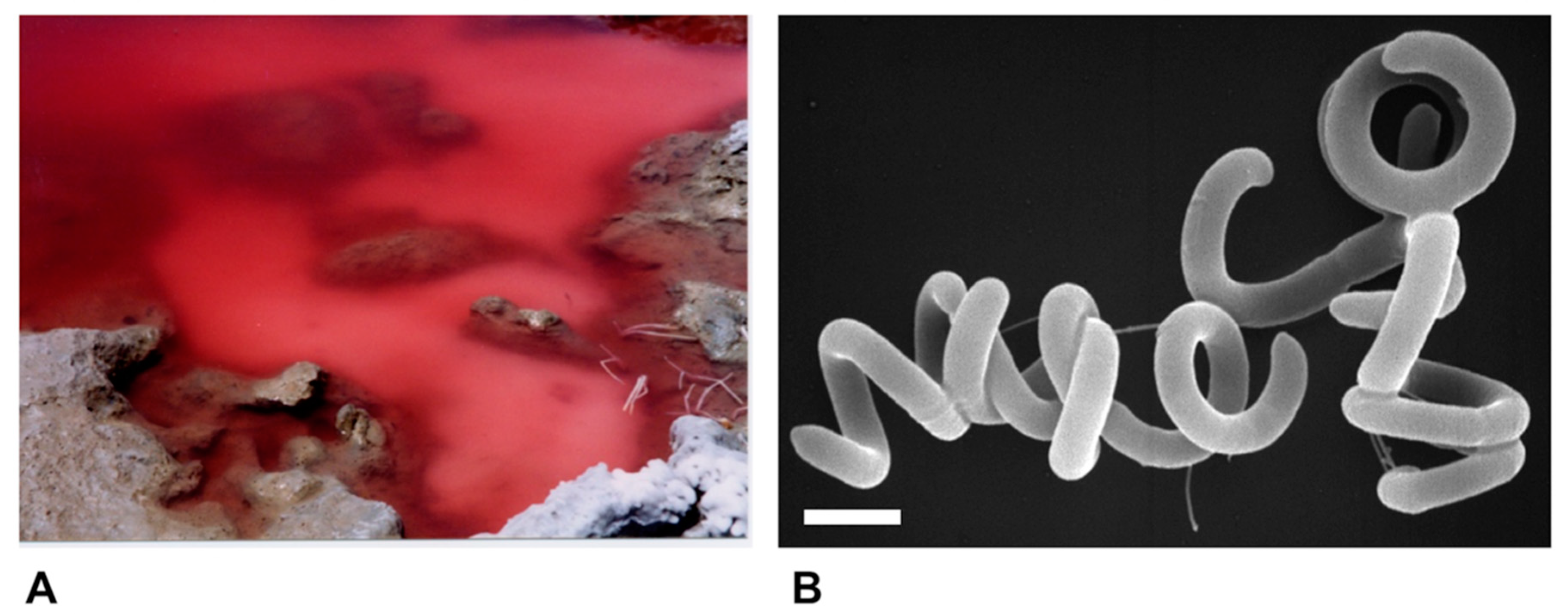
Species of alkaliphilic heliobacteria grow optimally between pH 8–9.5 and, unlike neutrophilic heliobacteria, are obligate photoheterotrophs, using light and organic compounds for growth but incapable of chemotrophic growth in darkness [19,20,21,22]. Consistent with other alkaliphilic heliobacteria originating from the soils and waters of soda lakes [19,20,22], Hrs. convoluta str. HHT was isolated from the shore of the alkaline (pH 10) Lake El Hamra (Figure 2A), located in the Wadi El Natroun region of northern Egypt [21]. In the past, the saline lakes of the Wadi El Natroun have also been a fertile source of alkaliphilic purple bacteria, yielding many extremely alkaliphilic (and in some cases also extremely halophilic) species, including in particular, new species of the genus Halorhodospira [23,24,25]. However, Hrs. convoluta is the first heliobacterium to originate from these unusual lakes. Experimental work with Hrs. convoluta revealed motile cells having an unusual tightly coiled morphology (Figure 2B) and displaying a mesophilic (optimal growth at 33 °C) and alkaliphilic (optimal growth at pH 8.5–9) physiology [21].
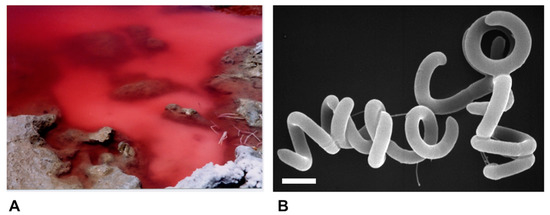
Figure 2.
Habitat and cells of Heliorestis convoluta strain HHT. (A) Red bloom of alkaliphilic Bacteria and Archaea on the shore of Lake El Hamra, Wadi Natroun, Egypt. M.T.M. sampled this bloom in May 2001, and enrichments for heliobacteria yielded Hrs. convoluta. The bloom is about 2 m in diameter.; (B) Scanning electron micrograph of cells of Hrs. convoluta strain HHT. A cell of Hrs. convoluta is about 0.5 μm in diameter and coils are of variable length. Scale bar = 1 μm.
To complement the analysis of the genome sequence of Hbt. modesticaldum [6,26], we present here a comparative analysis of the genome of Hrs. convoluta. Although a number of highly conserved genes encoding proteins that coordinate key processes in the cell (e.g., phototrophy and central carbon metabolism) are shared between these species, a close comparison of the two heliobacterial genomes revealed several genes encoding functions in carbon metabolism, biotin biosynthesis, nitrogen and sulfur assimilation, and carotenoid biosynthesis that are not held in common by these heliobacteria, which inhabit vastly different extreme environments. In addition, a comparative analysis of selected cytochrome and ATP synthase proteins in Hrs. convoluta revealed adaptations that likely facilitate its alkaliphilic lifestyle. The availability of a second heliobacterial genome, as well as the recent development of a genetic system in Hbt. modesticaldum [14], paves the way for increasing our understanding of the unique metabolism and physiology of heliobacteria.
2. Materials and Methods
Total genomic DNA from Hrs. convoluta str. HHT (ATCC BAA-1281 and DSMZ 19787) [21] was isolated through proteinase K treatment and subsequent phenol extraction. Complete genome sequencing was performed using a random shotgun approach, and reads were assembled using Velvet v. 2010 [27]. Pyrosequencing on a Roche-454 GS20 sequencer (Hoffman-La Roche AG, Basel, Switzerland) provided 14-fold genome coverage, and an additional 35-fold coverage was generated by the Illumina GAIIx platform.
Annotation of the Hrs. convoluta genome was performed in accordance with the Prokaryotic Annotation Pipeline of the University of Maryland School of Medicine’s Institute for Genome Sciences [28]. This pipeline employs Glimmer for gene identification and then searches the protein sequences with BLAST-extend-repraze (BER; a combination of BLAST and Smith–Waterman algorithms) to generate pairwise alignments, Hidden Markov Model (HMM), transmembrane (Tm) HMM, and SignalP predictions. An automated process employing the Pfunc evidence hierarchy is used to assign functional annotations. Manual verification of automated annotations was facilitated through the online tool Manatee [29] in conjunction with online databases including the Kyoto Encyclopedia of Genes and Genomes (KEGG), the Braunschweig Enzyme Database (BRENDA), MetaCyc, and Uniprot. The National Center for Biotechnology Information (NCBI) database was accessed to retrieve gene and protein sequences from related species for comparative analyses with corresponding genes in the genome of Hrs. convoluta.
The phylogenetic tree was generated as described in the legend to Figure 1. Genome statistics were compiled using the Pfam database v. 30.0 [30], the SignalP database v. 4.1 [31], the TMHMM database v. 2.0 [32], and CRISPRFinder v. 2.0 [33]. This complete genome sequence project has been deposited at DDBJ/EMBL/GenBank under accession number CP045875.
3. Results and Discussion
3.1. Genome Properties
The 3,218,981 base-pair (bp) genome of Heliorestis convoluta str. HHT is organized into a single circular chromosome with no plasmids (Table 1). The 43.1% GC content of Hrs. convoluta is among the lowest of all heliobacteria (41%–57.7%) and is typical of alkaliphilic species of this group of phototrophs [4]. Nearly 87% of the Hrs. convoluta genome content is protein-encoding, with a total of 3263 protein coding genes at an average length of 855 nucleotides (Table 1). The genome contains nine ribosomal RNA (rRNA) genes, including multiple copies each of 5S, 16S (two full and one partial), and 23S rRNA, which are distributed randomly on the chromosome. Nearly 11% of the open reading frames (ORFs) were of unknown enzyme specificity or function, and 28% of genes were annotated as hypothetical. The role category breakdown of protein-encoding genes of Hrs. convoluta is shown in Table 2.

Table 1.
Comparison of genome features of Heliorestis convoluta str. HHT and Heliobacterium modesticaldum str. Ice1T [6].

Table 2.
Functional role categories of Heliorestis convoluta str. HHT genes.
Genes encoding a total of 105 transfer RNAs (tRNAs) were identified in the Hrs. convoluta genome, as well as genes encoding all twenty common aminoacyl-tRNA synthetases except asparaginyl-tRNA synthetase, which could not be confirmed. However, genes encoding aspartyl/glutamyl-tRNA amidotransferase (gatABC) were identified in Hrs. convoluta and, as proposed for Hbt. modesticaldum [6], may encode a protein that compensates for the missing asparaginyl-tRNA synthetase by converting aspartyl-tRNA to asparaginyl-tRNA [34,35].
3.2. Central Carbon Metabolism
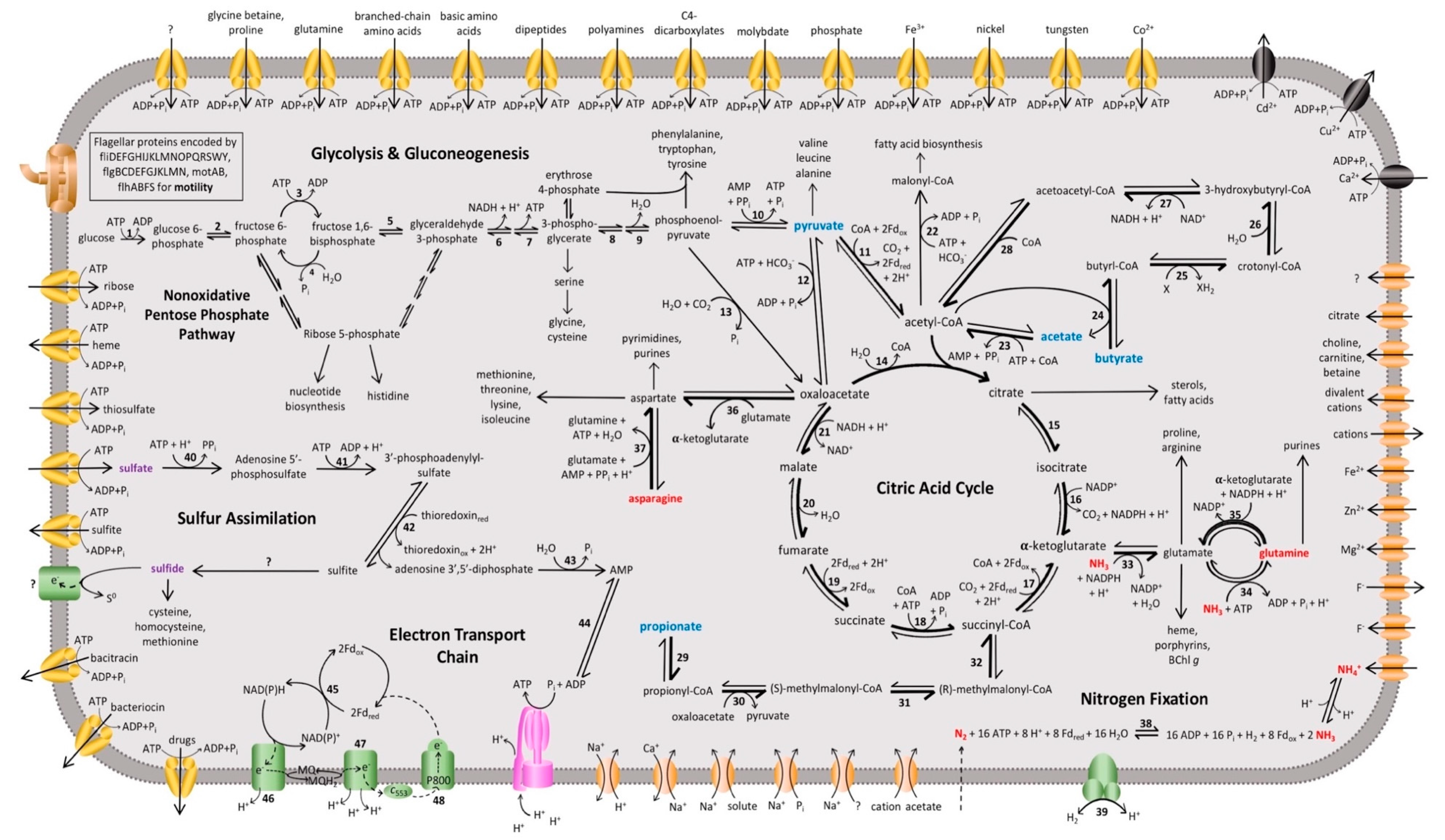
Analysis of the Hrs. convoluta genome confirmed culture-based observations of the limited set of carbon sources able to support light-driven growth of this species [21]. As an obligate photoheterotroph, Hrs. convoluta grows only in anoxic, light conditions when supplied with mineral media containing CO2 plus acetate, pyruvate, propionate, or butyrate as organic carbon sources [21]. Of the 12 described species of heliobacteria (Figure 1), only Heliorestis acidaminivorans, Heliorestis daurensis, and Hrs. convoluta are capable of propionate photoassimilation [4,19,21,22]. Genes encoding enzymes of the methylmalonyl pathway, which converts propionyl-coenzyme A (CoA) to succinyl-CoA for propionate assimilation, were identified in the Hrs. convoluta genome (Figure 3). Although a gene encoding propionyl-CoA carboxylase, which is thought to catalyze the first step in the proposed pathway [36,37], was not identified in the Hrs. convoluta genome, a gene predicted to encode methylmalonyl-CoA carboxyltransferase (FTV88_3237), which could circumvent this deficiency, was identified.
