Characterization of a Versatile Plant Growth-Promoting Rhizobacterium Pseudomonas mediterranea Strain S58
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Identification
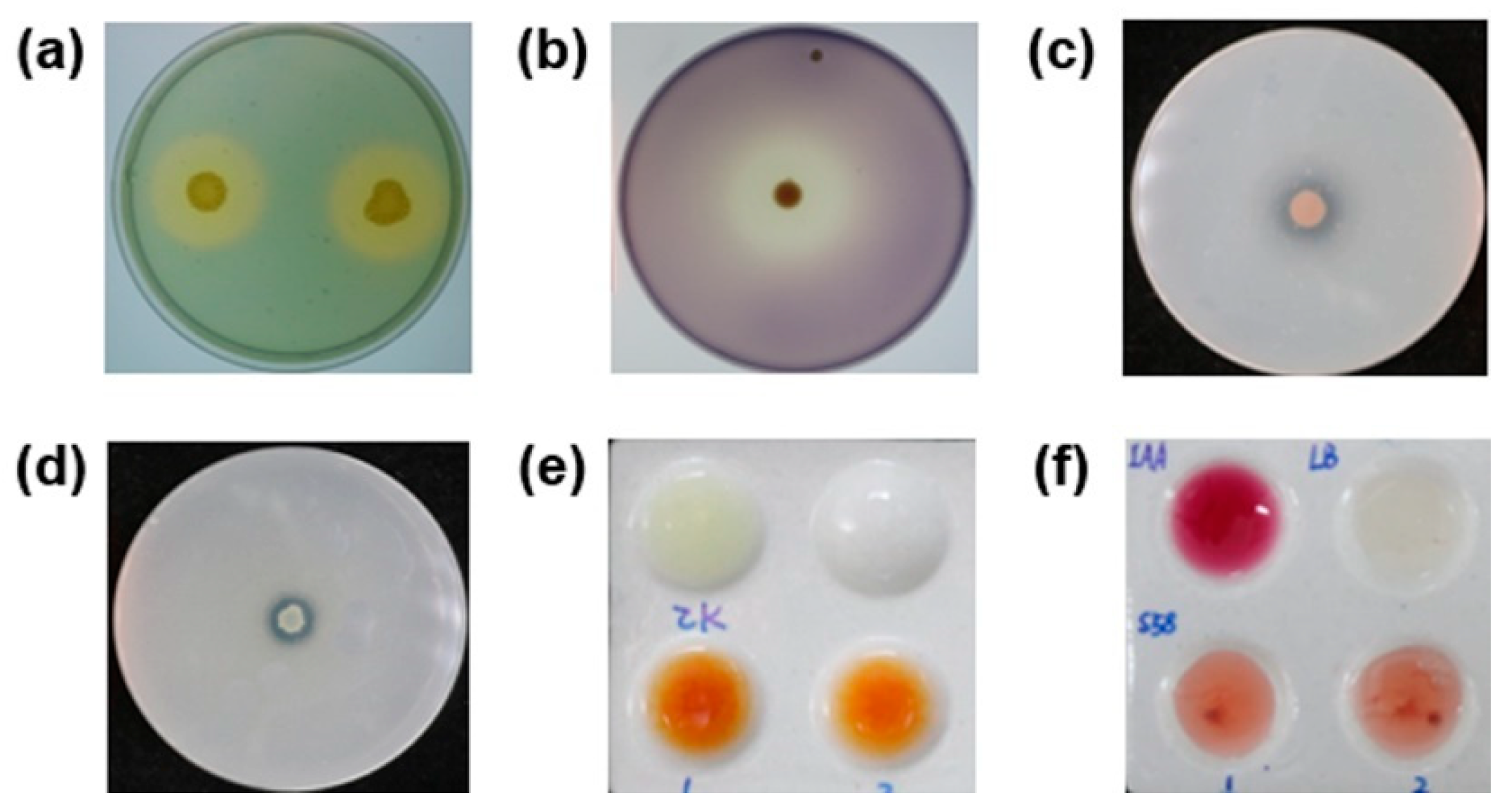
2.2. Antagonistic Test
2.3. Disease Control Assay
2.4. Identification of Plant Growth-Promoting Traits
2.5. Genome Sequencing and Comparative Genomic Analysis
2.6. Assay of Cell Death
2.7. Quantitative Reverse Transcription PCR
2.8. Statistical analysis
3. Results
3.1. Antagonism Against Phytopathogens
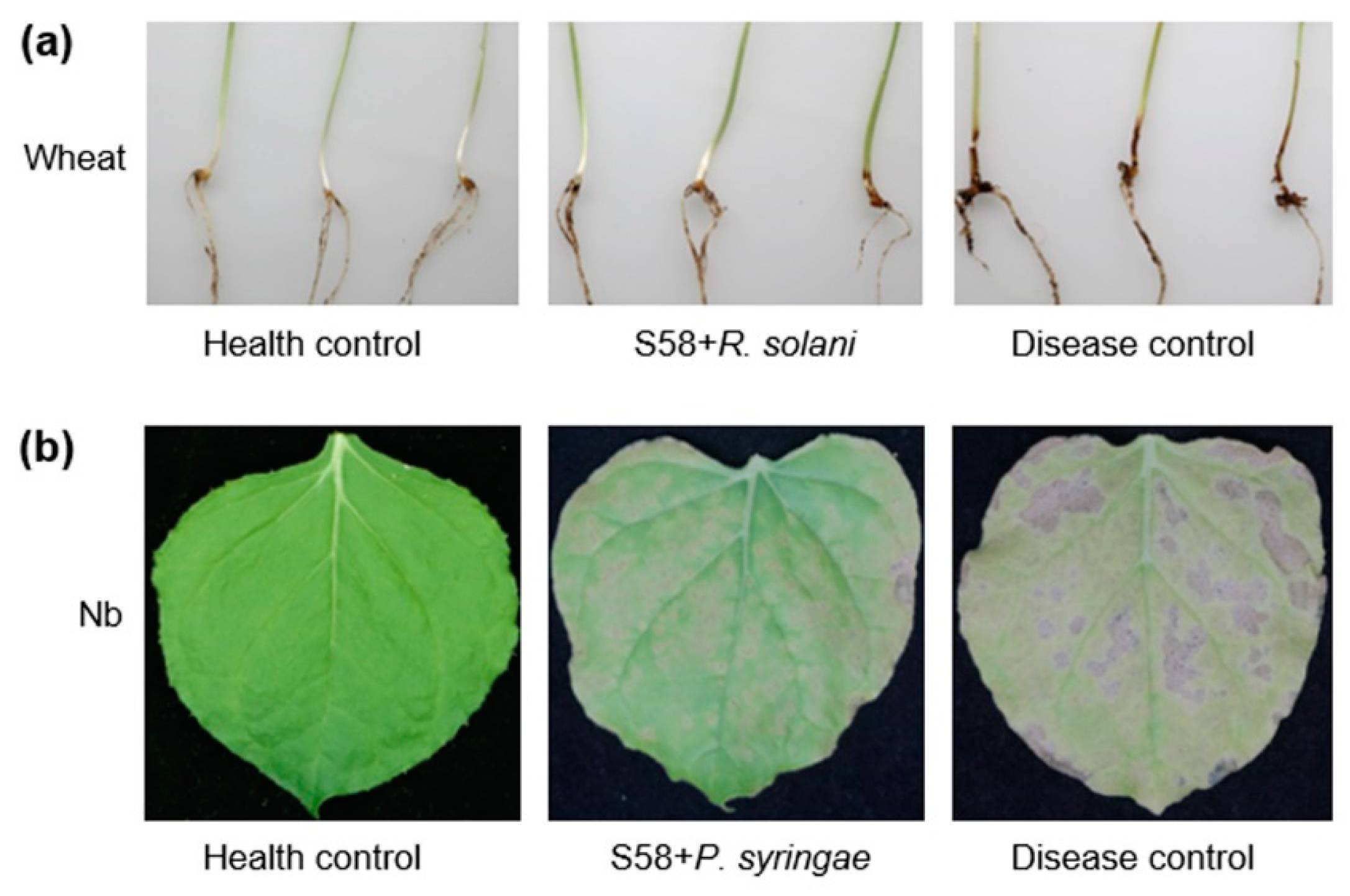
3.2. Biological Control of Plant Diseases
3.3. Plant-Growth Promoting Activities
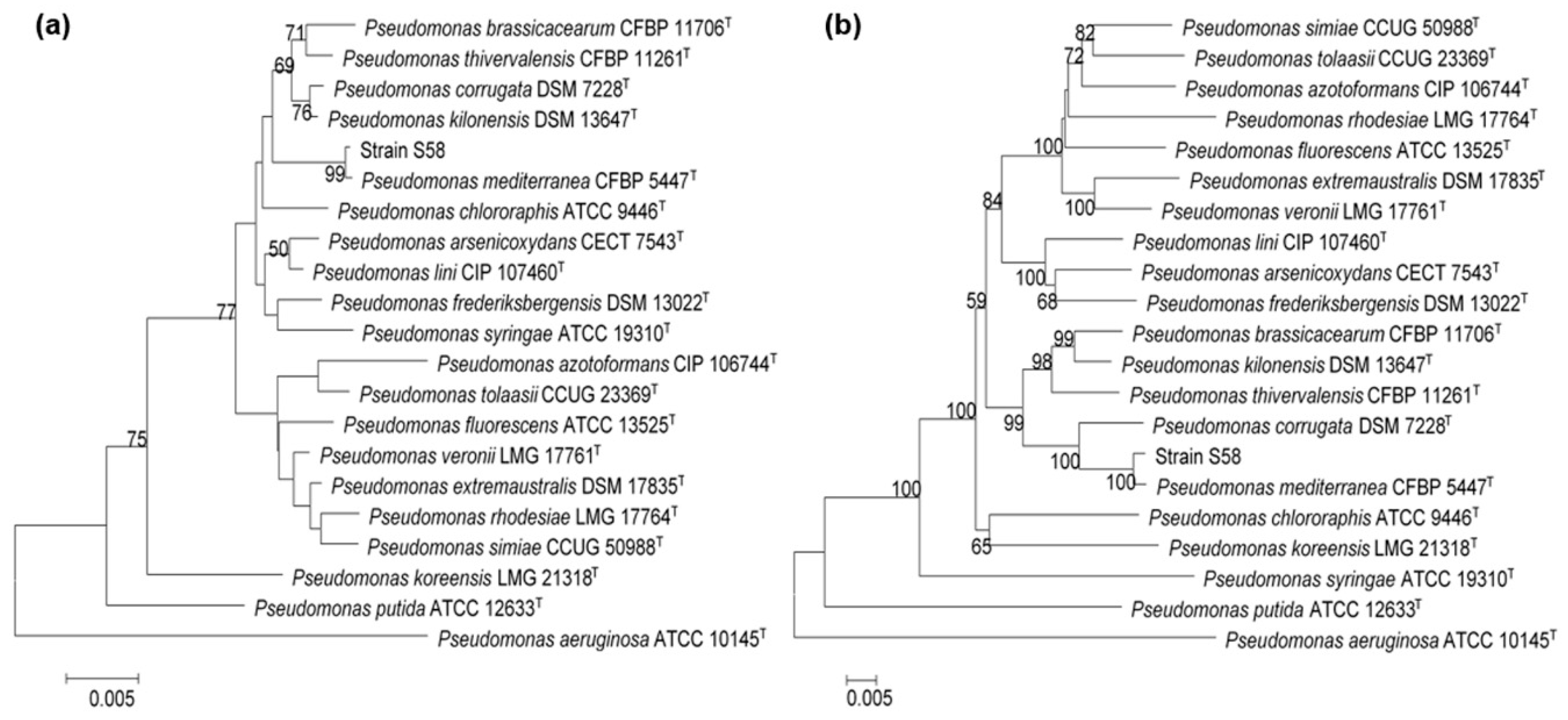
3.4. Identification of Strain S58
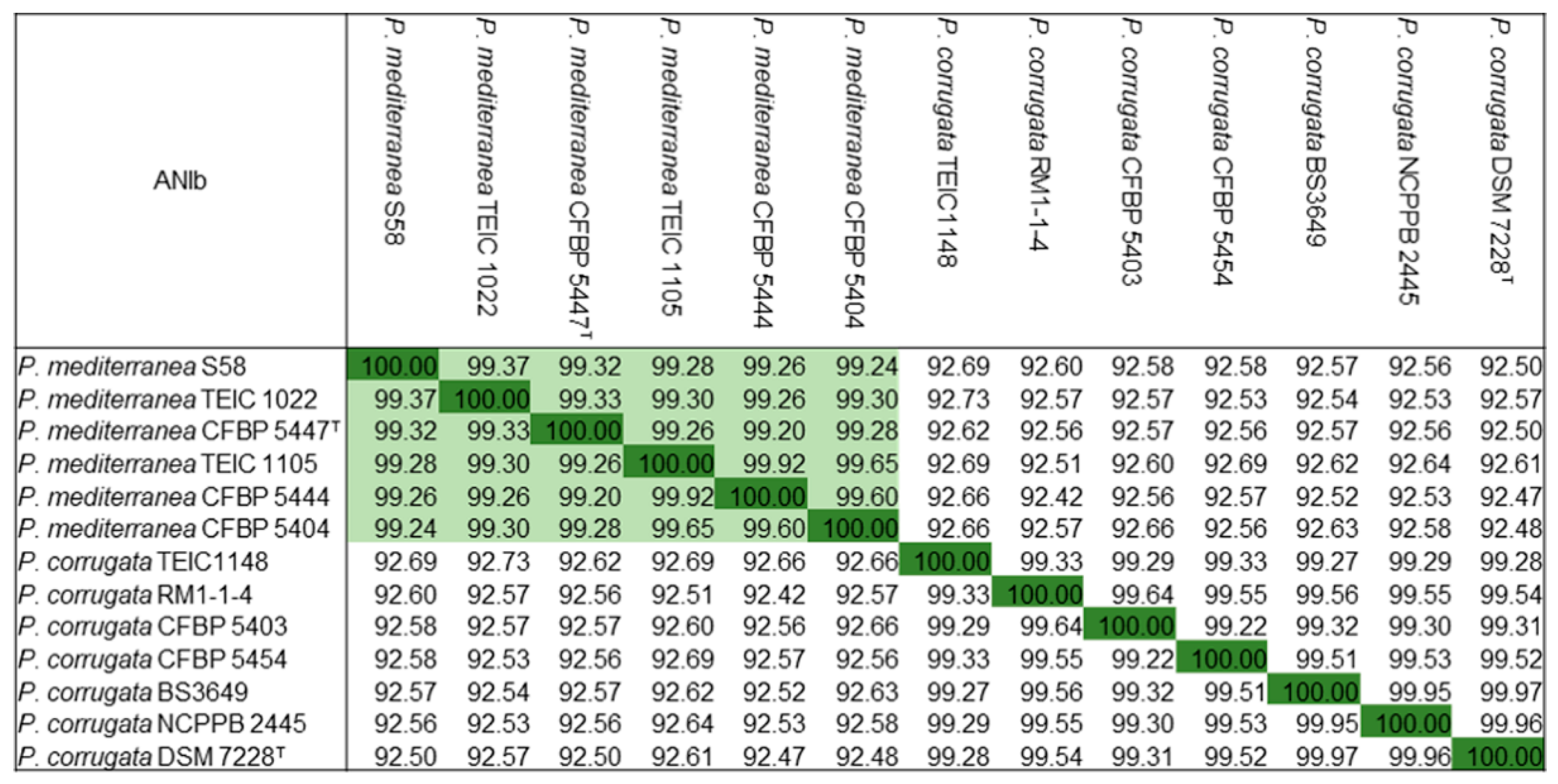
3.5. Comparative Genomic Analysis
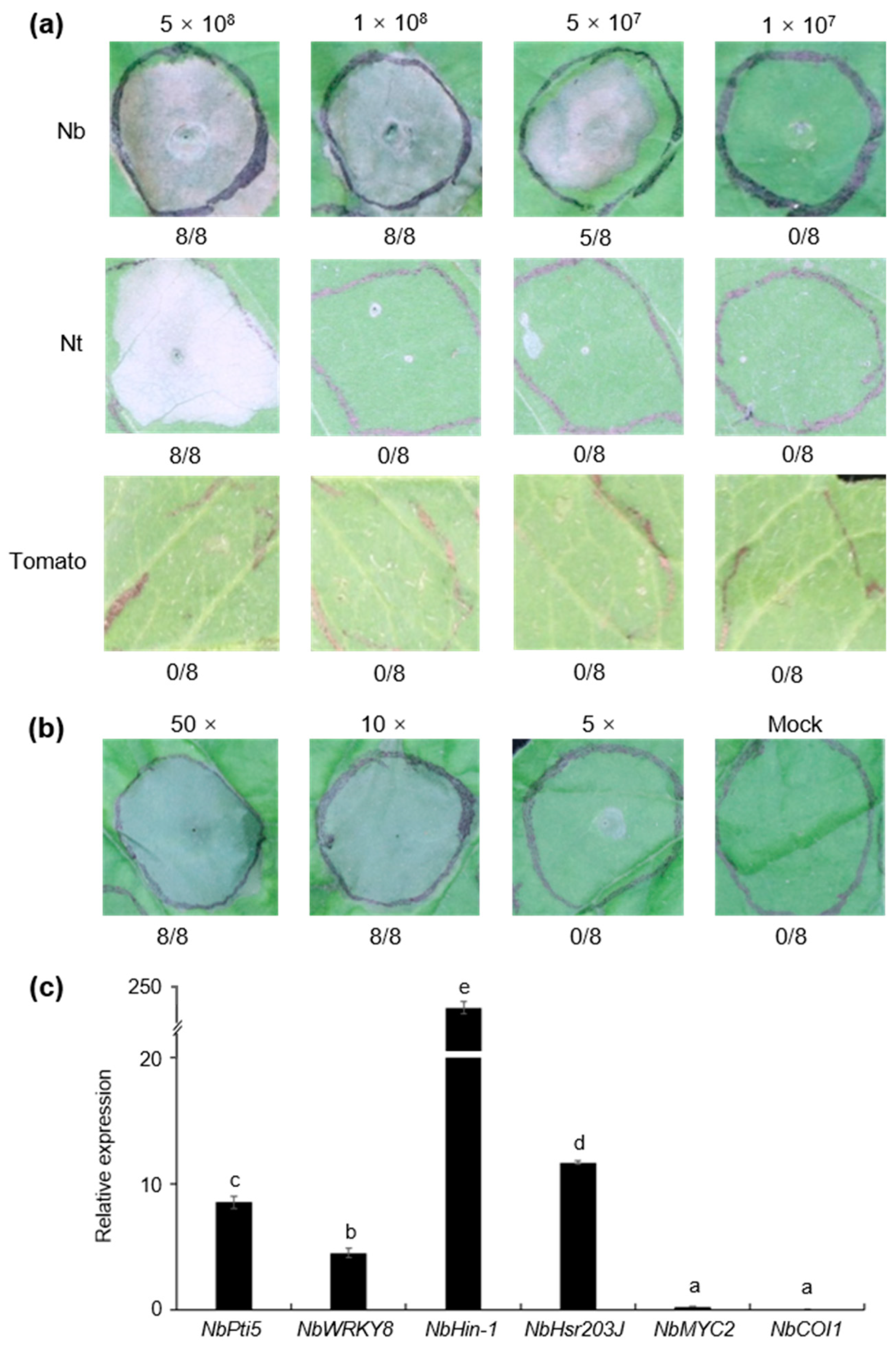
3.6. Strain S58 Triggered Cell Death and Plant Immunity
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Raaijmakers, J.M.; Weller, D.M.; Thomashow, L.S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 1997, 63, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Raguchander, T.; Samiyappan, R. Enhancing resistance of tomato and hot pepper to Pythium diseases by seed treatment with fluorescent pseudomonads. Eur. J. Plant Pathol. 2002, 108, 429–441. [Google Scholar] [CrossRef]
