Taxonomic Characterization and Secondary Metabolite Analysis of NEAU-wh3-1: An Embleya Strain with Antitumor and Antibacterial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Streptomyces-Like Strains
2.2. Screening of Strains with Antitumor Activity
2.3. Morphological and Physiological and Biochemical Characteristics of NEAU-wh3-1
2.4. Chemotaxonomic Analysis of NEAU-wh3-1
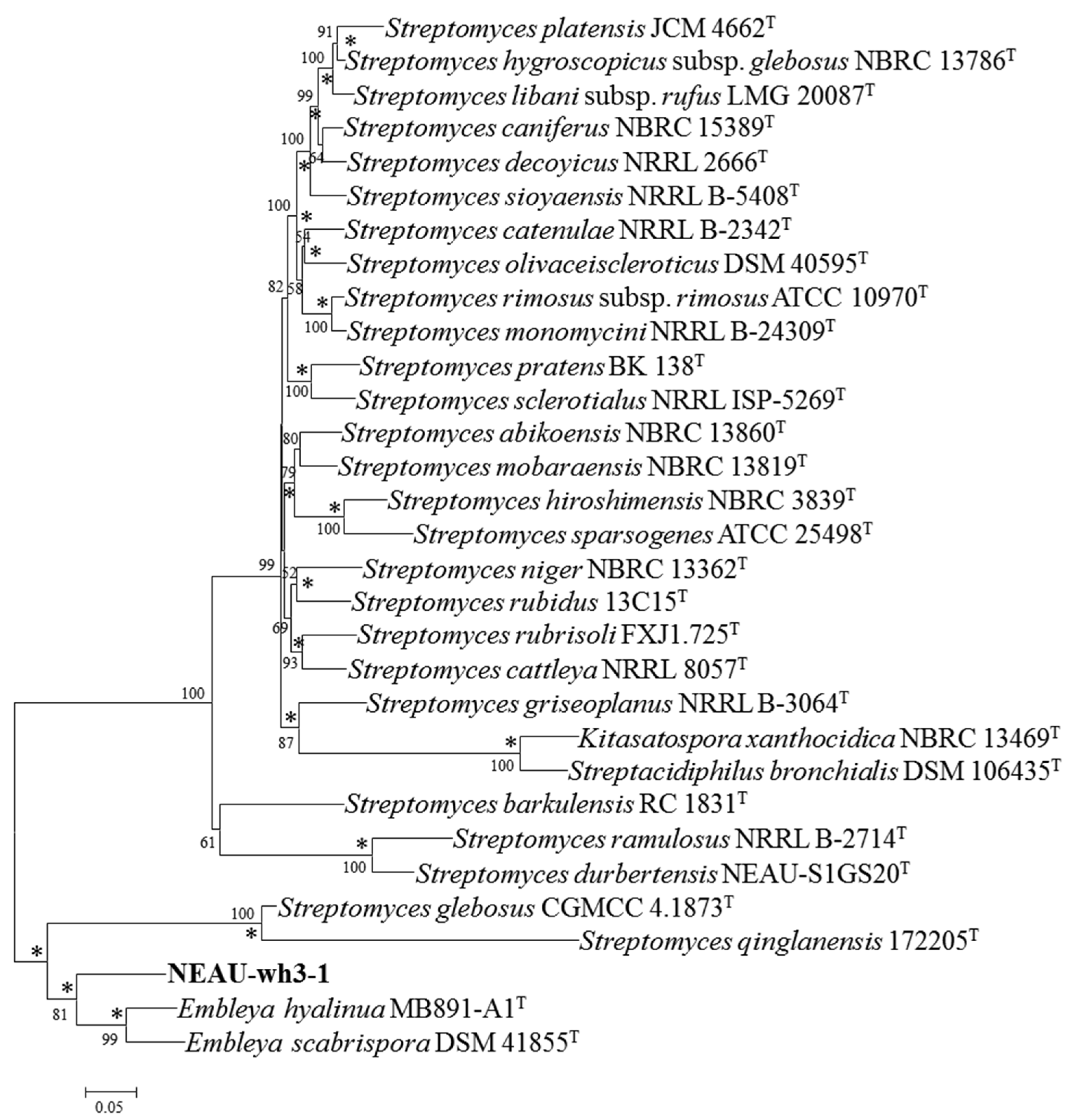
2.5. Phylogenetic Analysis of NEAU-wh3-1
2.6. Production
2.7. Extraction and Isolation
2.8. General Experimental Procedures
2.9. Biological Assays
3. Results
3.1. Isolation and Screening of an Antitumor Compound Producing Strains
3.2. Polyphasic Taxonomic Characterization of NEAU-wh3-1
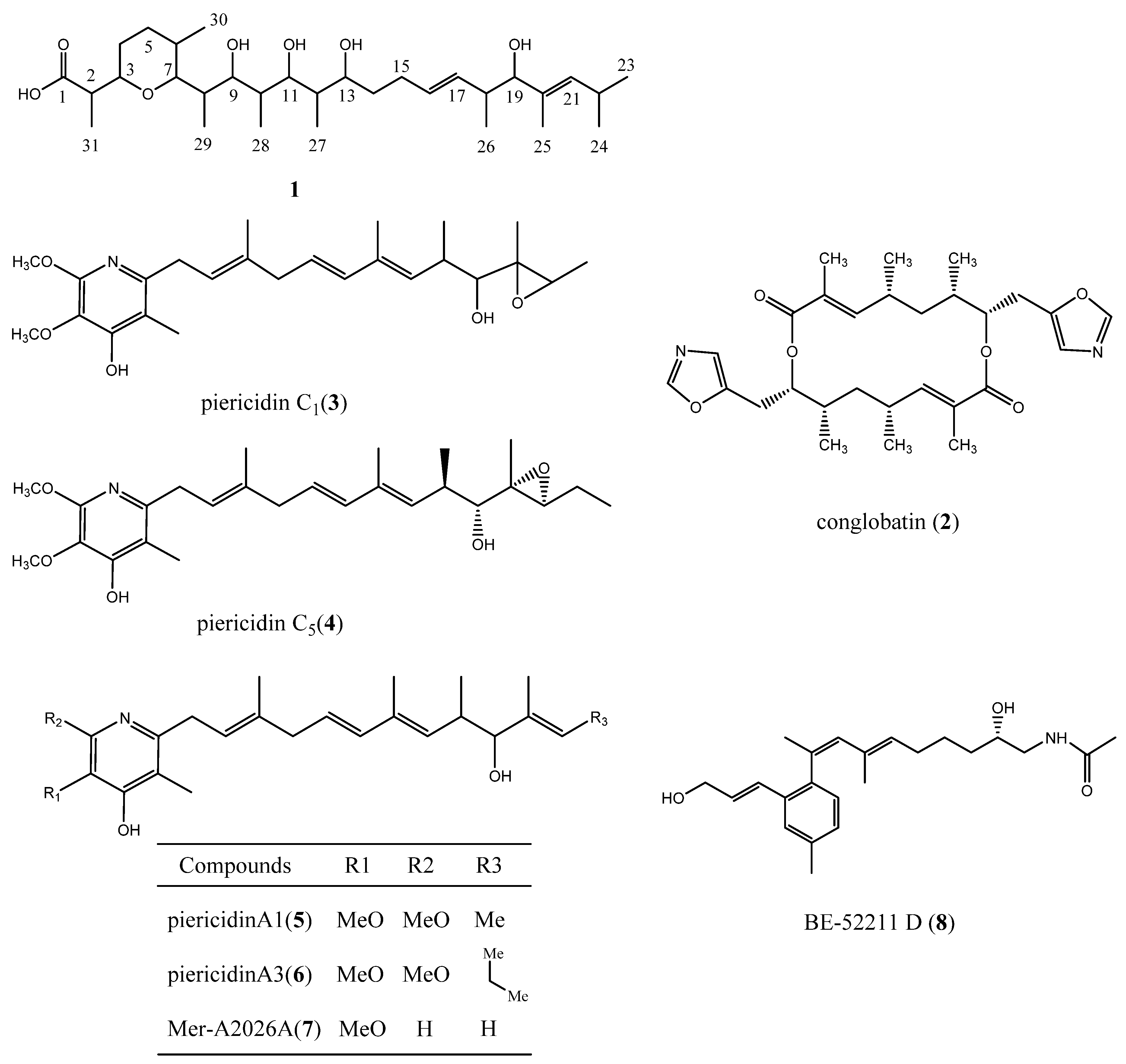
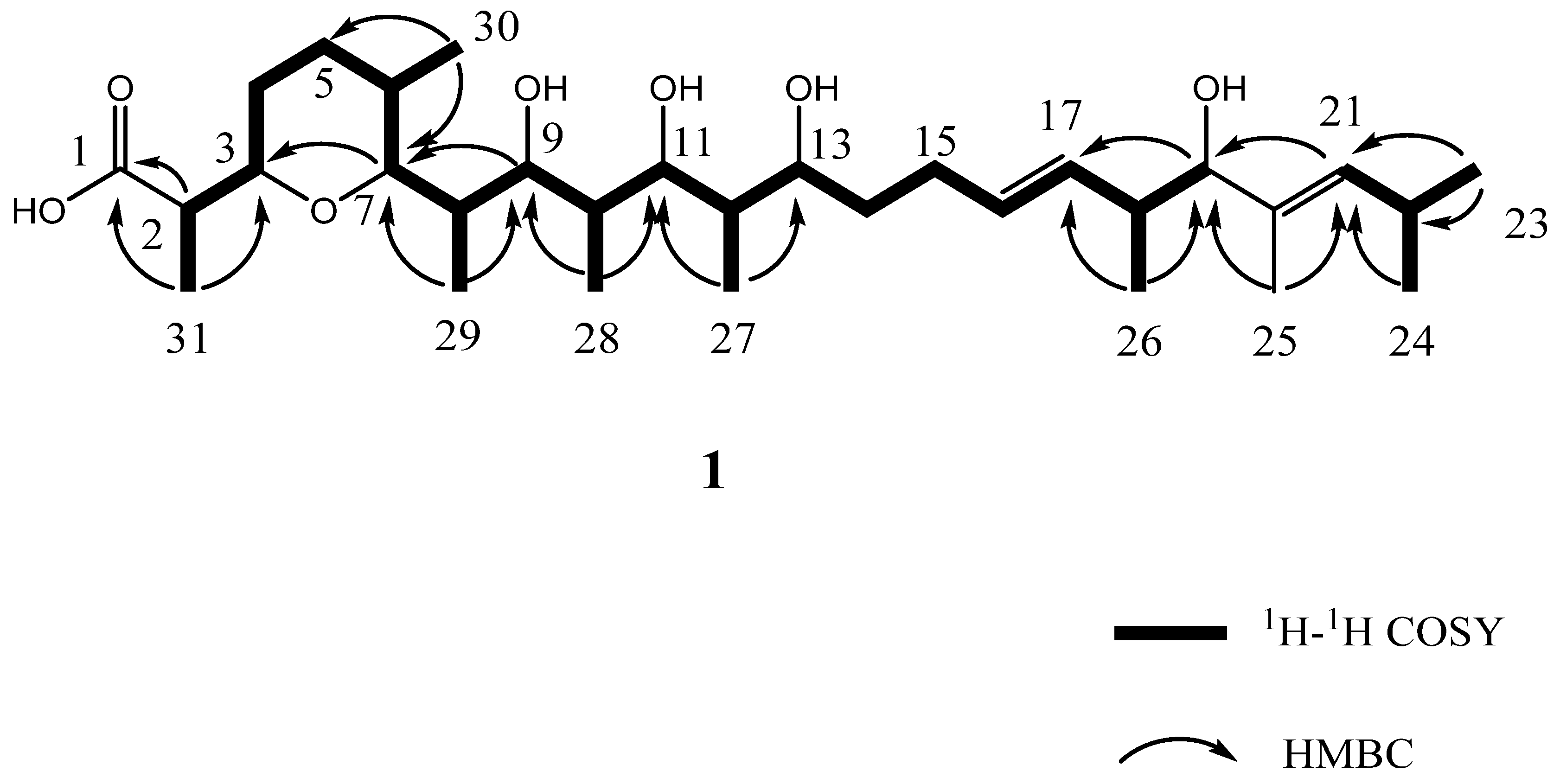
3.3. Structural Elucidation
3.4. Biological Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA. Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [Green Version]
- Commander, H.; Whiteside, G.; Perry, C. Vandetanib: First global approval. Drugs 2011, 71, 1355–1365. [Google Scholar] [CrossRef]
- Zhu, F.; Zhao, X.; Li, J.; Guo, L.; Bai, L.; Qi, X. A new compound Trichomicin exerts antitumor activity through STAT3 signaling inhibition. Biomed. Pharmacother. 2020, 121, 109608. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, X.; Wang, Z.; Wu, P.; Huang, J. Anthracyclines potentiate anti-tumor immunity: A new opportunity for chemoimmunotherapy. Cancer Lett. 2015, 369, 331–335. [Google Scholar] [CrossRef]
- Sznarkowska, A.; Kostecka, A.; Meller, K.; Bielawski, K.P. Inhibition of cancer antioxidant defense by natural compounds. Oncotarget 2017, 8, 15996–16016. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [Green Version]
- Cragg, G.M.; Newman, D.J.; Snader, K.M. Natural products in drug discovery and development. J. Nat. Prod. 1997, 60, 52–60. [Google Scholar] [CrossRef]
- Chin, Y.W.; Balunas, M.J.; Chai, H.B.; Kinghorn, A.D. Drug discovery from natural sources. Drug Addict. From Basic Res. Ther. 2008, 8, 17–39. [Google Scholar]
- Pettit, R.K. Culturability and Secondary Metabolite Diversity of Extreme Microbes: Expanding Contribution of Deep Sea and Deep-Sea Vent Microbes to Natural Product Discovery. Mar. Biotechnol. 2011, 13, 1–11. [Google Scholar] [CrossRef]
- Qin, S.; Li, J.; Chen, H.H.; Zhao, G.Z.; Zhu, W.Y.; Jiang, C.L.; Xu, L.H.; Li, W.J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna China. Appl. Environ. Microbiol. 2009, 75, 6176–6186. [Google Scholar] [CrossRef] [Green Version]
- Parkinson, D.R.; Arbuck, S.G.; Moore, T.; Pluda, J.M.; Christian, M.C. Clinical development of anticancer agents from natural products. Stem Cells 1994, 12, 30–43. [Google Scholar] [CrossRef]