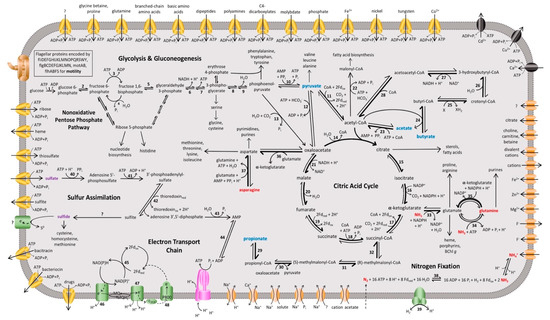
Figure 3.
Overview of the proposed metabolic pathways and membrane transporters in Heliorestis convoluta. Carbon (blue), nitrogen (red), and sulfur (purple) sources are catabolized or assimilated for phototrophic growth of Hrs. convoluta. The predicted dominant direction of metabolic flow is shown by bolded arrows. Numbers signify enzymes identified in the genome of Hrs. convoluta, whereas question marks indicate unidentified but anticipated enzymes catalyzing the respective reaction. Enzymes involved in glycolysis or gluconeogenesis include (1) glucokinase, (2) glucose-6-phosphate isomerase, (3) 6-phosphofructokinase, (4) fructose 1,6-bisphosphatase, (5) fructose-1,6-bisphosphate aldolase, (6) glyceraldehyde-3-phosphate dehydrogenase, (7) phosphoglycerate kinase, (8) phosphoglycerate mutase, (9) enolase, and (10) pyruvate-phosphate dikinase. CAC enzymes are (11) pyruvate:ferredoxin oxidoreductase, (12) pyruvate carboxylase, (13) phosphoenolpyruvate carboxylase, (14) citrate (re)-synthase, (15) aconitate hydratase, (16) NADP+-dependent isocitrate dehydrogenase, (17) 2-oxoglutarate synthase/2-oxoglutarate:ferredoxin oxidoreductase, (18) succinyl-CoA synthetase, (19) succinate dehydrogenase/fumarate reductase, (20) fumarate hydratase, and (21) NAD+-dependent malate dehydrogenase. Acetyl-CoA metabolism is carried out by (22) acetyl-CoA carboxylase and (23) acetyl-CoA synthetase. Butyrate metabolism enzymes include (24) CoA transferase, (25) acyl-CoA dehydrogenase, (26) enoyl-CoA hydratase, (27) 3-hydroxybutyryl-CoA dehydrogenase, and (28) acetyl-CoA C-acetyltransferase. Propionate metabolism is catabolized by (29) CoA transferase, (30) methylmalonyl-CoA carboxytransferase, (31) methylmalonyl-CoA epimerase, and (32) methylmalonyl-CoA mutase. Amino acid metabolism enzymes are (33) NADP+-specific glutamate dehydrogenase, (34) glutamine synthetase, (35) NADPH-dependent glutamate synthase, (36) pyridoxal phosphate-dependent aminotransferase, and (37) asparagine synthase. The enzymes (38) nitrogenase and (39) uptake [NiFe] hydrogenase catalyze nitrogen fixation and H2 oxidation, respectively, and sulfur assimilation is performed by (40) sulfate adenyltransferase, (41) adenylyl-sulfate kinase, (42) phosophoadenylyl-sulfate reductase, (43) bifunctional oligoribonuclease and PAP phosphatase, and (44) adenylate kinase. Finally, the electron transport chain includes (45) ferredoxin:NADP+ reductase, (46) NADH:quinone oxidoreductase, (47) cytochrome bc complex, and the (48) light-harvesting reaction center. Membrane proteins include ABC transporters (yellow), P-type ATPases (black), ATP synthase (pink), flagellar and motor proteins (brown), other transporters (orange), and other membrane proteins (green).
Unlike other Heliorestis species, Hrs. convoluta and a few other heliobacteria can use butyrate as a carbon source [19,20,21,38]. Analysis of the Hrs. convoluta genome revealed genes encoding enzymes that catabolize butyrate to acetyl-CoA for incorporation into the citric acid cycle (CAC) [39] (Figure 3). Genes encoding butyryl-CoA:acetate CoA transferase, which catalyzes the conversion of butyrate to butyryl-CoA in butyrate catabolism [39], and propionyl-CoA synthetase, which converts propionate to propionyl-CoA in propionate catabolism [37], were not identified in the genome of Hrs. convoluta. However, an experimentally characterized butyryl-CoA:acetate CoA transferase from Desulfosarcina cetonica [39] showed 47% amino acid sequence identity with 4-hydroxybutyrate CoA-transferase (FTV88_0224) from Hrs. convoluta. In addition, the product of a gene annotated as acetyl-coenzyme A synthetase (FTV88_0994) in Hrs. convoluta showed 37% sequence identity with propionyl-CoA synthetase from Salmonella enterica and contained the conserved lysine residue (Lys592) required in the initial reaction of propionate catabolism [40]. These findings suggest possible roles for 4-hydroxybutyrate CoA-transferase and acetyl-coenzyme A synthetase in butyrate and propionate catabolism, respectively, in Hrs. convoluta.
Although capable of growth on pyruvate, Hrs. convoluta str. HHT is unable to grow photoheterotrophically on lactate [21], a phenotype distinct from that of most other heliobacteria and the result of an underlying genetic deficiency. In this connection, a gene encoding a putative L-lactate dehydrogenase in Hbt. modesticaldum [6] showed no meaningful similarity to any genes in Hrs. convoluta. In addition to lactate, no growth was detected when alcohols of any kind were used as sole carbon source in cultures of strain HHT [21]. Despite this observation, genes encoding alcohol dehydrogenase and aldehyde dehydrogenase were annotated in the Hrs. convoluta genome and, thus, could potentially play a role in non-energetic processes, such as detoxification.
Although a full complement of genes encoding enzymes of the glycolytic and nonoxidative pentose phosphate pathways was present in the Hrs. convoluta genome (Figure 3), various common sugars did not support photoheterotrophic growth of strain HHT [21]. An inability to use sugars was also originally reported for Hbt. modesticaldum [16], but later experimentation showed that Hbt. modesticaldum utilized the glycolytic pathway when D-ribose, D-glucose, or D-fructose were supplied with low levels of yeast extract [41]. Although no gene encoding a hexose transporter was annotated in the Hrs. convoluta genome, a putative ribose ABC transporter complex (FTV88_0053, FTV88_0054, FTV88_0055) was identified and may allow for carbohydrate transport [41]. As genes encoding glycolytic pathway enzymes are present in the Hrs. convoluta genome, it is tempting to speculate that the alkaliphile can utilize sugars in a manner similar to Hbt. modesticaldum. The absence of genes encoding glucose 6-phosphate dehydrogenase and 6-phosphogluconolactonase suggest incomplete Entner-Doudoroff and oxidative pentose phosphate pathways, which was also the case for Hbt. modesticaldum [6].
It is likely that Hrs. convoluta can catalyze many of the steps in the CAC based on biochemical studies of Hbt. modesticaldum [42] and high sequence similarity of key CAC enzymes between the two species (Figure 3). However, since both Hrs. convoluta and Hbt. modesticaldum lack a gene encoding pyruvate dehydrogenase for oxidizing pyruvate to acetyl-CoA, this reaction in heliobacteria is likely catalyzed by the enzyme pyruvate:ferredoxin oxidoreductase (PFOR); the gene encoding PFOR in Hrs. convoluta (FTV88_3370) shares 61% sequence identity to an orthologous gene in Hbt. modesticaldum [6]. Furthermore, an unusual citrate synthase, citrate (re)-synthase, which specifically catalyzes the addition of the acetyl moiety from acetyl-CoA to the re face of the ketone carbon of oxaloacetate [a stereospecificity opposite to that of citrate (si)-synthase], has been identified in several clostridia and other strictly anaerobic Firmicutes, including Hbt. modesticaldum [42]. In Hrs. convoluta, a gene (FTV88_1447) having high amino acid sequence identity (81%) to the gene encoding citrate (re)-synthase (HM1_2993) in Hbt. modesticaldum supports the presence of citrate (re)-synthase in Hrs. convoluta and suggests this unusual form of citrate synthase is common to all heliobacteria.
In regards to photoautotrophic capacity, no genes encoding enzymes of any form of the Calvin-Benson cycle, including ribulose 1,5-bisphosphate carboxylase and phosphoribulokinase, were identified in the Hrs. convoluta genome. In addition, the lack of genes encoding key enzymes of other autotrophic pathways, such as malyl-CoA lyase (3-hydroxypropionate/4-hydroxybutyrate pathway) and acetyl-CoA synthase (Wood-Ljungdahl pathway), also prevents Hrs. convoluta from assimilating CO2 into organic carbon molecules for growth. The capacity for CO2 fixation by the reverse CAC, as observed in green sulfur bacteria [43], is apparently disrupted by the absence of a gene encoding ATP-citrate lyase. Although an ORF identified as a citrate lyase family protein (FTV88_0308) was annotated in the Hrs. convoluta genome based on sequence identities of approximately 50% with corresponding genes from other Firmicutes (but having no similarity to genes in Hbt. modesticaldum), biochemical analysis of this gene product would be required to assess its activity and role, if any, in metabolic pathways of Hrs. convoluta. Although anapleurotic CO2 assimilation has been shown in heliobacteria supplied with usable organic carbon sources [44], cultures of Hrs. convoluta strain HHT, like all other cultured heliobacteria, were unable to grow using CO2 as sole carbon source [21], thus supporting the premise that heliobacteria require an organic carbon source during phototrophic growth.
In addition to phototrophy, neutrophilic heliobacteria are able to grow chemotrophically in the dark by pyruvate fermentation [4]. Interestingly, however, the capacity for pyruvate fermentation has not been observed in any alkaliphilic heliobacterial isolate to date, including Hrs. convoluta [4,15,17,21]. Studies have suggested that the neutrophile Hbt. modesticaldum carries out substrate-level phosphorylation via acetyl-CoA conversion to acetate in dark, anoxic (fermentative) conditions through the activity of phosphotransacetylase (PTA) and acetate kinase (ACK) [15,17,41]. A gene encoding ACK (FTV88_2009) was annotated in the genome of Hrs. convoluta and has 67% sequence identity to a corresponding gene in Hbt. modesticaldum. However, a gene encoding PTA could not be identified in either Hrs. convoluta or Hbt. modesticaldum. Therefore, the genetic determinants that coordinate pyruvate fermentation in neutrophilic heliobacteria but are apparently absent from alkaliphilic heliobacteria remain unidentified.
Three Hrs. convoluta genes encoding acetyl-CoA synthetase (ACS) were identified in the genome, one of which showed 87% amino acid sequence identity with the corresponding gene in Hbt. modesticaldum. Activity of ACS in Hbt. modesticaldum cell extracts was detected only under phototrophic (light/anoxic) conditions, and expression levels of the ACS gene decreased when the bacterium was cultured in darkness [41], thus indicating that, although technically reversible, ACS activity is predominately skewed toward the production of acetyl-CoA from acetate (Figure 3). Activity of ACS therefore allows both Hbt. modesticaldum and Hrs. convoluta to grow photoheterotrophically using acetate as sole carbon source [16,21].
In contrast to all other heliobacteria, which require biotin for growth, Hrs. convoluta and close relative Hrs. acidaminivorans (Figure 1) have no growth factor requirements [21,22]. The presence of a full complement of genes (bioABCDF) encoding enzymes for biotin biosynthesis allows Hrs. convoluta to synthesize biotin, thereby supporting culture-based observations [21]. By contrast, analysis of the Hbt. modesticaldum genome revealed the absence of two key genes for biotin biosynthesis, bioC and bioF, thus explaining the absolute requirement for biotin in that species [16].