- Rosier, A.; Bishnoi, U.; Lakshmanan, V.; Sherrier, D.J.; Bais, H.P. A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 2016, 90, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.L.; Touraine, B.; Moenne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dye, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.A.; Zhang, H.; Pieterse, C.M.J.; Bolton, M.D.; de Jonge, R. Microbial small molecules—weapons of plant subversion. Nat. Prod. Rep. 2018, 35, 410–433. [Google Scholar] [CrossRef] [PubMed]
- Ghequire, M.G.; De Mot, R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 2014, 38, 523–568. [Google Scholar] [CrossRef] [PubMed]
- Geudens, N.; Martins, J.C. Cyclic lipodepsipeptides from Pseudomonas spp. - biological swiss-army knives. Front. Microbiol. 2018, 9, 1867. [Google Scholar] [CrossRef]
- Almario, J.; Bruto, M.; Vacheron, J.; Prigent-Combaret, C.; Moenne-Loccoz, Y.; Muller, D. Distribution of 2,4-diacetylphloroglucinol biosynthetic genes among the Pseudomonas spp. reveals unexpected polyphyletism. Front. Microbiol. 2017, 8, 1218. [Google Scholar] [CrossRef]
- Ramette, A.; Frapolli, M.; Fischer-Le Saux, M.; Gruffaz, C.; Meyer, J.M.; Défago, G.; Sutra, L.; Moënne-Loccoz, Y. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 2011, 34, 180–188. [Google Scholar] [CrossRef]
- Nowak-Thompson, B.; Gould, S.J.; Loper, J.E. Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene. 1997, 204, 17–24. [Google Scholar] [CrossRef]