- Berdy, J. Bioactive microbial metabolites. J Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, M.; Williams, S.T. Ecology of actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- Rabbani, A.; Finn, R.M.; Ausió, J. The anthracycline antibiotics: Antitumor drugs that alter chromatin structure. BioEssays 2005, 27, 50–56. [Google Scholar] [CrossRef]
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 Years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Brooks, H.A.; Gardner, D.; Poyser, J.P.; King, T.J. The structure and absolute stereochemistry of zincophorin (antibiotic m144255): a monobasic carboxylic acid ionophore having a remarkable specificity for divalent cations. J. Antibiot. 1984, 37(11), 1501–1504. [Google Scholar] [CrossRef] [Green Version]
- Walther, E.; Boldt, S.; Kage, H.; Lauterbach, T.; Martin, K.; Roth, M.; Hertweck, C.; Sauerbrei, A.; Schmidtke, M.; Nett, M. Zincophorin-biosynthesis in Streptomyces griseus and antibiotic properties. GMS Infect. Dis. 2016, 4, Doc08. [Google Scholar]
- Yang, S.X.; Gao, J.M.; Zhang, A.L.; Laatsch, H.S. A novel toxic macrolactam polyketide glycoside produced by actinomycete Streptomyces sannanensis. Bioorg. Med. Chem. Lett. 2011, 21, 3905–3908. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Culturable rare Actinomycetes: Diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 2013, 97, 9291–9321. [Google Scholar] [CrossRef]
- Heidari, B.; Mohammadipanah, F. Isolation and identification of two alkaloid structures with radical scavenging activity from Actinokineospora sp. UTMC 968, a new promising source of alkaloid compounds. Mol. Biol. Rep. 2018, 45, 2325–2332. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.Y.; Qi, H.; Li, J.S.; Zhang, H.; Zhang, J.; Wang, J.D.; Xiang, W.S. A new polysubstituted cyclopentene derivative from Streptomyces sp. HS-NF-1046. J. Antibiot. 2017, 70, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Peláez, F. The historical delivery of antibiotics from microbial natural products - Can history repeat? Biochem. Pharmacol. 2006, 71, 981–990. [Google Scholar] [CrossRef]
- Kekuda, P.; Onkarappa, R.; Jayanna, N. Characterization and Antibacterial Activity of a Glycoside Antibiotic from Streptomyces variabilis PO-178. Sci. Technol. Arts. Res. J. 2015, 3, 116. [Google Scholar] [CrossRef]
- Miao, V.; Davies, J. Actinobacteria: The good, the bad, and the ugly. Antonie van Leeuwenhoek 2010, 98, 143–150. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef]
- Ayed, A.; Slama, N.; Mankai, H.; Bachkouel, S.; ElKahoui, S.; Tabbene, O.; Limam, F. Streptomyces tunisialbus sp. nov., a novel Streptomyces species with antimicrobial activity. Antonie van Leeuwenhoek 2018, 111, 1571–1581. [Google Scholar] [CrossRef]