3.3. Nitrogen Metabolism
Hrs. convoluta is strongly diazotrophic [21], and as in Hbt. modesticaldum, genes for nitrogen fixation are grouped into a single nif gene cluster containing nifI1, nifI2, nifH, nifD, nifK, nifE, nifN, nifX, fdxB, nifB, and nifV [6]. Each of these genes shows between 63% and 93% sequence identity and analogous gene synteny to corresponding genes in Hbt. modesticaldum. A study with Paenibacillus sp. WLY78—also an endospore-former within the phylum Firmicutes—concluded that nine genes (nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA, nifV), which were grouped into a single gene cluster, are essential to synthesize a catalytically-active nitrogenase for dinitrogen assimilation [45]. All of these nitrogen fixation genes, except for hesA, were identified in the Hrs. convoluta and Hbt. modesticaldum genomes. Since HesA is proposed to play a role in metallocluster biosynthesis [45], it is possible that a gene (FTV88_2056) located outside of the nif gene cluster and encoding a putative dinitrogenase Fe/Mo cofactor biosynthesis protein fills this role in Hrs. convoluta. This encoded protein showed high (~64%) sequence identity to a corresponding protein in Hrs. acidaminivorans and over 50% sequence similarity to that from a variety of nonphototrophic Firmicutes, but it showed no significant similarity to proteins encoded by Hbt. modesticaldum.
Research on Hbt. modesticaldum revealed changes in expression levels of numerous genes essential for various metabolic, biosynthetic, and other cellular pathways when the organism was grown under N2-fixing conditions [46]. This diazotrophic effect likely exists in other heliobacteria as well, including Hrs. convoluta. In terms of regulation of nitrogen fixation genes, however, it is interesting that neither orf1 nor nifA, which encode regulatory proteins for the expression of nif structural genes [47,48], could be identified in the Hrs. convoluta genome. In Hbt. modesticaldum, the orf1 gene product likely regulates the expression of nif genes when levels of fixed nitrogen are too low to support non-diazotrophic growth of the organism [16,48]. It is possible that Hrs. convoluta lacks the orf1 and nifA regulatory genes and instead employs only nifI1 (FTV88_2453) and nifI2 (FTV88_2454) to coordinate post-translational regulation of nitrogenase [49].
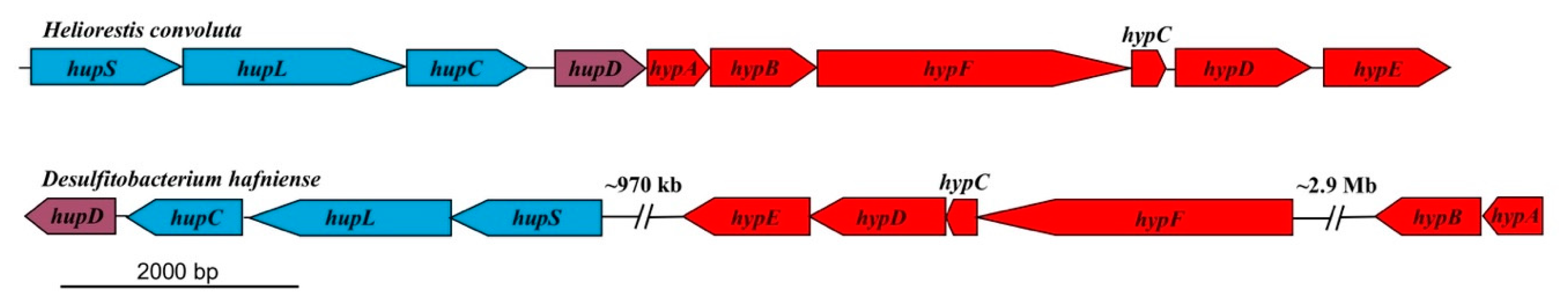
Hrs. convoluta and Hbt. modesticaldum both contain gene clusters (hypABCDEF and hupCDLS) that encode an uptake [NiFe] hydrogenase that can putatively catalyze the oxidation of H2 produced during nitrogen fixation [6] (Figure 3). The arrangement of these genes in Hrs. convoluta is identical to that reported for Hbt. modesticaldum [6], being organized into a single cluster instead of dispersed throughout different regions of the chromosome, as has been observed in the genomes of other Firmicutes (Figure 4).
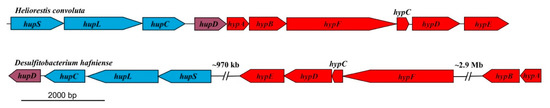
Figure 4.
Comparison of uptake [NiFe]-hydrogenase genes in related Firmicutes. The genes are concatenated within a single region in Heliorestis convoluta, but they are dispersed in different regions of the Desulfitobacterium hafniense chromosome. Colors: blue, [NiFe]-hydrogenase structural genes; purple, hydrogenase expression/formation; red, hydrogenase assembly/maturation. Adapted from Sattley et al. [6]. J. Bacteriol. 2008, 190, 4687–4696. Copyright 2008 American Society for Microbiology.
In addition to performing N2 fixation, cells of Hrs. convoluta strain HHT could assimilate ammonia, glutamine, and asparagine as nitrogen sources [21]. Accordingly, genes encoding the ammonium transporter protein Amt (FTV88_2595) and enzymes of the glutamine synthetase-glutamate synthase pathway, which incorporates ammonia in the formation of glutamine from glutamate [50,51] (Figure 3), were identified in the Hrs. convoluta genome. Following transport, glutamine can then be used for purine biosynthesis or, through the activity of NADPH-dependent glutamate synthase, can be condensed with α-ketoglutarate to yield two molecules of glutamate for other biosynthetic pathways [50,51]. In addition, a gene encoding NADP-specific glutamate dehydrogenase (FTV88_2506) enables Hrs. convoluta to assimilate ammonia when synthesizing glutamate directly from α-ketoglutarate (Figure 3). Finally, genes encoding a glutamine-hydrolyzing asparagine synthetase (FTV88_1161 and FTV88_3319), which converts asparagine and glutamate into aspartate and glutamine, respectively (Figure 3), allow for the use of asparagine as a nitrogen source. In contrast, aspartate and glutamate cannot serve as nitrogen sources for strain HHT [21]. Taken together, these findings suggest that, although the reactions are generally considered reversible, the enzymes catalyzing the conversion of asparagine to aspartate and glutamine to glutamate are physiologically unidirectional, strongly favoring the formation of aspartate and glutamate, respectively (shown as bolded arrows in Figure 3).
3.4. Assimilation of Sulfur
Growth studies indicate Hrs. convoluta is capable of assimilatory sulfate reduction [21]. Consistent with these observations, genomic analyses revealed that the pathway of assimilatory sulfate reduction in Hrs. convoluta begins with sulfate uptake using a sulfate/thiosulfate ABC transporter (cysAWTP). Typically, an enzyme encoded by cysD and cysN, sulfate adenyltransferase, catalyzes the assimilation of sulfate as adenosine phosphosulfate (APS) [52,53]. Hrs. convoluta lacks cysD, but genes encoding the bifunctional enzyme CysN/CysC (FTV88_1460 and FTV88_1458, respectively), which can also perform this function [54], are present. As shown in Figure 3, adenylyl-sulfate kinase (cysC) and phosphoadenylyl-sulfate reductase (cysH, FTV88_1461) catalyze the subsequent reaction to yield sulfite [52,53].
To produce sulfide for amino acid biosynthesis, sulfite must undergo further reduction through sulfite reductase [53]. However, a gene encoding sulfite reductase could not be identified, suggesting that Hrs. convoluta may employ an unusual reductase or an alternative mechanism to perform this reaction. Genes encoding all successive enzymes necessary to synthesize cysteine, homocysteine, and methionine from hydrogen sulfide were identified (data not shown). By comparison, the Hbt. modesticaldum genome lacked cysN, cysH, and sulfite reductase, supporting physiological studies indicating that Hbt. modesticaldum requires a reduced sulfur source for biosynthetic purposes [16].
Interestingly, cultures of Hrs. convoluta strain HHT were able to grow well in the presence of high levels of sulfide (10mM), with sulfide oxidation accompanied by the production of elemental sulfur globules during growth [21]. However, the pathway for this reaction remains unclear, as the Hrs. convoluta genome appears to lack genes encoding traditional sulfide oxidoreductases, such as the sulfide:quinone oxidoreductase (SQR) from the green sulfur bacterium Chlorobaculum (Chlorobium) tepidum that oxidizes H2S to S0 and reduces quinone [55], or sulfide:flavocytochrome c oxidoreductase from the purple sulfur bacterium Allochromatium vinosum that oxidizes sulfide to sulfur or polysulfides [56]. Thus, it is possible that Hrs. convoluta contains a novel sulfide oxidoreductase for this purpose.
3.5. Photosynthesis Genes and Pigment Biosynthesis
Heliobacteria synthesize bacteriochlorophyll (BChl) g, a pigment absorbing light maximally between 785 and 790 nm, for phototrophic growth [4]. Accordingly, genes encoding enzymes that catalyze the conversion of glutamic acid to divinyl protochlorophyllide (gltX, hemALBCDEN, and bchIDHME) for pigment biosynthesis (Figure 5) were annotated in Hrs. convoluta. However, as for Hbt. modesticaldum, neither of the genes encoding protoporphyrinogen oxidase (hemY or hemG), which catalyzes the oxidation of protoporphyrinogen to protoporphyrin, was identified in the Hrs. convoluta genome. Moreover, comparisons with hemG from Escherichia coli and hemY from Bacillus subtilis yielded no significant sequence identity to genes in the Hrs. convoluta genome. Due to the anaerobic nature of Hrs. convoluta, an alternative and unidentified enzyme likely acts as a dehydrogenase rather than an oxidase in this step of pigment biosynthesis. Studies with Desulfovibrio gigas, also a strict anaerobe, suggest that electron carriers, such as flavins and pyridine nucleotides, or electron-transport complexes, such as nitrite and fumarate reductases, do not use O2 as the electron acceptor in the conversion of protoporphyrinogen to protoporphyrin [57,58]. More recently, however, an alternative pathway that does not use protoporphyrin to synthesize heme has been described in Hbt. modesticaldum [59], and a similar mechanism likely exists in Hrs. convoluta.
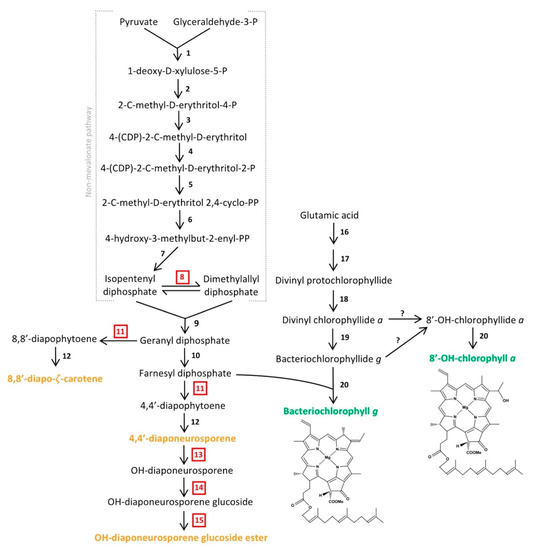
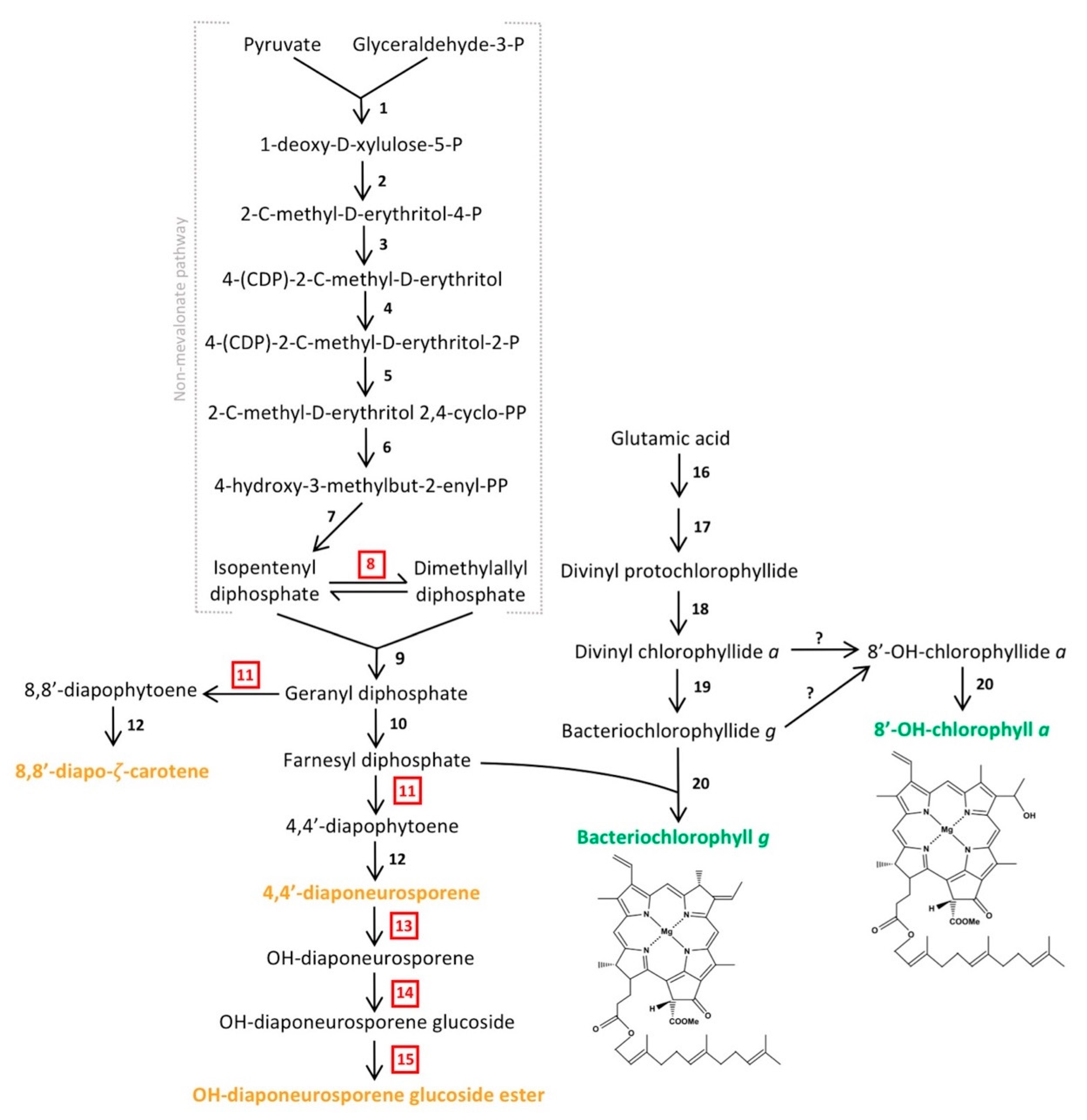
Figure 5.