- Wu, X.G.; Duan, H.M.; Tian, T.; Yao, N.; Zhou, H.Y.; Zhang, L.Q. Effect of the hfq gene on 2,4-diacetylphloroglucinol production and the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24. FEMS Microbiol. Lett. 2010, 309, 16–24. [Google Scholar] [PubMed]
- Berendsen, R.L.; van Verk, M.C.; Stringlis, I.A.; Zamioudis, C.; Tommassen, J.; Pieterse, C.M.J.; Bakker, P.A.H.M. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015, 16, 539. [Google Scholar] [CrossRef] [PubMed]
- Andrade, L.F.; de Souza, G.L.; Nietsche, S.; Xavier, A.A.; Costa, M.R.; Cardoso, A.M.; Pereira, M.C.; Pereira, D.F. Analysis of the abilities of endophytic bacteria associated with banana tree roots to promote plant growth. J. Microbiol. 2014, 52, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Xu, T.; Chen, L.; Chen, J.; Rao, C.; Xiao, X.; Wan, Y.; Zeng, G.; Long, F.; Liu, C.; et al. Endophyte-assisted promotion of biomass production and metal-uptake of energy crop sweet sorghum by plant-growth promoting endophyte Bacillus sp. SLS18. Appl. Microbiol. Biotechnol. 2012, 93, 1745–1753. [Google Scholar] [CrossRef]
- Taurian, T.; Anzuay, M.S.; Angelini, J.G.; Tonelli, M.L.; Ludueña, L.; Pena, D.; Ibáñez, F.; Fabra, A. Phosphate-solubilizing peanut associated bacteria: Screening for plant growth-promoting activities. Plant Soil. 2010, 329, 421–431. [Google Scholar] [CrossRef]