- Mccully, M.; Harper, J.D.I.; An, M.; Kent, J.H. The rhizosphere: the key functional unit in plant soil microbial interactions in the field implications for the understanding of allelopathic effects. Pol. J. Vet. Sci. 2005, 15, 493–498. [Google Scholar]
- Hiltner, L. Uber neuer erfahrungen und probleme auf dem gebiet der berücksichtigung unter besonderer berücksichtigung der gründüngung und brache. Arbeiten der Deustchen Landwirtschaftsgesellesschaft 1904, 32, 1405–1417. [Google Scholar]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends. Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Loper, J.E.; Gross, H. Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur. J. Plant Pathol. 2007, 119, 265–278. [Google Scholar] [CrossRef]
- Reiter, B.; Sessitsch, A.; Nowak, J.; Cle, C. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Society 2005, 71, 1685–1693. [Google Scholar]
- Lanteigne, C.; Gadkar, V.J.; Wallon, T.; Novinscak, A.; Filion, M. Production of DAPG and HCN by Pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology 2012, 102, 967–973. [Google Scholar] [CrossRef] [Green Version]
- Osei, E.; Kwain, S.; Mawuli, G.T.; Anang, A.K.; Owusu, K.B.A.; Camas, M.; Camas, A.S.; Ohashi, M.; Alexandru-Crivac, C.N.; Deng, H. Paenidigyamycin A, potent antiparasitic imidazole alkaloid from the ghanaian Paenibacillus sp. De2Sh. Mar. Drugs 2019, 17, 9. [Google Scholar] [CrossRef] [Green Version]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Malik, K.A. Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Brazilian J. Microbiol. 2019, 50, 85–97. [Google Scholar] [CrossRef]
- Orfali, R.; Perveen, S. Secondary metabolites from the Aspergillus sp. in the rhizosphere soil of Phoenix dactylifera (Palm tree). BMC Chem. 2019, 13, 1–6. [Google Scholar] [CrossRef]
- Wang, R.J.; Zhang, S.Y.; Ye, Y.H.; Yu, Z.; Qi, H.; Zhang, H.; Xue, Z.L.; Wang, J.D.; Wu, M. Three new isoflavonoid glycosides from the mangrove-derived actinomycete micromonospora aurantiaca 110B. Mar. Drugs 2019, 17, 294. [Google Scholar] [CrossRef] [Green Version]
- Nouioui, I.; Carro, L.; García-López, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Pukall, R.; Klenk, H.-P.; Goodfellow, M.; Göker, M. Genome-Based Taxonomic Classification of the Phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef] [Green Version]
- Oren, A.; Garrity, G.M. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2018, 68, 2707–2709. [Google Scholar] [CrossRef]
