Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the Pseudomonas fluorescens MFE01 Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Mucoid Phenotype and Swimming Motility
2.3. Disruption of the fliA Gene in P. Fluorescens MFE01 Strain
2.4. Construction of the Revertant Strain MFE01ΔtssC-R
2.5. Translational Fusion of Flag Sequence into the MFE01 flgM Gene
2.6. Insertion of fliA and flgM into Expression Vectors
2.7. Introduction of Plasmid into MFE01, MFE01 Mutants or MFN1032
2.8. Putative FliA Promoters and Consensus Motif
2.9. Extraction of Total RNA from P. Fluorescens MFE01
2.10. cDNA Amplification
2.11. Quantitative Reverse Transcription-PCR
2.12. Supernatant Protein Extraction
2.13. Intracellular Protein Extraction
2.14. SDS-PAGE Analysis
2.15. Western-Blot Analysis
2.16. Protein Identification by nanoLC-MS/MS
2.17. Flagellin and Hcp Proteins Mass Spectrometry Identification from SDS-PAGE
2.18. Transmission Electron Microscopy
3. Results and Discussion
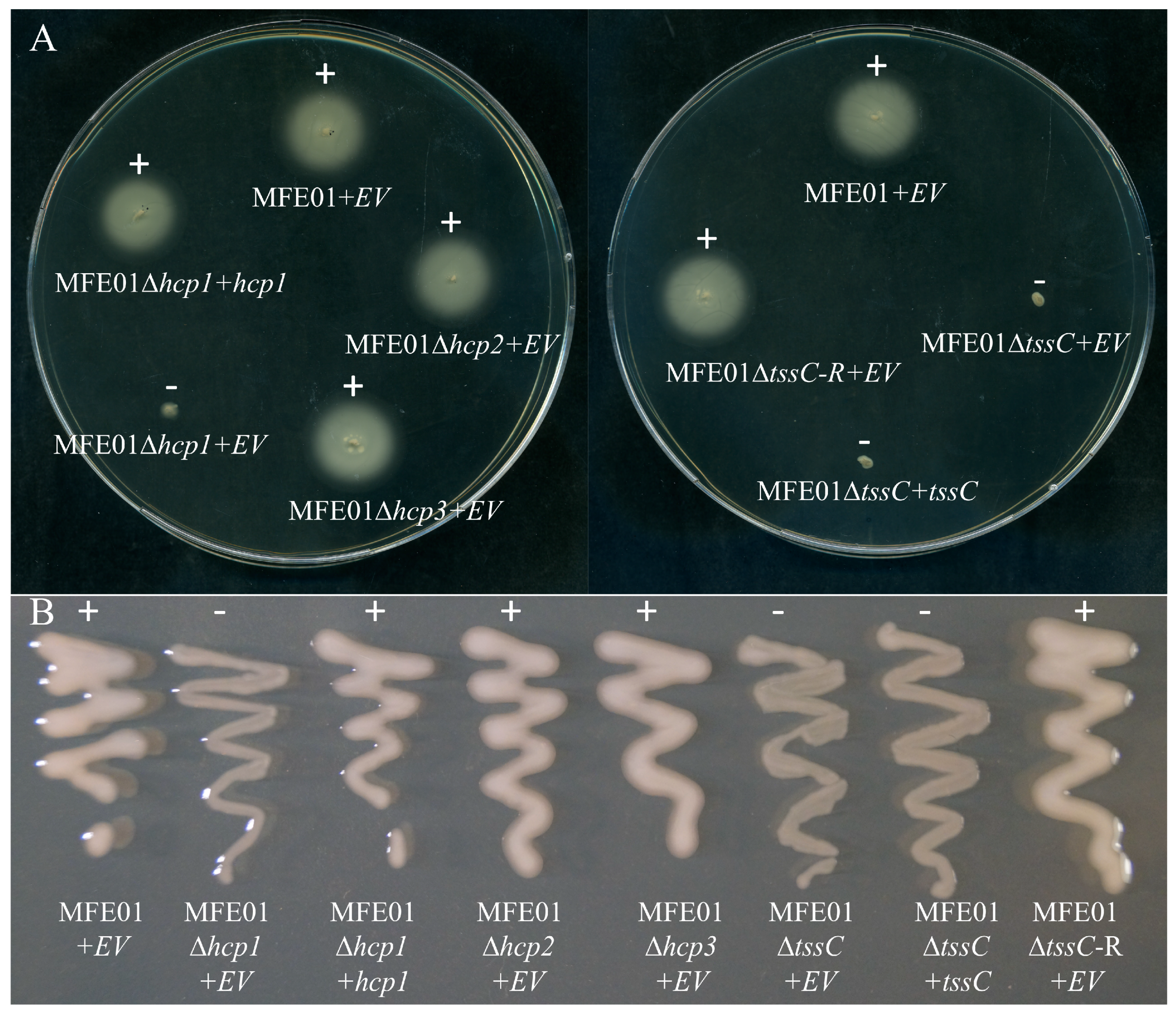
3.1. MFE01 Motility and Mucoidy are Specifically Dependent on Hcp1
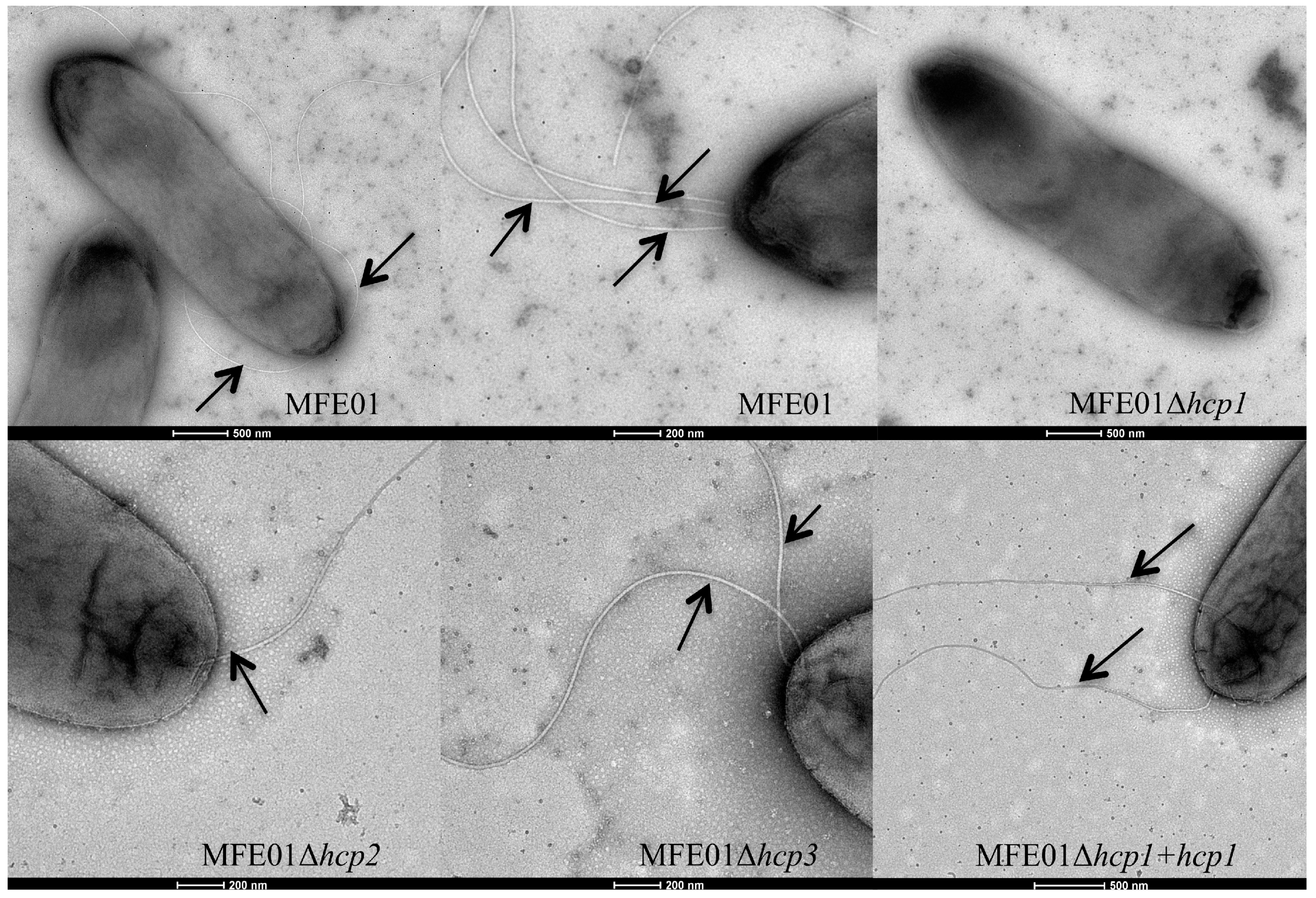
3.2. Disruption of the hcp1 Gene Results in the Lack of Flagella
3.3. FliA Controls Motility and the Mucoid Phenotype
3.4. FliA Activates the Transcription of Flagellar Class IV Genes in the MFE01 Strain
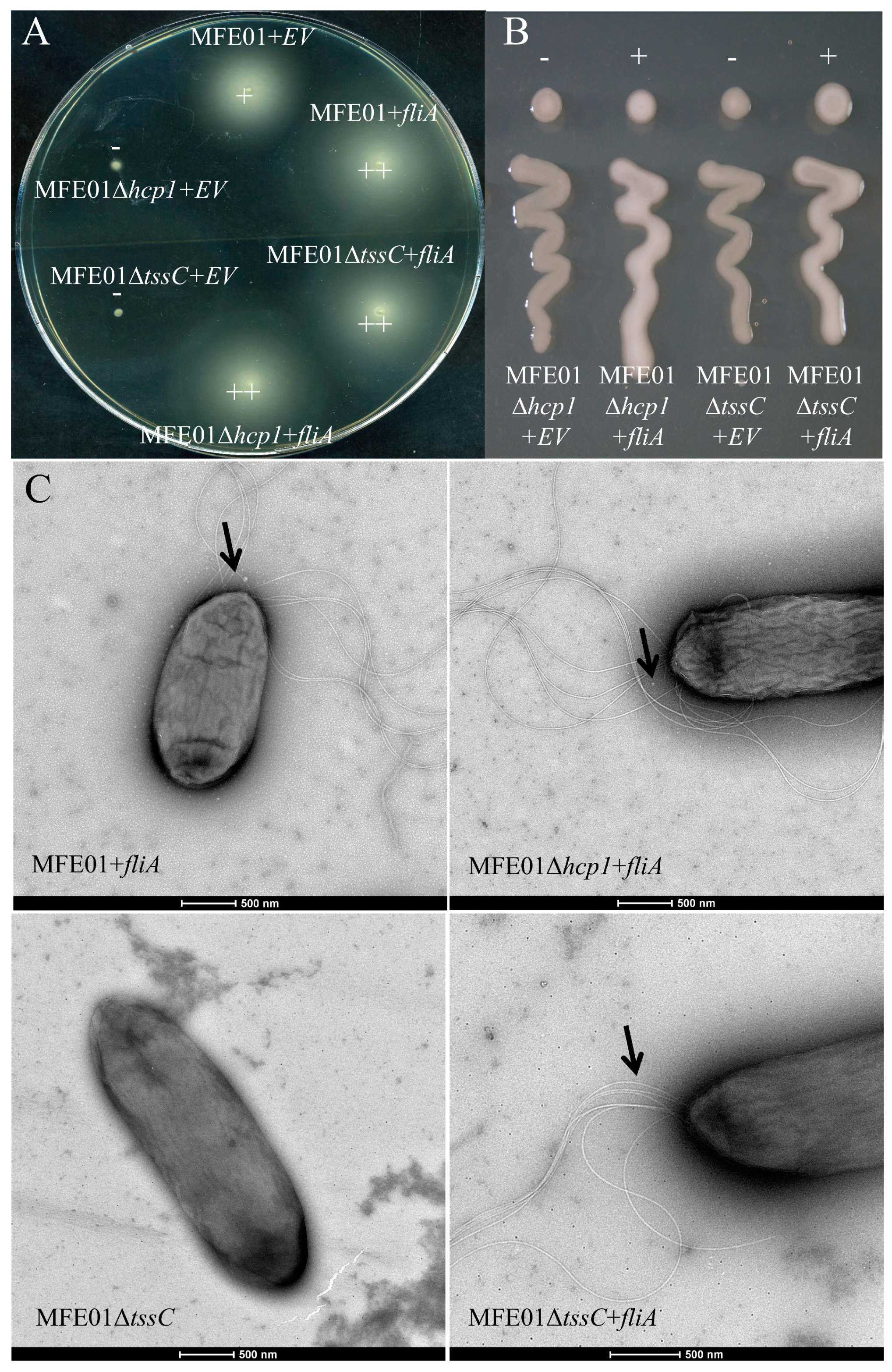
3.5. fliA Overexpression Restores Motility and Mucoidy in MFE01Δhcp1 and MFE01ΔtssC Mutants
3.6. Class IV Genes Expression is Affected in MFE01Δhcp1 but not in fliA Transcription
3.7. FlgM is not Secreted in MFE01Δhcp1
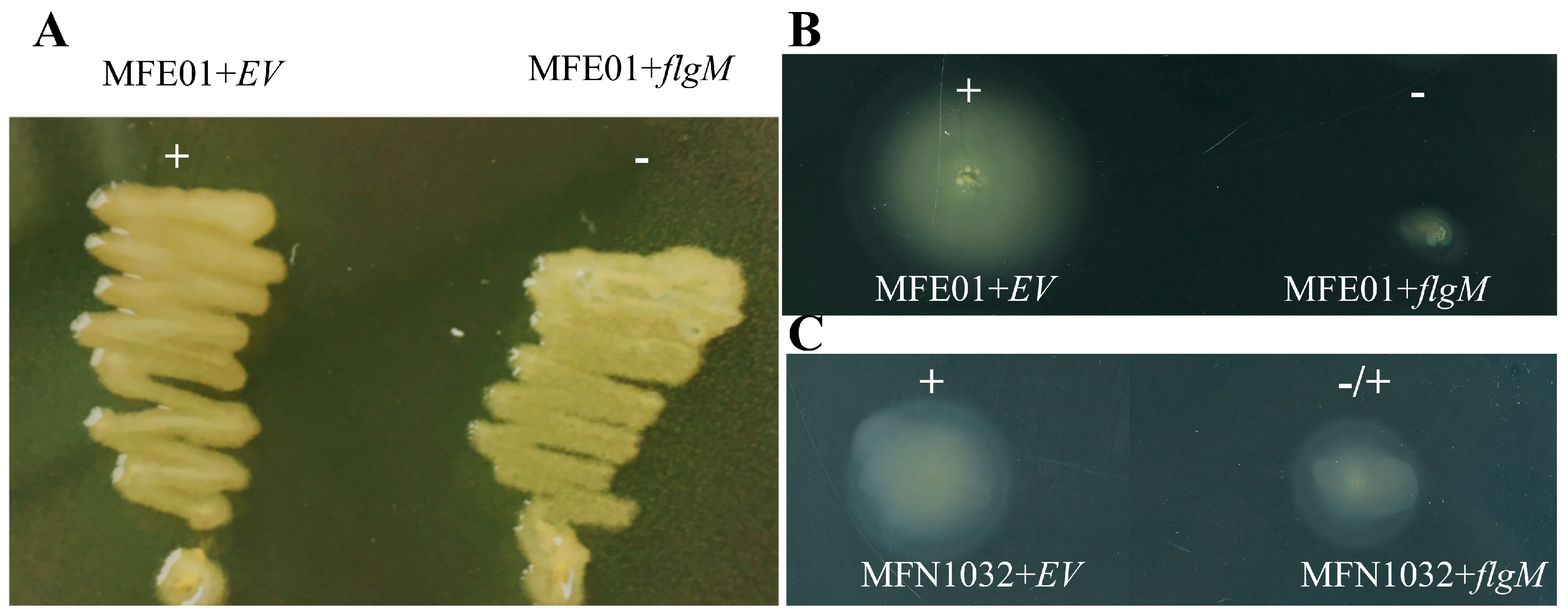