- Cavalcante, J.J.; Vargas, C.; Nogueira, E.M.; Vinagre, F.; Schwarcz, K.; Baldani, J.I.; Ferreira, P.C.; Hemerly, A.S. Members of the ethylene signalling pathway are regulated in sugarcane during the association with nitrogen-fixing endophytic bacteria. J. Exp. Bot. 2007, 58, 673–686. [Google Scholar] [CrossRef]
- Catara, V. Pseudomonas corrugata: Plant pathogen and/or biological resource? Mol. Plant Pathol. 2007, 8, 233–244. [Google Scholar] [CrossRef]
- Catara, V.; Sutra, L.; Morineau, A.; Achouak, W.; Christen, R.; Gardan, L. Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1749–1758. [Google Scholar]
- Ryder, M.H.; Rovira, A.D. Biological control of take-all of glass house-grown wheat using strains of Pseudomonas corrugata isolated from wheat field soil. Soil Biol. Biochem. 1993, 25, 311–320. [Google Scholar] [CrossRef]
- Schmidt, C.S.; Agostini, F.; Leifert, C.; Killham, K.; Mullins, C.E. Influence of inoculum density of the antagonistic bacteria Pseudomonas fluorescens and Pseudomonas corrugata on sugar beet seedling colonisation and suppression of Pythium damping off. Plant Soil. 2004, 265, 111–122. [Google Scholar] [CrossRef]