- Komaki, H.; Hosoyama, A.; Kimura, A.; Ichikawa, N.; Igarashi, Y.; Tamura, T. Classification of ‘Streptomyces hyalinum’ Hamada and Yokoyama as Embleya hyalina sp. nov., the second species in the genus Embleya, and emendation of the genus Embleya. Int. J. Syst. Evol. Microbiol. 2020, 1–5. [Google Scholar] [CrossRef]
- Kämpfer, P.; Genus, I. Streptomyces Waksman and Henrici 1943, 339 AL. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2012; pp. 1679–1680. [Google Scholar]
- Ping, X.; Takahashi, Y.; Seino, A.; Iwai, Y.; Omura, S. Streptomyces scarbrisporus sp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 577–581. [Google Scholar] [CrossRef]
- Omura, S.; Nakagawa, A.; Shibata, K.; Sano, H. The structure of hitachimycin, a novel macrocyclic lactam involving β-phenylalanine. Tetrahedron Lett. 1982, 23, 4713–4716. [Google Scholar] [CrossRef]
- Komiyama, K.; Edanami, K.I.; Yamamoto, H.; Umezawa, I. Antitumor activity of a new antitumor antibiotic, stubomycin. J. Antibiot. 1982, 35, 703–706. [Google Scholar] [CrossRef] [Green Version]
- Shibata, K.; Satsumabayashi, S.; Sano, H.; Komiyama, K.; Nakagawa, A.; Omura, S. Chemical modification of hitachimycin. Synthesis, antibacterial, cytocidal and in vivo antitumor activities of hitachimycin derivatives. J. Antibiot. 1988, 41, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Naganawa, H.; Wakashiro, T.; Yagi, A.; Kondo, S.; Takita, T.; Hamada, M.; Maeda, K.; Umezawa, H. Deoxynybomycin from a streptomyces. J. Antibiot. 1970, 23, 365–368. [Google Scholar] [CrossRef] [Green Version]
- Hiramatsu, K.; Igarashi, M.; Morimoto, Y.; Baba, T.; Umekita, M.; Akamatsu, Y. Curing bacteria of antibiotic resistance: Reverse antibiotics, a novel class of antibiotics in nature. Int. J. Antimicrob. Agents 2012, 39, 478–485. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, M.; Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 1987, 65, 501–509. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef] [Green Version]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Jin, L.; Zhao, Y.; Song, W.; Duan, L.; Jiang, S.; Wang, X.; Zhao, J.; Xiang, W. Streptomyces inhibens sp. Nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.). Int. J. Syst. Evol. Microbiol. 2019, 69, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.W.; Han, L.Y.; Yu, M.Y.; Cao, P.; Li, D.M.; Guo, X.W.; Liu, Y.Q.; Wang, X.J.; Xiang, W.S. Characterization of Streptomyces sporangiiformans sp. nov., a Novel Soil Actinomycete with Antibacterial Activity against Ralstonia solanacearum. Microorganisms 2019, 7, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology, 2nd ed.; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Gordon, R.E.; Barnett, D.A.; Handerhan, J.E.; Pang, C. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int. J. Syst. Bacteriol. 1974, 24, 54–63. [Google Scholar] [CrossRef] [Green Version]