3.8. FlgM Overexpression Perturbs MFE01 and MFN1032 Motility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ho, B.; Dong, T.G.; Mekalanos, J. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 2013, 15, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Ho, B.; Mekalanos, J. Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell 2013, 152, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulthurst, S.J. The Type VI secretion system—A widespread and versatile cell targeting system. Res. Microbiol. 2013, 164, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Hood, R.D.; Singh, P.; Hsu, F.; Güvener, T.; Carl, M.A.; Trinidad, R.R.S.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.; Plemel, R.L.; et al. A Type VI Secretion System of Pseudomonas aeruginosa Targets a Toxin to Bacteria. Cell Host Microbe 2010, 7, 25–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alteri, C.J.; Himpsl, S.D.; Pickens, S.R.; Lindner, J.R.; Zora, J.S.; Miller, J.; Arno, P.D.; Straight, S.W.; Mobley, H.L.T. Multicellular Bacteria Deploy the Type VI Secretion System to Preemptively Strike Neighboring Cells. PLoS Pathog. 2013, 9, e1003608. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, L.; Saak, C.; Gibbs, K.A. Two Proteins Form a Heteromeric Bacterial Self-Recognition Complex in Which Variable Subdomains Determine Allele-Restricted Binding. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenren, L.M.; Sullivan, N.L.; Cardarelli, L.; Septer, A.N.; Gibbs, K.A. Two Independent Pathways for Self-Recognition in Proteus mirabilis Are Linked by Type VI-Dependent Export. mBio 2013, 4, e00374-13. [Google Scholar] [CrossRef] [Green Version]
- Decoin, V.; Gallique, M.; Barbey, C.; Le Mauff, F.; Poc, C.D.; Feuilloley, M.; Orange, N.; Merieau, A. A Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility and bacterial competition. BMC Microbiol. 2015, 15, 72. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-J.; Kuo, T.-Y.; Hsieh, F.-C.; Chen, P.