- Licciardello, G.; Caruso, A.; Bella, P.; Gheleri, R.; Strano, C.P.; Anzalone, A.; Trantas, E.A.; Sarris, P.F.; Almeida, N.F.; Catara, V. The LuxR regulators PcoR and RfiA co-regulate antimicrobial peptide and alginate production in Pseudomonas corrugata. Front. Microbiol. 2018, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, G.; Strano, C.P.; Bertani, I.; Bella, P.; Fiore, A.; Fogliano, V.; Venturi, V.; Catara, V. N-acyl-homoserine-lactone quorum sensing in tomato phytopathogenic Pseudomonas spp. is involved in the regulation of lipodepsipeptide production. J. Biotechnol. 2012, 159, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, G.; Catara, A.F.; Catara, V. Production of polyhydroxyalkanoates and extracellular products using Pseudomonas corrugata and P. mediterranea: A review. Bioengineering 2019, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Bennasar, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. PseudoMLSA: A database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol. 2010, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Cattelan, A.J.; Hartel, P.G.; Fuhrmann, J.J. Screening of plant growth—Promoting rhizobacteria to promote early soybean growth. Soil Sci. Soc. Amer. 1999, 63, 1670–1680. [Google Scholar] [CrossRef]
- Cappuccino, J.C.; Sherman, N. Negative staining. In Microbiology: A Laboratory Manual, 3rd ed.; Cappuccino, J.C., Sherman, N., Eds.; Cumming Publ.: New York, NY, USA, 1992; pp. 125–179. [Google Scholar]