- Yokota, A.; Tamura, T.; Hasegawa; Huang, L.H. Catenuloplanes japonicas gen. nov., sp. nov., nom. rev., a new genus of the order Actinomycetales. Int. J. Syst. Bacteriol. 1993, 43, 805–812. [Google Scholar] [CrossRef] [Green Version]
- Lechevalier, M.P.; Lechevalier, H.A. The chemotaxonomy of actinomycetes. In Actinomycete taxonomy, 2nd ed.; Dietz, A., Thayer, D.W., Eds.; special publication for Society of Industrial Microbiology: Arlington, TX, USA, 1980; pp. 227–291. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Collins, M.D. Isoprenoid quinone analyses in bacterial classification and identification. In Chemical methods in bacterial systematics; Goodfellow, M., Minnikin, D.E., Eds.; Academic Press: London, UK, 1985; pp. 267–284. [Google Scholar]
- Shen, Y.; Sun, T.; Jiang, S.; Mu, S.; Li, D.; Guo, X.; Zhang, J.; Zhao, J.; Xiang, W. Streptomyces lutosisoli sp. nov., a novel actinomycete isolated from muddy soil. Antonie Van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2018, 111, 2403–2412. [Google Scholar] [CrossRef]
- Wu, C.; Lu, X.; Qin, M.; Wang, Y.; Ruan, J. Analysis of menaquinone compound in microbial cells by HPLC. Microbiology 1989, 16, 176–178. [Google Scholar]
- Kim, S.B.; Brown, R.; Oldfield, C.; Gilbert, S.C.; Iliarionov, S.; Goodfellow, M. Gordonia amicalis sp. nov., a novel dibenzothiophene-desulphurizing actinomycete. Int. J. Syst. Evol. Microbiol. 2000, 50, 2031–2036. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence Limits on Phylogenies: an Approach Using the Bootstrap. Evolution (N. Y). 1985, 39, 783–791. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Hatano, K.; Nishii, T.; Kasai, H. Taxonomic re-evaluation of whorl-forming Streptomyces (formerly Streptoverticillium) species by using phenotypes, DNA-DNA hybridization and sequences of gyrB, and proposal of Streptomyces luteireticuli (ex Katoh and Arai 1957) corrig., sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 2003, 53, 1519–1529. [Google Scholar] [CrossRef]
- Guo, Y.P.; Zheng, W.; Rong, X.Y.; Huang, Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: Use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 2008, 58, 149–159. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; Ying, L.; Wang, C.; Jiang, N.; Zhou, Y.; Wang, H.; Bai, H. Five new epothilone metabolites from Sorangium cellulosum strain So0157-2. J. Antibiot. 2009, 62, 483–487. [Google Scholar] [CrossRef] [Green Version]
- Wayne, P.A. Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition. Clin. Lab. Standards Institute 2012, 32, 2. [Google Scholar]