-Y.; Wang, C.-S.; Shih, Y.L.; Lai, Y.-M.; Liu, J.-R.; Yang, Y.-L.; Shih, M.-C. Involvement of type VI secretion system in secretion of iron chelator pyoverdine in Pseudomonas taiwanensis. Sci. Rep. 2016, 6, 32950. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Zhang, W.; Cheng, J.; Yang, X.; Zhu, K.; Wang, Y.; Wei, G.; Qian, P.-Y.; Luo, Z.-Q.; Shen, X. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 2017, 8, 14888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Zhao, C.; Burkinshaw, B.; Zhang, B.; Wei, D.; Wang, Y.; Dong, T.G.; Shen, X. Manganese scavenging and oxidative stress response mediated by type VI secretion system inBurkholderia thailandensis. Proc. Natl. Acad. Sci. USA 2017, 114, E2233–E2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Wang, Y.; Zhang, B.; Zhao, C.; Kang, Y.; Bai, H.; Wei, D.; Zhu, L.; Zhang, L.; Dong, T.G.; et al. The Type VI Secretion System Engages a Redox-Regulated Dual-Functional Heme Transporter for Zinc Acquisition. Cell Rep. 2017, 20, 949–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Si, M.; Song, Y.; Zhu, W.; Gao, F.; Wang, Y.; Zhang, L.; Zhang, W.; Wei, G.; Luo, Z.-Q.; et al. Type VI Secretion System Transports Zn2+ to Combat Multiple Stresses and Host Immunity. PLoS Pathog. 2015, 11, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallique, M.; Bouteiller, M.; Merieau, A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Fichant, G.; Berthod, J.; Vandenbrouck, Y.; Attrée, I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genom. 2009, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef] [Green Version]
- Durand, E.; Nguyen, V.S.; Zoued, A.; Logger, L.; Pehau-Arnaudet, G.; Aschtgen, M.-S.; Spinelli, S.; Desmyter, A.; Bardiaux, B.; Dujeancourt, A.; et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 2015, 523, 555–560. [Google Scholar] [CrossRef]