Predicted biosynthetic pathway of major pigments in Heliorestis convoluta. The non-mevalonate pathway shows the synthesis of farnesyl diphosphate for either conversion into carotenoids (orange) or incorporation into the final chlorophyll (green) structures. The enzymes that catalyze each individual numbered reaction are (1) 1-deoxy-d-xylulose-5-P synthase, (2) 1-deoxy-d-xylulose-5-P reductoisomerase, (3) 4-(CDP)-2-C-methyl-d-erythritol synthase, (4) 4-(CDP)-2-C-methyl-d-erythritol kinase, (5) 2-C-methyl-d-erythritol 2,4-cyclo-PP synthase, (6) 4-hydroxy-3-methylbut-2-enyl-PP synthase, (7) 4-hydroxy-3-methylbut-2-enyl-PP reductase, (8) isomerase, (9) geranyl diphosphate synthase, (10) farnesyl diphosphate synthase, (11) 4,4′-diapophytoene synthase, (12) diapophytoene dehydrogenase, (13) hydratase, (14) glucosyl transferase, (15) esterase, (16) enzymes encoded by gltx and hemALBCDENYG genes, (17) enzymes encoded by bchIDHME genes, (18) protochlorophyllide reductase (bchLNB), (19) chlorophyllide reductase (bchXYZ), and (20) bacteriochlorophyll synthase (bchG). Red, boxed numbers represent enzymes not yet identified in the Hrs. convoluta genome but are proposed based on the predicted pathway. Adapted from Takaichi et al. [63], Arch. Microbiol. 2003, 179, 95–100. Copyright 2002 Springer Nature; Dubey et al. [64] J. Biosci. 2003, 28, 637–646. Copyright 2003 Springer Nature; Sattley et al. [6] J. Bacteriol. 2008, 190, 4687–4696. Copyright 2008 American Society for Microbiology; Sattley and Swingley [7], Adv. Bot. Res. 2013, 66, 67–97, Copyright 2013 Elsevier Ltd.; and Tsukatani et al. [60], Biochim. Biophys. Acta 2013, 1827, 1200–1204. Copyright 2013 Elsevier Ltd.
Following the synthesis of divinyl protochlorophyllide in Hrs. convoluta, genes encoding protochlorophyllide reductase (bchLNB), chlorophyllide reductase (bchXYZ), and bacteriochlorophyll synthase (bchG) are present to facilitate catalysis of subsequent reactions and produce BChl g. Previous work with Hbt. modesticaldum suggested the need for an isomerase in the interconversion between 8-vinyl bacteriochlorophyllide a and bacteriochlorophyllide g [6], but more recent experimental work with this species revealed the ability of chlorophyllide reductase to perform both reduction and isomerization of divinyl chlorophyllide a and circumvent the need for a separate isomerase in the biosynthesis of bacteriochlorophyllide g [60,61].
Heliobacteria also contain an alternative form of chlorophyll (Chl) a, 81-OH-Chl a, which was observed as a smaller absorption peak at 672 nm in spectrophotometric studies of Hrs. convoluta [21]. Whereas BChl g, a bacteriochlorin-type chlorophyll, is reduced at the C-7 and C-8 bond and has an ethylidene functional group at C-8 [5], 81-OH-Chl a, a chlorin, has a double bond connecting C-7 and C-8 with a hydroxyethyl group at C-8 [44]. BChl g and 81-OH-Chl a are putatively synthesized from a common precursor, divinyl chlorophyllide a [7,60].
Hydration of the C-8 vinyl group of divinyl chlorophyllide a is catalyzed by 8-vinyl chlorophyllide hydratase, and bacteriochlorophyll synthase catalyzes the addition of a farnesyl group to produce the mature 81-OH-Chl a [7,60]. However, a gene encoding chlorophyllide hydratase or an analogous enzyme was not identified in the genomes of Hrs. convoluta or Hbt. modesticaldum [6]. Hence, a possible alternative mechanism for 81-OH-Chl a synthesis includes steps of dehydrogenation and subsequent hydroxygenation of bacteriochlorophyllide g to produce 81-OH-chlorophyllide a [60], but genes encoding enzymes for this reaction were not identified in either Hrs. convoluta or Hbt. modesticaldum. Yet another possible mechanism for 81-OH-Chl a synthesis would require the irreversible conversion of BChl g into 81-OH-Chl a upon exposure to O2 and light [62]. However, as strict anaerobes, the viability of heliobacteria is compromised upon exposure to O2, and therefore this mechanism is unlikely as the major pathway for 81-OH-Chl a production [3,62].
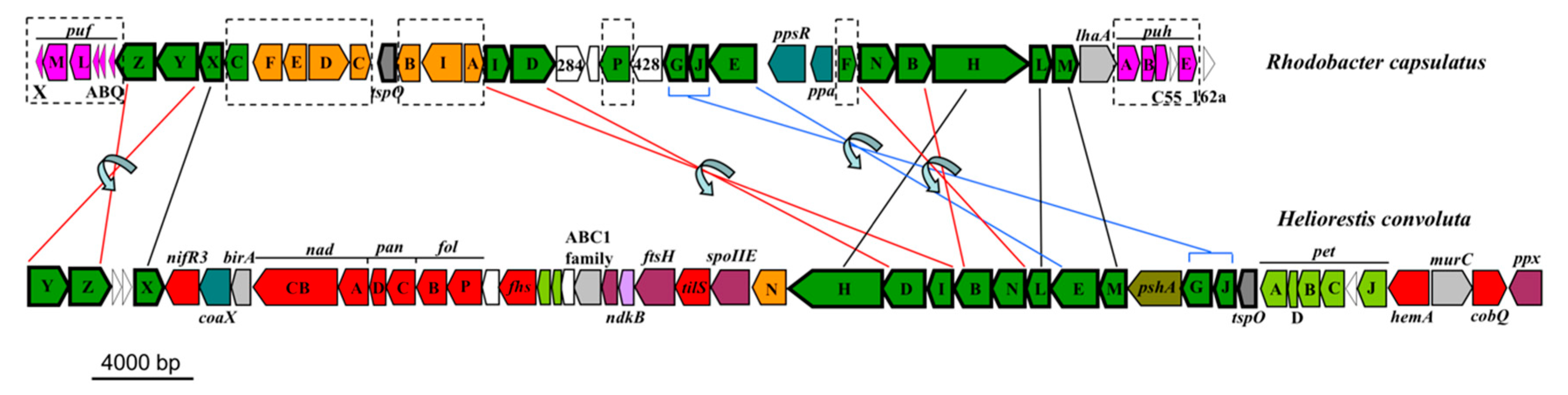
Due to high sequence identity between genes allowing for phototrophic growth (data not shown), a mechanism similar to BChl g and 81-OH-Chl a biosynthesis in Hbt. modesticaldum [6,7,60] is predicted for Hrs. convoluta (Figure 5). Many of the genes encoding enzymes required for pigment biosynthesis are grouped into a single photosynthesis gene cluster (PGC) in heliobacteria. The PGCs of Hrs. convoluta and Hbt. modesticaldum were nearly identical and displayed a shared gene synteny in all key genes, including those associated with pigment and cofactor biosynthesis, electron transport, and light harvesting, suggesting that a common genetic architecture—one that differs substantially from the PGCs present in the genomes of purple bacteria—defines the heliobacterial PGC (Figure 6).
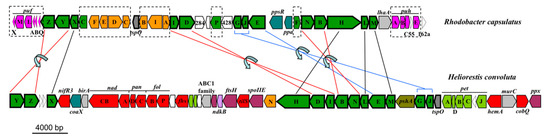
Figure 6.
Photosynthesis gene clusters from Heliorestis convoluta and the purple bacterium Rhodobacter capsulatus. Shared genes are outlined in bold. Lines indicate gene synteny: black, single gene rearrangements; red, inverted genes; and blue, inverted genes with a gene insertion. Dashed boxes show Rba. capsulatus photosynthesis genes absent from Hrs. convoluta. Colors: green, bacteriochlorophyll biosynthesis (bch); orange, carotenoid biosynthesis (crt); pink, proteobacterial reaction centers (puf) and light harvesting complexes (puh); olive, heliobacterial reaction center (psh); teal, regulatory proteins; light green, electron transport (pet); red, cofactor biosynthesis; purple, cell division and sporulation; light blue, nitrogen fixation; grey, transcription; light grey, other nonphotosynthesis genes; and white, uncharacterized genes. Adapted from Sattley et al. [6]. J. Bacteriol. 2008, 190, 4687–4696. Copyright 2008 American Society for Microbiology.
Like BChls c, d, and e of green sulfur bacteria, both BChl g and 81-OH-Chl a of heliobacteria are esterified with farnesol [63]. A non-mevalonate pathway is employed to synthesize the esterifying alcohol, farnesyl diphosphate, of heliobacterial pigments [64]. As was noted for Hbt. modesticaldum, Hrs. convoluta contained the complete complement of genes for this pathway, beginning with pyruvate and glyceraldehyde-3-phosphate and proceeding to an unidentified but predicted isomerase that could catalyze the interconversion of isopentenyl diphosphate and dimethylallyl diphosphate [6,64] (Figure 5). Following this, farnesyl diphosphate can either be incorporated into the final structures of BChl g and 81-OH-Chl a or further transformed into the major carotenoids found in Hrs. convoluta [63] (Figure 5). The high specificity of BchG for incorporation of a farnesol moiety over longer alcohol groups, such as phytol, has been demonstrated in studies of pigment biosynthesis in Hbt. modesticaldum [61], and the high sequence identity (68%) of BchG from Hrs. convoluta to that of Hbt. modesticaldum suggests a similar activity in the alkaliphile.
Experimental work and pigment extraction from Hrs. convoluta, Hrs. daurensis, and Hrs. baculata revealed that the major carotenoid in alkaliphilic heliobacteria is OH-diaponeurosporene glucoside C16:0 ester, followed by 4,4′-diaponeurosporene, OH-diaponeurosporene glucoside C16:1 ester, and 8,8′-zeta-carotene [21,63]. These novel glucoside esters in alkaliphilic heliobacteria were not found in neutrophilic heliobacteria, in which 4,4′-diaponeurosporene was the major carotenoid [63,65]. The synthesis of these C30 carotenoids [66] is complicated by the apparent absence of a gene (crtM) encoding 4,4′-diapophytoene synthase in both Hbt. modesticaldum [6] and Hrs. convoluta. Presumably, the presence of an enzyme with 4,4′-diapophytoene synthase activity is essential in the proposed biosynthetic pathway for each of the carotenoids found in heliobacteria [63] (Figure 5). Although crtM was not identified, two nonidentical copies of crtN (FTV88_2648 and FTV88_3059) having a sequence identity of 71% were annotated in the Hrs. convoluta genome, and it is possible that one of their gene products exhibits CrtM-like activity.