- Liu, C.; Han, C.; Jiang, S.; Zhao, X.; Tian, Y.; Yan, K.; Wang, X.; Xiang, W. Streptomyces lasii sp. nov., a Novel Actinomycete with Antifungal Activity Isolated from the Head of an Ant (Lasius flavus). Curr. Microbiol. 2018, 75, 353–358. [Google Scholar] [CrossRef]
- Rong, X.; Huang, Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 2012, 35, 7–18. [Google Scholar] [CrossRef]
- Westley, J.W.; Liu, C.M.; Evans, R.H.; Blount, J.F. Conglobatin, a novel macrolide dilactone from Streptomyces conglobatus ATCC 31005. J. Antibiot. 1979, 32, 874–877. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Yoneyama, K.; Shiraishi, S.; Watanabe, A.; Takahashi, N. Chemical structures of new piericidins produced by Streptomyces pactum. Agric. Biol. Chem. 1977, 41, 855–862. [Google Scholar] [CrossRef]
- Kubota, N.K.; Ohta, E.; Ohta, S.; Koizumi, F.; Suzuki, M.; Ichimura, M.; Ikegami, S. Piericidins C5 and C6: New 4-pyridinol compounds produced by Streptomyces sp. and Nocardioides sp. Bioorganic Med. Chem. 2003, 11, 4569–4575. [Google Scholar] [CrossRef]
- Tamura, S.; Takahashi, N.; Miyamoto, S.; Mori, R.; Suzuki, S.; Nagatsu, J. Isolation and physiological activities of piericidin A, a natural insecticide produced by Streptomyces. Agr. Biol. Chem. 1963, 27, 576–582. [Google Scholar] [CrossRef]
- Kominato, K.; Watanabe, Y.; Hirano, S.I.; Kioka, T.; Tone, H. Mer-a2026a and b, novel piericidins with vasodilating effect. ii. physico-chemical properties and chemical structures. J. Antibiot. 1994, 48, 103–105. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhao, X.L.; Gao, Y.H.; Qi, H.; Zhang, H.; Xiang, W.S.; Wang, J.D.; Wang, X.J. Two new cytotoxic metabolites from Streptomyces sp. HS-NF-813. J. Asian. Nat. Prod. Res. 2018, 22, 249–256. [Google Scholar] [CrossRef]
- Hall, C.; Wu, M.; Crane, F.L.; Takahashi, H.; Tamura, S.; Folkers, K. Piericidin A: A new inhibitor of mitochondrial electron transport. Biochem. Biophys. Res. Commun. 1966, 25, 373–377. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Helmke, E.; Kelter, G.; Fiebig, H.H.; Laatsch, H. Glucopiericidin C: A cytotoxic piericidin glucoside antibiotic produced by a marine Streptomyces isolate. J. Antibiot. 2011, 64, 205–209. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Ye, M.; Zhang, L.; Wu, Q.; Zhang, M.; Xu, J.; Zheng, W. FW-04-806 inhibits proliferation and induces apoptosis in human breast cancer cells by binding to N-terminus of Hsp90 and disrupting Hsp90-Cdc37 complex formation. Mol. Cancer 2014, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Wu, Q.D.; Zhang, M.; Kong, Y.L.; Cao, P.R.; Zheng, W.; Xu, J.H.; Ye, M. Novel Hsp90 inhibitor FW-04-806 displays potent antitumor effects in HER2-positive breast cancer cells as a single agent or in combination with lapatinib. Cancer Lett. 2015, 356, 862–871. [Google Scholar] [CrossRef]