- Lemanceau, P.; Corberand, T.; Gardan, L.; Latour, X.; Laguerre, G.; Boeufgras, J.; Alabouvette, C. Effect of Two Plant Species, Flax (Linum usitatissinum L.) and Tomato (Lycopersicon esculentum Mill.), on the Diversity of Soilborne Populations of Fluorescent Pseudomonads. Appl. Environ. Microbiol. 1995, 61, 1004–1012. [Google Scholar] [CrossRef] [Green Version]
- Bergsma-Vlami, M.; Prins, M.E.; Raaijmakers, J.M. Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol. Ecol. 2005, 52, 59–69. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Genet. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Latour, X.; Delorme, S.; Mirleau, P.; Lemanceau, P. Identification of Traits Implicated in the Rhizosphere Competence of Fluorescent Pseudomonads: Description of a Strategy Based on Population and Model Strain Studies. In Sustainable Agriculture; Springer Science and Business Media LLC: New York, NY, USA, 2009; pp. 285–296. [Google Scholar]
- Bergeau, D.; Mazurier, S.; Barbey, C.; Merieau, A.; Chane, A.; Goux, D.; Bernard, S.; Driouich, A.; Lemanceau, P.; Vicré, M.; et al. Unusual extracellular appendages deployed by the model strain Pseudomonas fluorescens C7R12. PLoS ONE 2019, 14, e0221025. [Google Scholar] [CrossRef] [PubMed]
- Cole, B.J.; Feltcher, M.E.; Waters, R.J.; Wetmore, K.M.; Mucyn, T.S.; Ryan, E.M.; Wang, G.; Ul-Hasan, S.; McDonald, M.; Yoshikuni, Y.; et al. Genome-wide identification of bacterial plant colonization genes. PLoS Boil. 2017, 15, e2002860. [Google Scholar] [CrossRef] [PubMed]