- Zamioudis, C.; Mastranesti, P.; Dhonukshe, P.; Blilou, I.; Pieterse, C.M.J. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 2013, 162, 304–318. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Ikeda-Ohtsubo, W.; Nagata, Y.; Tsuda, M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.M.; Kwon, S.J.; Chun, J.A. large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA—An ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Licciardello, G.; Bella, P.; Devescovi, G.; Strano, C.P.; Sarris, P.F.; Catara, A.F.; Venturi, V.; Catara, V. Draft genome sequence of Pseudomonas mediterranea strain CFBP 5447T, a producer of filmable medium-chain-length polyhydroxyalkanoates. Genome Announc. 2014, 2, e01260-14. [Google Scholar] [CrossRef]
- Trantas, E.A.; Licciardello, G.; Almeida, N.F.; Witek, K.; Strano, C.P.; Duxbury, Z.; Ververidis, F.; Goumas, D.E.; Jones, J.D.; Guttman, D.S.; et al. Comparative genomic analysis of multiple strains of two unusual plantpathogens: Pseudomonas corrugata and Pseudomonas mediterranea. Front. Microbiol. 2015, 6, 811. [Google Scholar] [CrossRef]
- Zachow, C.; Müller, H.; Laireiter, C.M.; Tilcher, R.; Berg, G. Complete genome sequence of Pseudomonas corrugata strain RM1-1-4, a stress protecting agent from the rhizosphere of an oilseed rape bait plant. Stand. Genomic Sci. 2017, 12, 66. [Google Scholar] [CrossRef]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef]