In alkaliphilic heliobacteria, a proposed CrtC-like hydratase catalyzes the formation of OH-diaponeurosporene from 4,4′-diaponeurosporene, followed by synthesis of OH-diaponeurosporene glucoside by a CrtX-like glucosyl transferase, with a putative esterase making the final conversion to the mature glucoside ester [63]. Genes encoding the enzymes catalyzing the final three steps of OH-diaponeurosporene glucoside ester synthesis were not identified in Hrs. convoluta (Figure 5), but genes encoding two carotenoid biosynthesis proteins (FTV88_0301 and FTV88_0302) were annotated. These genes showed no significant sequence similarity to genes of the neutrophilic Hbt. modesticaldum, and they may be candidates for encoding proteins to perform the final steps of carotenoid biosynthesis in alkaliphilic heliobacteria.
3.6. Reaction Center and Electron Transport Chain
Heliobacteria possess a type I (Fe–S type) photosynthetic reaction center (RC) imbedded in the cytoplasmic membrane [4,67,68]. As the simplest known and perhaps most ancient extant (bacterio)chlorophyll-binding photochemical apparatus [69], the heliobacterial RC is a symmetrical homodimer consisting of the PshA polypeptide and the novel, single-transmembrane helix PshX polypeptide [70]. PshA of Hrs. convoluta (encoded by pshA, FTV88_2638) showed 71% sequence identity to PshA of Hbt. modesticaldum but nearly 96% identity to PshA of Hrs. acidaminivorans (GenBank accession WBXO01000000, unpublished). As is the case in Hbt. modesticaldum, pshX (FTV88_2551) is situated outside of the PGC in Hrs. convoluta and encodes a protein consisting of just 31 amino acids. The PshX RC subunit from Hrs. convoluta showed a 74% sequence identity to that of Hbt. modesticaldum.
The crystal structure of the Hbt. modesticaldum RC revealed the presence of 54 BChl g molecules, two 81-OH-Chl a molecules, two carotenoids (4,4′-diaponeurosporene), four BChl g′ molecules (a C-13 epimer of BChl g that functions as the primary electron donor, P800) [68,71,72], two lipids, and one [4Fe–4S] cluster [70]. Experimental data on the structure of the Hrs. convoluta RC are not yet available. However, with their highly similar PshA and PshX proteins, the geometry and pigment composition of the Hrs. convoluta RC should closely resemble that of the Hbt. modesticaldum RC [69]. Nevertheless, some distinctions may materialize considering the alternative carotenoids produced by alkaliphilic heliobacteria and their inherently alkaline habitat [63].
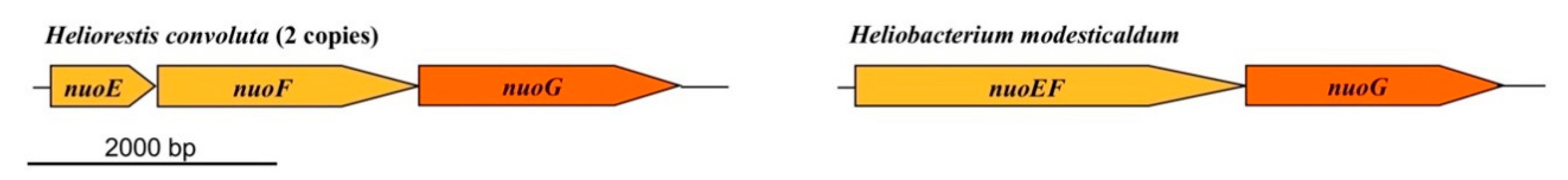
Proteins of the electron transport chain (ETC) of Hrs. convoluta exhibited high sequence similarity to those from Hbt. modesticaldum, and thus the overarching mechanism of light-driven energy conservation is likely to be highly conserved across all heliobacterial taxa. Although not experimentally confirmed, it is likely that electrons first enter the chain by the activity of either NADH:quinone oxidoreductase (Figure 3), a 14-subunit protein complex embedded in the cytoplasmic membrane and encoded by nuoABCDEFGHIJKLMN, or perhaps a complex having ferredoxin:menaquinone oxidoreductase activity. As observed in Hbt. modesticaldum, the nuoEFG genes in Hrs. convoluta are not co-localized within the same operon as the other nuo genes. However, unlike in Hbt. modesticaldum, in which nuoEF are fused, nuoE and nuoF exist as individual genes (present in duplicate copies) in Hrs. convoluta (Figure 7). With the exception of this distinction, all nuo genes show high sequence identity (62–79%) between the two species. As for Hbt. modesticaldum, menaquinone is predicted to shuttle electrons from Complex I to the cytochrome bc complex (PetABCD), and electron transfer through these complexes drives translocation of H+ to the periplasmic space (Figure 3), forming a proton motive force (PMF) [6,73].

Figure 7.
Comparison of nuoEFG genes in Heliorestis convoluta and Heliobacterium modesticaldum. Whereas in Hbt. modesticaldum (and Heliobacillus mobilis) nuoE and nuoF are fused, these genes are independent in Hrs. convoluta. In both species, however, nuoEFG are separated from other nuo genes on the chromosome. In addition, unlike in Hbt. modesticaldum, Hrs. convoluta contains two copies of nuoEFG, as well as the hydEFG maturase genes (FTV88_1003–1005) that may impart [FeFe] hydrogenase activity. Colors: gold, NADH dehydrogenase subunits; orange, structural genes.
Cytochrome bc1 complexes, which are found in a variety of anoxygenic phototrophs and also in eukaryotic mitochondria, consist of a minimum of three protein subunits: cytochrome b, cytochrome c1, and the Rieske iron-sulfur protein [74]. In contrast, the related cytochrome b6f complex, which is present in cyanobacteria and chloroplasts, is comprised of cytochrome f (PetA), cytochrome b6 (PetB), the Rieske iron-sulfur protein (PetC), and subunit IV (PetD) [74]. Having similar functions but distinct structural properties, cytochrome b contains eight transmembrane helices, whereas cytochrome b6 and its associated subunit IV contain four and three transmembrane helices, respectively [74]. Cytochrome b6 shows homology to the N-terminal half of cytochrome b, and subunit IV is homologous to the C-terminal half of cytochrome b [74].
An analysis of the cytochrome bc complex of Hrs. convoluta indicated that it resembles a hybrid of the cytochrome b6f complex and the cytochrome bc1 complex. A comparison of cytochrome b6 and subunit IV proteins from Hrs. convoluta and the model cyanobacterium Synechocystis PCC 6803 showed 48% and 42% amino acid sequence identity, respectively. However, cytochrome b6 and subunit IV from Hrs. convoluta also showed 36% and 30% amino acid identity, respectively, to the N-terminal and C-terminal halves of cytochrome b from the purple bacterium Rhodobacter sphaeroides. Furthermore, whereas subunit IV from Hrs. convoluta is predicted to contain the usual three transmembrane helices, cytochrome b6 from Hrs. convoluta contained a predicted five transmembrane regions instead of the four typically observed in the b6f complex. Notably, cytochrome b6 from Hbt. modesticaldum is predicted to contain the conventional four transmembrane helices. Therefore, considering the above sequence analyses and their total of eight predicted transmembrane helices, the PetB and PetD proteins of Hrs. convoluta may represent a structural and evolutionary intermediate between cytochrome b and cytochrome b6/subunit IV proteins, a distinction perhaps not shared with neutrophilic heliobacteria.
The PetA protein in heliobacteria is also of interest because it functions as a diheme cytochrome c (as opposed to the typical monoheme protein) and shows no sequence or structural similarity to cytochrome f [74]. Although unusual among the Firmicutes, the diheme cytochrome c has been identified in all heliobacteria studied thus far and is likely a universal feature of these phototrophs. PetA from Hrs. convoluta showed high sequence identity with PetA from Hrs. acidaminivorans (79%), and sequence identities to PetA from neutrophilic heliobacteria (e.g., Hbt. modesticaldum, Hbt. gestii, Heliobacillus mobilis, and Heliophilum fasciatum) were all near 50%. Based on similarities in its N- and C-terminal domains, the heliobacterial diheme cytochrome c may have been the result of a past gene duplication and subsequent fusion [75].
A single operon containing all eight genes encoding the subunits of ATP synthase (atpABCDEFGH) was identified in the genome of Hbt. modesticaldum [6,26], and the encoded ATP synthase itself has since been biochemically characterized [76]. The composition and arrangement of ATP synthase genes in Hrs. convoluta was identical to that in the Hbt. modesticaldum genome. Kinetic studies with Hba. mobilis and Hbt. modesticaldum and physiological similarity to photosystem I of cyanobacteria suggest that a PMF established by cyclic electron flow drives photophosphorylation in heliobacteria [73,77,78]. For overviews of electron transfer reactions in heliobacteria, see Sattley and Swingley [7], Kondo et al. [79], and, more recently, Kashey et al. [73].
3.7. Endosporulation
A likely universal trait of heliobacteria is the ability to form endospores [11], differentiated and largely dormant cells that are highly resistant to environmental extremes, such as heat and desiccation. Genomic comparisons of Hrs. convoluta and Hbt. modesticaldum revealed high similarity between endosporulation genes in each species. For example, genes encoding key sporulation sigma factors (σH, σE, σF, σG, σK) in Hbt. modesticaldum were also identified in the Hrs. convoluta genome. Like Hbt. modesticaldum, Hrs. convoluta lacked the spo0M gene functioning to regulate stage 0 development of endosporulation [80] and the spoIIB gene necessary for robust sporulation in B. subtilis [81]. This may help explain the sporadic (as opposed to consistent) production of endospores in serially subcultured cells of Hrs. convoluta strain HHT [21], as the deletion of either spo0M or spoIIB in B. subtilis results in impairment of endosporulation [80,81]. Additionally, the 20 cot genes encoding proteins that comprise the protective spore coat for B. subtilis, including cotH required for spore coat assembly [82], did not show significant similarity to genes in Hbt. modesticaldum [6] or Hrs. convoluta. Likewise, key proteins that coordinate spore coat assembly and composition in Clostridioides (Clostridium) difficile, including CotA and CotB [83], showed no sequence similarity to genes in Hrs. convoluta. Despite these deficiencies, cells of Hrs. convoluta strain HHT were still capable of forming heat-resistant endospores, even if sporadically [21]. These findings suggest shared biosynthetic and regulatory mechanisms governing endosporulation in Hbt. modesticaldum and Hrs. convoluta that differ in some respects from those that govern endosporulation in species of Bacillus and Clostridium.
3.8. Molecular Adaptations to Alkaliphily in Heliorestis convoluta
Alkaliphilic bacteria employ several mechanisms to maintain intracellular pH homeostasis in their highly alkaline environments. Experimental work conducted with alkaliphiles revealed that these organisms maintain a lower cytoplasmic pH than their external environment—up to a 2.3 pH unit difference—for optimal enzyme activity and cellular functioning [84,85]. Despite its optimal growth pH of 8.5–9 and ability to grow slowly at pH 10 [21], it is likely that Hrs. convoluta maintains a cytoplasmic pH at or below pH 8, as is true from studies of several alkaliphilic strains of Bacillus [84,86].
Cytoplasmic pH homeostasis in Hrs. convoluta is likely supported by the presence of a Na+/H+ antiporter encoded by nhaA (FTV88_0116). To maintain cytoplasmic pH at homeostatic levels, the Na+/H+ antiporter operates in an electrogenic manner, facilitating the import of twice as many H+ as Na+ exported [87,88]. The inward movement of protons through the antiporter acidifies the cytoplasm to maintain a pH closer to neutral [87,88,89]. The NhaA protein from Hrs. convoluta was found to be 87% identical in amino acid sequence to NhaA from Heliorestis acidaminivorans—also an alkaliphile—but only 50% identical to NhaA from the neutrophile Hbt. modesticaldum. The NhaA enzyme may, therefore, be a good candidate to study which amino acid residues facilitate antiporter activity in alkaline versus neutral environments.
In addition to its role in cytoplasmic pH maintenance, the Na+/H+ antiporter generates a sodium motive force (SMF) that has been shown to be important for secondary active transport of various substrates [89,90,91] (see Figure 3 for examples in Hrs. convoluta). The use of Na+-coupling for transport is potentially more important in Hrs. convoluta than in its neutrophilic relative, Hbt. modesticaldum, as genes encoding multiple Na+-dependent transporters (FTV88_2418 and FTV88_1400) and a Na+/Ca+ antiporter (FTV88_2739) in Hrs. convoluta showed little to no significant similarity with genes in Hbt. modesticaldum.