- Barahona, E.; Navazo, A.; Garrido-Sanz, D.; Muriel, C.; Martínez-Granero, F.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Pseudomonas fluorescens F113 Can Produce a Second Flagellar Apparatus, Which Is Important for Plant Root Colonization. Front. Microbiol. 2016, 7, 611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capdevila, S.; Martínez-Granero, F.M.; Sanchez-Contreras, M.; Rivilla, R.; Martin, M.; Palma, R.R. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 2004, 150, 3889–3897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Weger, L.A.; Van der Vlugt, C.I.; Wijfjes, A.H.M.; Bakker, P.A.; Schippers, B.; Lugtenberg, B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 1987, 169, 2769–2773. [Google Scholar] [CrossRef] [Green Version]
- Jahn, C.E.; Willis, D.K.; Charkowski, A. The Flagellar Sigma Factor FliA Is Required forDickeya dadantiiVirulence. Mol. Plant-Microbe Interact. 2008, 21, 1431–1442. [Google Scholar] [CrossRef] [Green Version]
- Evans, L.; Hughes, C.; Fraser, G. Building a flagellum outside the bacterial cell. Trends Microbiol. 2014, 22, 566–572. [Google Scholar] [CrossRef] [Green Version]
- Das, C.; Mokashi, C.; Mande, S.S.; Saini, S. Dynamics and Control of Flagella Assembly in Salmonella typhimurium. Front. Microbiol. 2018, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, N.; Wolfgang, M.C.; Goodman, A.L.; Arora, S.K.; Jyot, J.; Lory, S.; Ramphal, R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 809–824. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Romero, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Garrido-Sanz, D.; Ramos-González, M.-I.; Martín, M.; Rivilla, R. Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci. Rep. 2018, 8, 13145. [Google Scholar] [CrossRef] [PubMed]