- McClerklin, S.A.; Lee, S.G.; Harper, C.P.; Nwumeh, R.; Jez, J.M.; Kunkel, B.N. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 2018, 14, e1006811. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W., II; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.L.; Collmer, A. Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Mol. Plant Pathol. 2018, 19, 1779–1794. [Google Scholar]
- Scaloni, A.; Dalla Serra, M.; Amodeo, P.; Mannina, L.; Vitale, R.M.; Segre, A.L.; Cruciani, O.; Lodovichetti, F.; Greco, M.L.; Fiore, A.; et al. Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: Cormycin A. Biochem. J. 2004, 384, 25–36. [Google Scholar] [CrossRef] [PubMed]



| Features | Chromosome |
|---|---|
| Size (bp) | 6,486,667 |
| G+C content (%) | 61.06 |
| Number of total CDSs | 5594 |
| Number of genes | 5312 |
| Pseudogenes | 282 |
| tRNAs | 67 |
| rRNA genes | 15 |
| ncRNAs | 4 |
| Contigs | 1 |
| Total CDSs size (bp) | 5,683,341 |
| Coding % | 87.62 |
| Average CDS length (nt) | 992.90 |
| Cluster Type/Metabolites | S58 | RM1-1-4 1 | |
|---|---|---|---|
| Location | Gene ID | ||
| NRPS-like/Fragin | 177591-219516 | E3Z27_00825-E3Z27_00960 | + |
| Arylpolyene/APE Vf | 476416-520027 | E3Z27_02080-E3Z27_02290 | + |
| Bacteriocin | 1422090-1432977 | E3Z27_06235-E3Z27_06295 | + |
| NAGGN | 1834296-1849084 | E3Z27_08105-E3Z27_08160 | + |