- Song, Z.; Lohse, A.G.; Hsung, R.P. Challenges in the synthesis of a unique mono-carboxylic acid antibiotic, (+)-zincophorin. Cheminform 2009, 26, 560–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dirlam, J.P.; Belton, A.M.; Chang, S.P.; Cullen, W.P.; Huang, L.H.; Kojima, Y.; Maeda, H.; Nishiyama, S.; Oscarson, J.R.; Sakakibara, T.; et al. Cp-78,545, a new monocarboxylic acid ionophore antibiotic related to zincophorin and produced by a streptomyces. J. Antibiot. 1989, 42, 1213–1220. [Google Scholar] [CrossRef] [PubMed]



| Characteristic | 1 | 2a,c | 3b | 4c |
|---|---|---|---|---|
| Decomposition of | ||||
| Cellulose | − | − | ND | + |
| Tween 20 | − | + | ND | + |
| Tween 40 | + | + | ND | + |
| Tween 80 | − | + | ND | + |
| Liquefaction of gelatin | − | − | + | + |
| Growth temperature (°C) | 4–37 | 18–36 | 10–28 | 15–38 |
| pH range for growth | 5–12 | 4–10 | 6–11 | 5–11 |
| NaCl tolerance range (w/v, %) | 0–3 | 0–3 | 0–1 | 0–2.5 |
| Milk coagulation | − | w | − | + |
| Nitrate reduction | − | + | − | − |
| Starch hydrolysis | − | w | − | − |
| Carbon source utilization | ||||
| l-arabinose | + | ± | − | − |
| Dulcitol | + | w | − | W |
| d-Fructose | + | − | + | − |
| d-Galactose | + | − | + | − |
| d-Glucose | + | + | + | + |
| Inositol | − | + | + | − |
| Lactose | + | + | − | + |
| d-Maltose | + | − | ± | − |
| d-Mannitol | + | − | − | − |
| d-Mannose | + | − | + | − |
| d-Raffinose | + | − | − | + |
| d-Ribose | − | + | ND | − |
| d-Sorbitol | + | − | − | − |
| d-Sucrose | + | ± | W | + |
| d-Xylose | + | + | − | − |
| l-Rhamnose | − | + | + | − |
| Nitrogen source utilization | ||||
| l-Alanine | + | W | ND | + |
| l-Arginine | + | W | ND | + |
| l-Asparagine | + | W | ND | − |
| l-Aspartic acid | + | + | ND | + |
| Creatine | + | w | ND | − |
| l-Glutamic acid | + | w | ND | + |
| l-Glutamine | + | w | ND | + |
| Glycine | + | w | ND | + |
| l-Proline | − | + | ND | + |
| l-Serine | + | − | ND | − |
| l-Threonine | + | w | ND | + |
| l-Tyrosine | + | + | ND | + |
| Phospholipids | DPG, PE, PI, UL | PE, PGL | DPG, PE, PI | DPG, PME, PI, PIM, GL |
| Menaquinones | MK-9(H4), MK-9(H6), MK-9(H8) | MK-9(H2), MK-9(H4), MK-9(H6) | MK-9(H4), MK-9(H6), MK-9(H8) | MK-9(H4), MK-9(H6), MK-9(H8) |
| Whole cell-wall sugars | Arabinose, glucose, ribose | Arabinose | Arabinose, glucose | Glucose, ribose |
| No. | δH (J in Hz) | δC (p.p.m) | No. | δH (J in Hz) | δC (p.p.m) |
|---|---|---|---|---|---|
| 1 | 175.6 | 16 | 5.52 m | 132.4 | |
| 2 | 3.28 m | 37.1 | 17 | 5.36 m | 134.5 |
| 3 | 4.07 m | 74.0 | |||
| 4 | 1.67 m | 24.9 | 18 | 2.27 m | 42.2 |