The more neutral cytoplasm compared to the alkaline extracellular milieu would seemingly create an outward-directed bulk PMF rather than the inward-directed PMF needed to drive ATP synthesis [89,90,91]. Despite this, most alkaliphiles, including Hrs. convoluta, still employ a PMF rather than a SMF to power ATP synthase [89,92]. Alkaliphilic bacteria must therefore have mechanisms in place to prevent H+ equilibration with the external environment so that an effective local PMF can be established. To this end, carotenoids, which are produced in large quantities by alkaliphilic heliobacteria [93], have been proposed to play a role in organizing proton pumps close to ATP synthases in the membrane, thus facilitating more efficient ATP generation [91,94,95]. In addition, cardiolipin, a glycerophospholipid that assists in membrane domain organization, may also help prevent H+ equilibration by functioning as a proton sink for the H+-coupled ATP synthase [96,97,98]. By functioning in this capacity, cardiolipin allows for the retention of H+ near the surface of the cell membrane so that they are unable to spontaneously diffuse into the alkaline environment. Notably, a gene encoding cardiolipin synthase was present in Hrs. convoluta (FTV88_2523), but no corresponding homolog was identified in Hbt. modesticaldum.
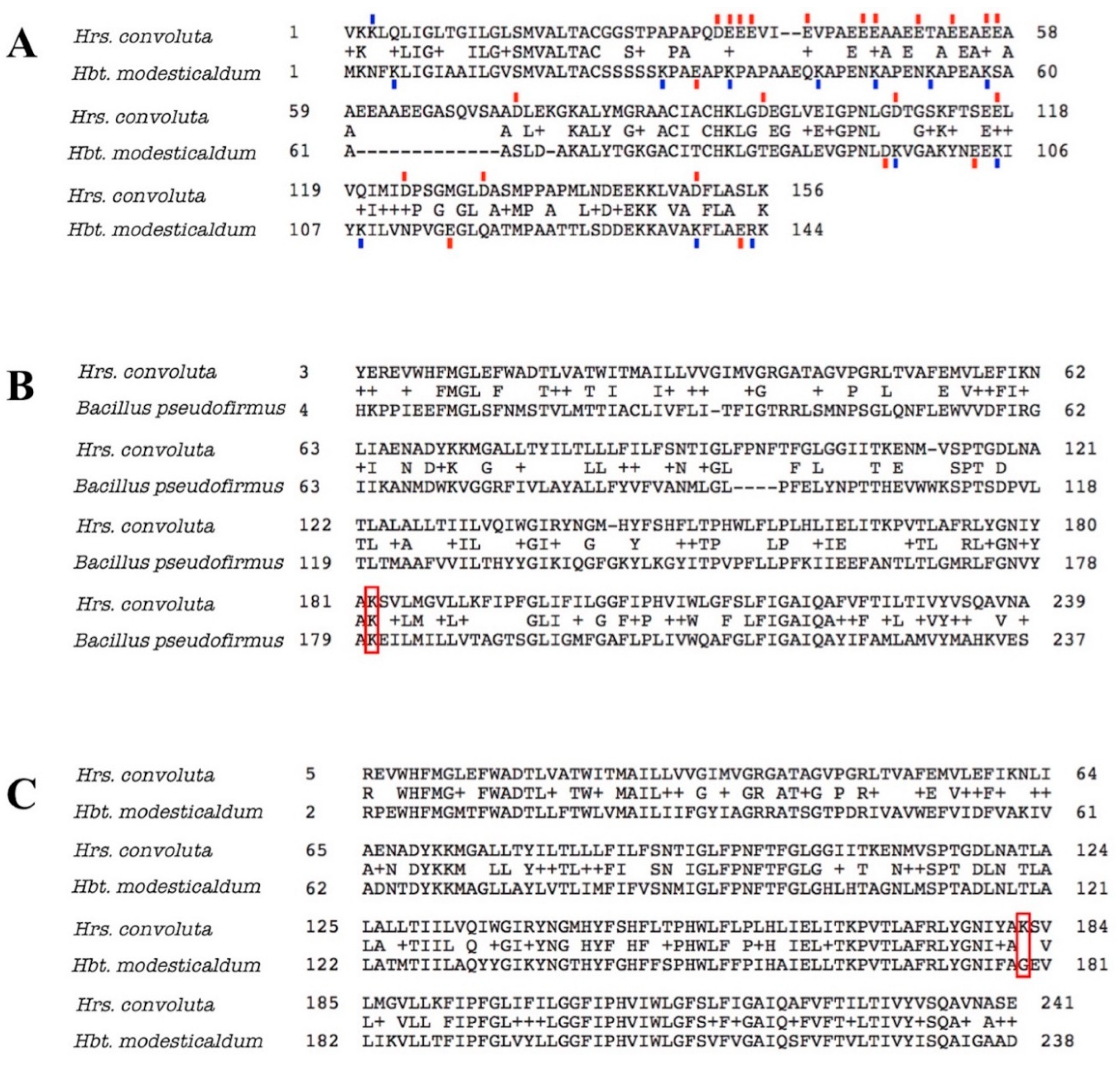
In addition to producing proteins and other molecules that counteract the pH difference between the cytoplasm and environment and the consequences thereof, homologous proteins also have amino acid substitutions that optimize the functioning of normal processes for the alkaline environment. In alkaliphiles, the portions of extracellular enzymes that are exposed to the external environment tend to have decreased numbers of basic residues (arginine, histidine, or lysine), with acidic amino acids (aspartate or glutamate) or neutral residues in their place [99,100,101]. In a noteworthy example, the amino acid sequence for cytochrome c553 (PetJ) of Hrs. convoluta contained 13 more acidic amino acid residues and 11 fewer basic residues than PetJ of Hbt. modesticaldum (Figure 8A). In line with previous discussion, the elevated number of acidic residues and corresponding decrease in basic residues in the externally-functioning Hrs. convoluta PetJ should contribute to OH− repulsion and H+ attraction near the membrane surface and help maintain the PMF [89]. Although additional investigation of the cell surface of Hrs. convoluta is required to confirm its electrochemical nature, genomic data suggest that this phototroph can sequester H+ near the cell surface to create an effective PMF for ATP synthesis and flagellar motility.
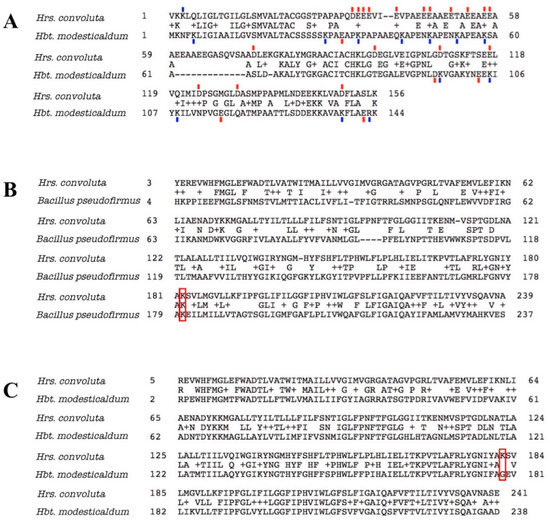
Figure 8.
Amino acid sequence alignments for cytochrome c553 and ATP synthase Fo alpha subunit of Heliorestis convoluta with related species. (A) The sequence alignment for cytochrome c553 of Hrs. convoluta and Heliobacterium modesticaldum. Acidic amino acid residues (red), aspartate (D) and glutamate (E), and basic amino acid residues (blue), arginine (R) and lysine (K), that differed between each species were indicated by a colored dash directly above or below the residue. Acidic or basic amino acids in the gap (–) regions were not marked. (B) Sequence alignment for ATP synthase Fo alpha subunit of Hrs. convoluta and Bacillus pseudofirmus OF4. The lysine residue of interest at position 180 in B. pseudofirmus aligns with Lys182 in Hrs. convoluta (red box). (C) Sequence alignment for ATP synthase Fo alpha subunit of Hrs. convoluta and Hbt. modesticaldum. The lysine residue of interest at position 182 in Hrs. convoluta aligns with Gly179 in Hbt. modesticaldum (red box). All sequence alignments were generated using the BLAST algorithm.
In a similar way, several key amino acid residues and motifs in ATP synthase have been found to contribute to optimal functioning of the enzyme at different pH levels [89,91,102,103]. For example, a lysine residue found at position 180 in the Fo alpha subunit of ATP synthase in Bacillus pseudofirmus OF4 was determined to favor H+-powered ATP synthesis at an alkaliphilic pH due to its basic properties [102,104,105,106]. As expected, a corresponding Lys182 in the Hrs. convoluta Fo alpha subunit (Figure 8B) can presumably capture protons optimally from the alkaline environment and release them into the rotor subunit of ATP synthase at an external basic pH near the high pKa of the side chain [102]. The lysine residue would be detrimental to ATP synthesis in a neutral pH range, as H+ would be retained on the residue side chain at ~pH 7 (below the side chain pKa). This highlights the significance of a glycine residue at the corresponding position in the Fo alpha subunit in neutrophilic bacteria, including Hbt. modesticaldum [102] (Figure 8C).
Several alkaliphilic bacteria use a SMF to power flagellar motor proteins, thus reserving the valuable and limited PMF for ATP production [107,108]. Research conducted with alkaliphilic Bacillus spp. concluded that a highly conserved valine residue is present in H+-driven (MotB) flagellar motor protein sequences, whereas a leucine residue takes the place of this valine in Na+-driven (MotS) motor protein sequences [107,108]. The alkaliphilic Bacillus spp. contained MotS with the conserved leucine amino acid, allowing these bacteria to use a SMF to power Na+-coupled flagellar motility [107,108]. Interestingly, MotB—with its conserved valine—was identified in both Hrs. convoluta and Hbt. modesticaldum, suggesting that a PMF is used to power motility in both alkaliphilic and neutrophilic heliobacteria. Genomic analyses confirmed the presence of a core set of 24 genes (fliCDEFGHIMNPQR, flgBCDEFGKL, motAB, flhAB) in Hrs. convoluta that are essential for PMF-driven swimming motility in numerous flagellated bacteria [109].
4. Conclusions
The analysis of the complete genome sequence of Hrs. convoluta has provided further insight into the photoheterotrophic metabolism, nitrogen utilization, sulfur assimilation, and pigment biosynthesis pathways of heliobacteria, as well as molecular adaptations to an alkaliphilic existence. Further biochemical and genetic experimentation with alkaliphilic heliobacteria, including Hrs. convoluta, is necessary to confirm genomics-based predictions regarding the roles of specific genes and the apparent absence of specific enzyme activities.
Author Contributions
J.W.T., R.E.B., M.T.M., E.D.D., and W.M.S. conceived and designed the experiments. All authors performed the experiments and analyzed the data, with the bulk of the analysis done by E.D.D., L.M.S., B.M.B., K.N.S., J.M.B., and W.M.S. The culture of Heliorestis convoluta str. HHT was provided by M.T.M. Submission and accessioning of the Hrs. convoluta genome with GenBank was initiated and overseen by S.N. and M.G.G. The manuscript was prepared by E.D.D., W.M.S., and M.T.M., with other authors providing reviews and edits for the final version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the U.S. National Science Foundation Phototrophic Prokaryotes Sequencing Project as an award to J.W.T., R.E.B., and M.T.M. (Evolutionary Diversification of Photosynthesis and the Anoxygenic to Oxygenic Transition; NSF Grant #0950550).