| NRPS/Fengycin | 2221816-2245051 | E3Z27_09835-E3Z27_-09940 | + |
| NRPS/Crochelin A | 2691862-2766373 | E3Z27_12005-E3Z27_12180 | - |
| NRPS-like/Entolysin | 2967333-3015201 | E3Z27_12950-E3Z27_13125 | + |
| NRPS/Orfamide B | 3271309-3337076 | E3Z27_14180-E3Z27_14400 | - |
| NRPS/Siderophore | 3444389-3463297 | E3Z27_14930-E3Z27_15000 | + |
| NRPS/Syringomycin | 3672567-3850583 | E3Z27_15910-E3Z27_16205 | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Wang, J.; Xia, Z.; Wei, H.-L. Characterization of a Versatile Plant Growth-Promoting Rhizobacterium Pseudomonas mediterranea Strain S58. Microorganisms 2020, 8, 334. https://doi.org/10.3390/microorganisms8030334
Gu Y, Wang J, Xia Z, Wei H-L. Characterization of a Versatile Plant Growth-Promoting Rhizobacterium Pseudomonas mediterranea Strain S58. Microorganisms. 2020; 8(3):334. https://doi.org/10.3390/microorganisms8030334
Chicago/Turabian StyleGu, Yilin, Jing Wang, Zhenyuan Xia, and Hai-Lei Wei. 2020. "Characterization of a Versatile Plant Growth-Promoting Rhizobacterium Pseudomonas mediterranea Strain S58" Microorganisms 8, no. 3: 334. https://doi.org/10.3390/microorganisms8030334
APA StyleGu, Y., Wang, J., Xia, Z., & Wei, H.-L. (2020). Characterization of a Versatile Plant Growth-Promoting Rhizobacterium Pseudomonas mediterranea Strain S58. Microorganisms, 8(3), 334. https://doi.org/10.3390/microorganisms8030334