| 19 | 3.58 d (9.2) | 81.7 | |||
| 5a | 1.28 m | 27.1 | 20 | 132.3 | |
| 5b | 1.40 m | 21 | 5.20 d (8.9) | 136.6 | |
| 6 | 1.52 m | 26.1 | 22 | 2.59 m | 26.1 |
| 7 | 3.77 d (8.9) | 76.1 | 23 | 0.97 d (6.6) | 22.9 |
| 8 | 2.04 m | 33.0 | 24 | 0.98 d (6.5) | 22.9 |
| 9 | 3.72 dd | 84.3 | 25 | 1.63 s | 10.1 |
| (9.6, 2.1) | |||||
| 10 | 2.01 m | 36.5 | 26 | 0.86 d (6.7) | 16.7 |
| 11 | 3.49 m | 83.3 | 27 | 1.15 d (7.2) | 10.8 |
| 12 | 1.75 m | 36.0 | 28 | 0.70 d (6.7) | 12.3 |
| 29 | 1.11 d (7.0) | 11.3 | |||
| 13 | 4.06 m | 69.4 | 30 | 0.81 d (6.5) | 17.1 |
| 14a | 1.35 m | 34.2 | 31 | 1.18 d (7.1) | 15.4 |
| 14b | 1.77 m | ||||
| 15a | 2.13 m | 28.6 | |||
| 15b | 2.18 m |
| Compound | IC50 (μg/mL) | ||
|---|---|---|---|
| K562 | HCT-116 | HepG2 | |
| 1 | 8.8 ± 1.5 | 9.5 ± 0.8 | 9.6 ± 5.6 |
| 2 3 4 5 6 7 | 57.1 ± 7.3 — — 28.3 ± 1.1 36.6 ± 2.4 — | 75.42 ± 2.1 68.39 ± 3.3 36.8 ± 5.6 14.3 ± 1.6 21.6 ± 4.1 112.3 ± 5.7 | — 53.78 ± 6.7 17.5 ± 1.9 27.3 ± 5.8 79.7 ± 5.9 — |
| 8 | 11.42 ± 3.05 | 15.13 ± 1.76 | 10.83 ± 3.47 |
| Doxorubicin | 1.1 ± 0.1 | 0.9 ± 0.3 | 2.1 ± 0.2 |
| Compounds | MIC (μg/mL) | ||||
|---|---|---|---|---|---|
| Gram-Positive Bacteria | Gram-Negative Bacteria | ||||
| Staphylococcus aureus | Sarcina lutea | Bacillus subtilis | Klebsiella pneumonie | Escherichia coli | |
| 1 | 31.0 ± 2.5 | 44.0 ± 5.8 | 3.5 ± 0.5 | 25.0 ± 1.5 | — |
| 2–7 | — | — | — | — | — |
| 8 | 210.0 ± 20.0 | 190.0 ± 15.0 | — | — | — |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Sun, T.; Song, W.; Guo, X.; Cao, P.; Xu, X.; Shen, Y.; Zhao, J. Taxonomic Characterization and Secondary Metabolite Analysis of NEAU-wh3-1: An Embleya Strain with Antitumor and Antibacterial Activity. Microorganisms 2020, 8, 441. https://doi.org/10.3390/microorganisms8030441
Wang H, Sun T, Song W, Guo X, Cao P, Xu X, Shen Y, Zhao J. Taxonomic Characterization and Secondary Metabolite Analysis of NEAU-wh3-1: An Embleya Strain with Antitumor and Antibacterial Activity. Microorganisms. 2020; 8(3):441. https://doi.org/10.3390/microorganisms8030441
Chicago/Turabian StyleWang, Han, Tianyu Sun, Wenshuai Song, Xiaowei Guo, Peng Cao, Xi Xu, Yue Shen, and Junwei Zhao. 2020. "Taxonomic Characterization and Secondary Metabolite Analysis of NEAU-wh3-1: An Embleya Strain with Antitumor and Antibacterial Activity" Microorganisms 8, no. 3: 441. https://doi.org/10.3390/microorganisms8030441
APA StyleWang, H., Sun, T., Song, W., Guo, X., Cao, P., Xu, X., Shen, Y., & Zhao, J. (2020). Taxonomic Characterization and Secondary Metabolite Analysis of NEAU-wh3-1: An Embleya Strain with Antitumor and Antibacterial Activity. Microorganisms, 8(3), 441. https://doi.org/10.3390/microorganisms8030441