Acknowledgments
Support for participation of E.D.D., L.M.S., B.M.B., K.N.S., and W.M.S. in this research was provided by Hodson Research Institute (Indiana Wesleyan University) grants to W.M.S. We thank Dr. Marie Asao for her detailed studies of the basic physiology of Hrs. convoluta str. HHT.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gest, H.; Favinger, J.L. Heliobacterium chlorum, an anoxygenic brownish-green photosynthetic bacterium containing a “new” form of bacteriochlorophyll. Arch. Microbiol. 1983, 136, 11–16. [Google Scholar] [CrossRef]
- Stevenson, A.K.; Kimble, L.K.; Woese, C.R.; Madigan, M.T. Characterization of new phototrophic heliobacteria and their habitats. Photo. Res. 1997, 53, 1–12. [Google Scholar] [CrossRef]
- Madigan, M.T.; Ormerod, J.G. Taxonomy, physiology, and ecology of heliobacteria. In Anoxygenic Photosynthetic Bacteria; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; Volume 2, pp. 17–30. [Google Scholar]
- Sattley, W.M.; Madigan, M.T. The Family Heliobacteriaceae. In The Prokaryotes—Firmicutes and Tenericutes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 185–196. [Google Scholar]
- Brockmann, H.; Lipinski, A. Bacteriochlorophyll g. A new bacteriochlorophyll from Heliobacterium chlorum. Arch. Microbiol. 1983, 136, 17–19. [Google Scholar] [CrossRef]
- Sattley, W.M.; Madigan, M.T.; Swingley, W.D.; Cheung, P.C.; Clocksin, K.M.; Conrad, A.L.; Dejesa, L.C.; Honchak, B.M.; Jung, D.O.; Karbach, L.E.; et al. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J. Bacteriol. 2008, 190, 4687–4696. [Google Scholar] [CrossRef] [PubMed]
- Sattley, W.M.; Swingley, W.D. Properties and evolutionary implications of the heliobacterial genome. In Genome Evolution of Photosynthetic Bacteria; Beatty, T.J., Ed.; Academic Press, Elsevier Inc.: San Diego, CA, USA, 2013; Volume 66, pp. 67–98. [Google Scholar]
- Woese, C.R.; Debrunner-Vossbrinck, B.A.; Oyaizu, H.; Stackebrandt, E.; Ludwig, W. Gram-positive bacteria: Possible photosynthetic ancestry. Science 1984, 229, 762–765. [Google Scholar] [CrossRef]
- Miller, K.R.; Jacob, J.S.; Smith, U.; Kolaczkowski, S.; Bowman, M.K. Heliobacterium chlorum: Cell organization and structure. Arch. Microbiol. 1986, 146, 111–114. [Google Scholar] [CrossRef]
- Pickett, M.W.; Weiss, N.; Kelly, D.J. Gram-positive cell wall structure of the A3γ type in heliobacteria. FEMS Microbiol. Lett. 1994, 122, 7–12. [Google Scholar] [CrossRef]
- Kimble-Long, L.K.; Madigan, M.T. Molecular evidence that the capacity for endosporulation is universal among phototrophic heliobacteria. FEMS Microbiol. Lett. 2001, 199, 191–195. [Google Scholar] [CrossRef]
- Grégoire, D.S.; Lavoie, N.C.; Poulain, A.J. Heliobacteria reveal fermentation as a key pathway for mercury reduction in anoxic environments. Environ. Sci. Technol. 2018, 52, 4145–4153. [Google Scholar] [CrossRef]
- Lavoie, N.C.; Grégoire, D.S.; Stenzler, B.R.; Poulain, A.J. Reduced sulphur sources favour HgII reduction during anoxygenic photosynthesis by heliobacteria. Geobiology. 2020, 18, 70–79. [Google Scholar] [CrossRef]
- Baker, P.L.; Orf, G.S.; Khan, Z.; Espinoza, L.; Leung, S.; Kevershan, K.; Redding, K.E. A molecular biology tool kit for the phototrophic Firmicute Heliobacterium modesticaldum. Appl. Environ. Microbiol. 2019, 85, e01287-19. [Google Scholar] [CrossRef] [PubMed]
- Kimble, L.K.; Stevenson, A.K.; Madigan, M.T. Chemotrophic growth of heliobacteria in darkness. FEMS Microbiol. Lett. 1994, 115, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Kimble, L.K.; Mandelco, L.; Woese, C.R.; Madigan, M.T. Heliobacterium modesticaldum, sp. nov., a thermophilic heliobacterium of hot springs and volcanic soils. Arch. Microbiol. 1995, 163, 259–267. [Google Scholar] [CrossRef]
- Pickett, M.W.; Williamson, M.P.; Kelly, D.J. An enzyme and 13CNMR study of carbon metabolism in heliobacteria. Photosynth. Res. 1994, 41, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Bruno, W.J.; Nicholas, D.S.; Halpern, A.L. Weighted neighbor joining: A likelihood-based approach to distance-based phylogeny reconstruction. Mol. Biol. and Evol. 2000, 17, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Bryantseva, I.A.; Gorlenko, V.M.; Kompantseva, E.I.; Achenbach, L.A.; Madigan, M.T. Heliorestis daurensis, gen. nov. sp. nov., an alkaliphilic rod-to-coiled-shaped phototrophic heliobacterium from a Siberian soda lake. Arch. Microbiol. 1999, 172, 167–174. [Google Scholar] [CrossRef]
- Bryantseva, I.A.; Gorlenko, V.M.; Kompantseva, E.I.; Tourova, T.P.; Kuznetsov, B.B.; Osipov, G.A. Alkaliphilic heliobacterium Heliorestis baculata sp. nov., and emended description of the genus Heliorestis. Arch. Microbiol. 2000, 174, 283–291. [Google Scholar] [CrossRef]
- Asao, M.; Jung, D.O.; Achenbach, L.A.; Madigan, M.T. Heliorestis convoluta sp. nov., a coiled, alkaliphilic heliobacterium from the Wadi El Natroun, Egypt. Extremophiles 2006, 10, 403–410. [Google Scholar] [CrossRef]
- Asao, M.; Takaichi, S.; Madigan, M.T. Amino acid-assimilating phototrophic heliobacteria from soda lake environments: Heliorestis acidaminivorans sp. nov. and ‘Candidatus Heliomonas lunata’. Extremophiles 2012, 16, 585–595. [Google Scholar] [CrossRef]
- Imhoff, J.F. Anoxygenic phototrophic bacteria from extreme environments. In Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects; Hallenbeck, P.C., Ed.; Springer: Dordrecht, The Netherlands, 2017; Chapter 13; pp. 427–480. [Google Scholar]
- Imhoff, J.F.; Hashwa, F.; Trüper, H.G. Isolation of extremely halophilic phototrophic bacteria from the alkaline Wadi Natrun, Egypt. Arch. Hydrobiol. 1978, 84, 381–388. [Google Scholar]
- Imhoff, J.F.; Sahl, H.G.; Soliman, G.S.H.; Trüper, H.G. The Wadi Natrun: Chemical composition and microbial mass developments in alkaline brines of eutrophic desert lakes. Geomicrobiol. J. 1979, 1, 219–234. [Google Scholar] [CrossRef]
- Sattley, W.M.; Blankenship, R.E. Insights into heliobacterial photosynthesis and physiology from the genome of Heliobacterium modesticaldum. Photosynth. Res. 2010, 104, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinform. 2010, 31, 11.5.1–11.5.12. [Google Scholar] [CrossRef] [PubMed]
- Galens, K.; Orvis, J.; Daugherty, S.; Creasy, H.H.; Angiuoli, S.; White, O.; Wortman, J.; Mahurkar, A.; Giglio, M.G. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci. 2011, 4, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Manatee. Available online: http://manatee.sourceforge.net (accessed on 24 February 2020).
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krogh, A.; Larrson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Curnow, A.W.; Tumbula, D.L.; Pelaschier, J.T.; Min, B.; Söll, D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 12838–12843. [Google Scholar] [CrossRef]
- Akochy, P.-M.; Bernard, D.; Roy, P.H.; Lapointe, J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004, 186, 767–776. [Google Scholar] [CrossRef]
- Savvi, S.; Warner, D.F.; Kana, B.D.; McKinney, J.D.; Mizrahi, V.; Dawes, S.S. Functional characterization of vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: Implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 2008, 190, 3884–3895. [Google Scholar] [CrossRef] [PubMed]
- Suvorova, I.A.; Ravcheev, D.A.; Gelfand, M.S. Regulation and evolution of malonate and propionate catabolism in proteobacteria. J. Bacteriol. 2012, 194, 3234–3240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ormerod, J.G.; Kimble, L.K.; Nesbakken, T.; Torgersen, T.A.; Woese, C.R.; Madigan, M.T. Heliophilum fasciatum gen. nov. et sp. nov., and Heliobacterium gestii sp. nov. endospore-forming heliobacteria from rice field soils. Arch. Microbiol. 1996, 165, 226–234. [Google Scholar]
- Janssen, P.H.; Schink, B. Pathway of butyrate catabolism by Desulfobacterium cetonicum. J. Bacteriol. 1995, 177, 3870–3872. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Horswill, A.R.; Escalante-Semerena, J.C. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: Residue Lys592 is required for propionyl-AMP synthesis. Biochemistry 2002, 41, 2379–2387. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.H.; Yue, H.; Blankenship, R.E. Energy metabolism of Heliobacterium modesticaldum during phototrophic and chemotrophic growth. BMC Microbiol. 2010, 10, 150. [Google Scholar] [CrossRef]
- Tang, K.H.; Feng, X.; Zhuang, W.Q.; Alvarez-Cohen, L.; Blankenship, R.E.; Tang, Y.J. Carbon flow of heliobacteria is related more to clostridia than to the green sulfur bacteria. J. Biol. Chem. 2010, 285, 35104–35112. [Google Scholar] [CrossRef]
- Evans, M.C.; Buchanan, B.B.; Arnon, D.I. New cyclic process for carbon assimilation by a photosynthetic bacterium. Science 1966, 152, 673. [Google Scholar] [CrossRef]
- Sattley, W.M.; Asao, M.; Tang, J.K.H.; Collins, A.M. Energy conservation in heliobacteria: Photosynthesis and central carbon metabolism. In The Structural Basis of Biological Energy Generation, Advances in Photosynthesis and Respiration; Hohmann-Marriott, M.F., Ed.; Springer: Dordrecht, The Netherlands, 2014; Volume 39, pp. 231–247. [Google Scholar]
- Wang, L.; Zhang, L.; Liu, Z.; Zhao, D.; Liu, X.; Zhang, B.; Xie, J.; Hong, Y.; Li, P.; Chen, S.; et al. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet. 2013, 9, e1003865. [Google Scholar] [CrossRef]
- Sheehy, D.; Lu, Y.-K.; Osman, F.; Alattar, Z.; Flores, C.; Sussman, H.; Zaare, S.; Dooling, M.; Meraban, A.; Baker, P.; et al. Genome-wide Transcriptional Response during the Shift to N2-fixing Conditions in Heliobacterium modesticaldum. J. Proteomics Bioinform. 2018, 11, 143–160. [Google Scholar] [CrossRef]
- Merrick, M.J. Regulation of nitrogen fixation in free-living diazotrophs. In Genetics and regulation of nitrogen fixation in free-living bacteria; Klipp, W.B., Masepohl, B., Gallon, J.R., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2004; Volume 2, pp. 197–223. [Google Scholar]
- Enkh-Amgalan, J.; Kawasaki, H.; Oh-oka, H.; Seki, T. Cloning and characterization of a novel gene involved in nitrogen fixation in Heliobacterium chlorum: A possible regulatory gene. Arch. Microbiol. 2006, 186, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Dodsworth, J.A.; Leigh, J.A. Regulation of nitrogenase by 2-oxoglutarate reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc. Natl. Acad. Sci. USA 2006, 103, 9779–9784. [Google Scholar] [CrossRef] [PubMed]
- Vanoni, M.A.; Curti, B. Glutamate synthase: A complex iron-sulfur flavoprotein. Cell Mol. Life Sci. 1999, 55, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S. Nitrogen fixation in the clostridia. In Genetics and Regulation of Nitrogen Fixation in Free-living Bacteria; Klipp, W., Masepohl, B., Gallon, J.R., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2004; Volume 2, pp. 53–64. [Google Scholar]
- Peck, H.D. Enzymatic basis for assimilatory and dissimilatory sulfate reduction. J. Bacteriol. 1961, 82, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Sekowska, A.; Kung, H.-F.; Danchin, A. Sulfur metabolism in Escherichia coli and related bacteria: Facts and fiction. J. Mol. Microbiol. Biotechnol. 2000, 2, 145–177. [Google Scholar] [PubMed]
- Schwedock, J.S.; Liu, C.; Leyh, T.S.; Long, S.R. Rhizobium meliloti NodP and NodQ form a multifunctional sulfate-activating complex requiring GTP for activity. J. Bacteriol. 1994, 176, 7055–7064. [Google Scholar] [CrossRef]
- Eisen, J.A.; Nelson, K.E.; Paulsen, I.T.; Heidelberg, J.F.; Wu, M.; Dodson, R.J.; Deboy, R.; Gwinn, M.L.; Nelson, W.C.; Haft, D.H.; et al. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 2002, 99, 9509–9514. [Google Scholar] [CrossRef]
- Weissgerber, T.; Zigann, R.; Bruce, D.; Chang, Y.-J.; Detter, J.C.; Han, C.; Hauser, L.; Jeffries, C.D.; Land, M.; Munk, A.C.; et al. Complete genome sequence of Allochromatium vinosum DSM 180T. Stand. Genomic Sci. 2011, 5, 311–330. [Google Scholar] [CrossRef]
- Klemm, D.J.; Barton, L.L. Oxidation of protoporphyrinogen in the obligate anaerobe Desulfovibrio gigas. J. Bacteriol. 1985, 164, 316–320. [Google Scholar] [CrossRef]
- Klemm, D.J.; Barton, L.L. Purification and properties of protoporphyrinogen oxidase from an anaerobic bacterium, Desulfovibrio gigas. J. Bacteriol. 1987, 169, 5209–5215. [Google Scholar] [CrossRef]
- Dailey, H.A.; Gerdes, S.; Dailey, T.A.; Burch, J.S.; Phillips, J.D. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc. Natl. Acad. Sci. USA 2015, 112, 2210–2215. [Google Scholar] [CrossRef] [PubMed]
- Tsukatani, Y.; Yamamoto, H.; Mizoguchi, T.; Fujita, Y.; Tamiaki, H. Completion of biosynthetic pathways for bacteriochlorophyll g in Heliobacterium modesticaldum: The C8-ethylidene group formation. Biochim. Biophys. Acta 2013, 1827, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ramos, M.; Canniffe, D.P.; Radle, M.I.; Hunter, C.N.; Bryant, D.A.; Golbeck, J.H. Engineered biosynthesis of bacteriochlorophyll gF in Rhodobacter sphaeroides. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 501–509. [Google Scholar] [CrossRef] [PubMed]
- van de Meent, E.J.; Kobayashi, M.; Erkelens, C.; van Veelen, P.A.; Amesz, J.; Watanabe, T. Identification of 81-hydroxychlorophyll a as a functional reaction center pigment in heliobacteria. Biochim. Biophys. Acta 1991, 1058, 356–362. [Google Scholar] [CrossRef]
- Takaichi, S.; Oh-oka, H.; Maoka, T.; Jung, D.O.; Madigan, M.T. Novel carotenoid glucoside esters from alkaliphilic heliobacteria. Arch. Microbiol. 2003, 179, 95–100. [Google Scholar] [CrossRef]
- Dubey, V.S.; Bhalla, R.; Luthra, R. An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 2003, 28, 637–646. [Google Scholar] [CrossRef]
- Takaichi, S.; Inoue, K.; Akaike, M.; Kobayashi, M.; Oh-oka, H.; Madigan, M.T. The major carotenoid in all known species of heliobacteria is the C30 carotenoid 4,4′-diaponeurosporene, not neurosporene. Arch. Microbiol. 1997, 168, 277–281. [Google Scholar] [CrossRef]
- Takaichi, S. Carotenoids and carotenogenesis in anoxygenic photosynthetic bacteria. In The Photochemistry of Carotenoids: Applications in Biology; Frank, H.A., Cogdell, R.J., Young, A., Britton, G., Eds.; Springer: Dordrecht, The Netherlands, 1999; Volume 8, pp. 39–69. [Google Scholar]
- Trost, J.T.; Brune, D.C.; Blankenship, R.E. Protein sequences and redox titrations indicate that the electron acceptors in reaction centers from heliobacteria are similar to Photosystem I. Photosynth. Res. 1992, 32, 11–22. [Google Scholar] [CrossRef]
- Neerken, S.; Amesz, J. The antenna reaction center complex of heliobacteria: Composition, energy conversion and electron transfer. Biochim. Biophys. Acta 2001, 1507, 278–290. [Google Scholar] [CrossRef][Green Version]
- Orf, G.S.; Gisriel, C.; Redding, K.E. Evolution of photosynthetic reaction centers: Insights from the structure of the heliobacterial reaction center. Photosynth. Res. 2018, 138, 11–37. [Google Scholar] [CrossRef]
- Gisriel, C.; Sarrou, I.; Ferlez, B.; Golbeck, J.H.; Redding, K.E.; Fromme, R. Structure of a symmetric photosynthetic reaction center–photosystem. Science 2017, 357, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; van de Meent, E.J.; Erkelens, C.; Amesz, J.; Ikegami, I.; Watanabe, T. Bacteriochlorophyll g epimer as a possible reaction center component of heliobacteria. Biochim. Biophys. Acta 1991, 1057, 89–96. [Google Scholar] [CrossRef]
- Kobayashi, M.; Watanabe, T.; Ikegami, I.; van de Meent, E.J.; Amesz, J. Enrichment of bacteriochlorophyll g′ in membranes of Heliobacterium chlorum by ether extraction: Unequivocal evidence for its existence in vivo. FEBS Lett. 1991, 284, 129–131. [Google Scholar] [CrossRef]
- Kashey, T.S.; Luu, D.D.; Cowgill, J.C.; Baker, P.L.; Redding, K.E. Light-driven quinone reduction in heliobacterial membranes. Photosynth. Res. 2018, 138, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis, 2nd ed.; Wiley Blackwell: Oxford, UK, 2014. [Google Scholar]
- Yue, H.; Kang, Y.; Zhang, H.; Gao, X.; Blankenship, R.E. Expression and characterization of the diheme cytochrome c subunit of the cytochrome bc complex in Heliobacterium modesticaldum. Arch. Biochem. Biophys. 2012, 517, 131–137. [Google Scholar] [CrossRef]
- Yang, J.-H.; Sarrou, I.; Martin-Garcia, J.M.; Zhang, S.; Redding, K.E.; Fromme, P. Purification and biochemical characterization of the ATP synthase from Heliobacterium modesticaldum. Protein. Expr. Purif. 2015, 114, 1–8. [Google Scholar] [CrossRef]
- Kramer, D.M.; Schoepp, B.; Liebl, U.; Nitschke, W. Cyclic electron transfer in Heliobacillus mobilis involving a menaquinol-oxidizing cytochrome bc complex and an RCI-type reaction center. Biochemistry 1997, 36, 4203–4211. [Google Scholar] [CrossRef]
- Heinnickel, M.; Shen, G.; Golbeck, J.H. Identification and characterization of PshB, the dicluster ferredoxin that harbors the terminal electron acceptors FA and FB in Heliobacterium modesticaldum. Biochemistry 2007, 46, 2530–2536. [Google Scholar] [CrossRef]
- Kondo, T.; Itoh, S.; Matsuoka, M.; Azai, C.; Oh-oka, H. Menaquinone as the secondary electron acceptor in the type I homodimeric photosynthetic reaction center of Heliobacterium modesticaldum. J. Phys. Chem. B. 2015, 119, 8480–8489. [Google Scholar] [CrossRef]
- Han, W.-D.; Kawamoto, S.; Hosoya, Y.; Fujita, M.; Sadaie, Y.; Suzuki, K.; Ohashi, Y.; Kawamura, F.; Ochi, K. A novel sporulation-control gene (spo0M) of Bacillus subtilis with a σH-regulated promoter. Gene 1998, 217, 31–40. [Google Scholar] [CrossRef]
- Margolis, P.S.; Driks, A.; Losick, R. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 1993, 175, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Naclerio, G.; Baccigalupi, L.; Zilhao, R.; De Felice, M.; Ricca, E. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 1996, 178, 4375–4380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Permpoonpattana, P.; Phetcharaburanin, J.; Mikelsone, A.; Dembek, M.; Tan, S.; Brisson, M.C.; La Ragione, R.; Brisson, A.R.; Fairweather, N.; Hong, H.A.; et al. Functional characterization of Clostridium difficile spore coat proteins. J. Bacteriol. 2013, 195, 1492–1503. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, K. Alkaliphiles. Proc. Jpn. Acad. Ser. B 2004, 80, 166–178. [Google Scholar] [CrossRef]
- Hicks, D.B.; Liu, J.; Fujisawa, M.; Krulwich, T.A. F1Fo-ATP synthases of alkaliphilic bacteria: Lessons from their adaptations. Biochim. Biophys. Acta. 2010, 1797, 1362–1377. [Google Scholar] [CrossRef]
- Sturr, M.G.; Guffanti, A.A.; Krulwich, T.A. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J. Bacteriol. 1994, 176, 3111–3116. [Google Scholar] [CrossRef]
- Macnab, R.M.; Castle, A.M. A variable stoichiometry model for pH homeostasis in bacteria. Biophys. J. 1987, 52, 637–647. [Google Scholar] [CrossRef]
- Padan, E.; Schuldiner, S. Bacterial Na+/H+ antiporters: Molecular biology, biochemistry and physiology. In Handbook of Biological Physics; Konings, W.N., Kaback, H.R., Lolkema, J.S., Eds.; Elsevier Science: Amsterdam, The Netherlands, 1996; Volume 2, pp. 501–531. [Google Scholar]
- Krulwich, T.A.; Liu, J.; Morino, M.; Fujisawa, M.; Ito, M.; Hicks, D.B. Adaptive mechanisms of extreme alkaliphiles. In Extremophiles Handbook; Horikoshi, K., Ed.; Springer: Tokyo, Japan, 2011; pp. 119–139. [Google Scholar]
- Peddie, C.J.; Cook, G.M.; Morgan, H.W. Sucrose transport by the alkaliphilic, thermophilic Bacillus sp. strain TA2.A1 is dependent on a sodium gradient. Extremophiles 2000, 4, 291–296. [Google Scholar] [CrossRef]
- Preiss, L.; Hicks, D.B.; Suzuki, S.; Meier, T.; Krulwich, T.A. Alkaliphilic bacteria with impact on industrial applications, concepts of early life forms, and bioenergetics of ATP synthesis. Front. Bioeng. Biotechnol. 2015, 3, 1–16. [Google Scholar] [CrossRef]
- von Ballmoos, C.; Cook, G.M.; Dimroth, P. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 2008, 37, 43–64. [Google Scholar] [CrossRef]
- Asao, M.; Madigan, M.T. Family IV Heliobacteriaceae Madigan 2001, 625. In Bergey’s Manual of Systematic Bacteriology (The Firmicutes), 2nd ed.; De Vos, P., Garrity, G.M., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.-H., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2009; Volume 3, pp. 923–931. [Google Scholar]
- Aono, R.; Horikoshi, K. Carotenes produced by alkaliphilic yellow-pigmented strains of Bacillus. Agric. Biol. Chem. 1991, 55, 2643–2645. [Google Scholar] [CrossRef]
- Steiger, S.; Perez-Fons, L.; Cutting, S.M.; Fraser, P.D.; Sandmann, G. Annotation and functional assignment of the genes for the C30 carotenoid pathways from the genomes of two bacteria: Bacillus indicus and Bacillus firmus. Microbiology 2015, 161, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Haines, T.H.; Dencher, N.A. Cardiolipin: A proton trap for oxidative phosphorylation. FEBS Lett. 2002, 528, 35–39. [Google Scholar] [CrossRef]
- Dowhan, W.; Bogdanov, M.; Mileykovskaya, E. Functional roles of lipids in membranes. In Biochemistry of Lipids. Lipoproteins and Membranes, 5th ed.; Vance, D.E., Vance, J.E., Eds.; Elsevier Press: Amsterdam, The Netherlands, 2008; pp. 1–37. [Google Scholar]
- Schlame, M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 2008, 49, 1607–1620. [Google Scholar] [CrossRef]
- Kang, S.-K.; Kudo, T.; Horikoshi, K. Molecular cloning and characterization of an alkalophilic Bacillus sp. C125 gene homologous to Bacillus subtilis sec Y. J. Gen. Microbiol. 1992, 138, 1365–1370. [Google Scholar] [CrossRef][Green Version]
- Quirk, P.G.; Hicks, D.B.; Krulwich, T.A. Cloning of the cta operon from alkaliphilic Bacillus firmus OF4 and characterization of the pH-regulated cytochrome caa3 oxidase it encodes. J. Biol. Chem. 1993, 268, 678–685. [Google Scholar]
- Krulwich, T.A. Alkaliphiles: ‘basic’ molecular problems of pH tolerance and bioenergetics. Mol. Microbiol. 1995, 15, 403–410. [Google Scholar] [CrossRef]
- McMillan, D.G.; Keis, S.; Dimroth, P.; Cook, G.M. A specific adaptation in the α subunit of thermoalkaliphilic F1Fo-ATP synthase enables ATP synthesis at high pH but not at neutral pH values. J. Biol. Chem. 2007, 282, 17395–17404. [Google Scholar] [CrossRef]
- Liu, J.; Fujisawa, M.; Hicks, D.B.; Krulwich, T.A. Characterization of the functionally critical AXAXAXA and PXXEXXP motifs of the ATP synthase c-subunit from an alkaliphilic Bacillus. J. Biol. Chem. 2009, 284, 8714–8725. [Google Scholar] [CrossRef]
- Ivey, D.M.; Krulwich, T.A. Organization and nucleotide sequence of the atp genes encoding the ATP synthase from alkaliphilic Bacillus firmus OF4. Mol. Gen. Genet. 1991, 229, 292–300. [Google Scholar] [CrossRef]
- Ivey, D.M.; Krulwich, T.A. Two unrelated alkaliphilic Bacillus species possess identical deviations in sequence from those of other prokaryotes in regions of Fo proposed to be involved in proton translocation through the ATP synthase. Res. Microbiol. 1992, 143, 467–470. [Google Scholar] [CrossRef]
- Wang, Z.; Hicks, D.B.; Guffanti, A.A.; Baldwin, K.; Krulwich, T.A. Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and nonfermentative growth at pH 10.5. J. Biol. Chem. 2004, 279, 26546–26554. [Google Scholar] [CrossRef] [PubMed]
- Terahara, N.; Krulwich, T.A.; Ito, M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl. Acad. Sci. USA 2008, 105, 14359–14364. [Google Scholar] [CrossRef] [PubMed]
- Fujinami, S.; Terahara, N.; Krulwich, T.A.; Ito, M. Motility and chemotaxis in alkaliphilic Bacillus species. Future Microbiol. 2009, 4, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ochman, H. Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. USA 2007, 104, 7116–7121. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).