- Decoin, V.; Barbey, C.; Bergeau, D.; Latour, X.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. A Type VI Secretion System Is Involved in Pseudomonas fluorescens Bacterial Competition. PLoS ONE 2014, 9, e89411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallique, M.; Decoin, V.; Barbey, C.; Rosay, T.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. Contribution of the Pseudomonas fluorescens MFE01 Type VI Secretion System to Biofilm Formation. PLoS ONE 2017, 12, e0170770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapalain, A.; Rossignol, G.; Lesouhaitier, O.; Merieau, A.; Gruffaz, C.; Guerillon, J.; Meyer, J.-M.; Orange, N.; Feuilloley, M. Comparative study of 7 fluorescent pseudomonad clinical isolates. Can. J. Microbiol. 2008, 54, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Rietsch, A.; Vallet-Gely, I.; Dove, S.L.; Mekalanos, J.J. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2005, 102, 8006–8011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sayed, A.; Hothersall, J.; Thomas, C.M. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586 The GenBank accession numbers for the sequences determined in this work are AF318063 (mupA), AF318064 (mupR) and AF318065 (mupI). Microbiology 2001, 147, 2127–2139. [Google Scholar] [CrossRef] [Green Version]
- Newman, J.R.; Fuqua, C. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 1999, 227, 197–203. [Google Scholar] [CrossRef]
- Frisk, A.; Jyot, J.; Arora, S.K.; Ramphal, R. Identification and Functional Characterization of flgM, a Gene Encoding the Anti-Sigma 28 Factor in Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 1514–1521. [Google Scholar] [CrossRef] [Green Version]
- Bouffartigues, E.; Gicquel, G.; Bazire, A.; Bains, M.; Maillot, O.; Vieillard, J.; Feuilloley, M.G.J.; Orange, N.; Hancock, R.E.; Dufour, A.; et al. Transcription of the oprF Gene of Pseudomonas aeruginosa Is Dependent Mainly on the SigX Sigma Factor and Is Sucrose Induced. J. Bacteriol. 2012, 194, 4301–4311. [Google Scholar] [CrossRef] [Green Version]
- Guyard-Nicodème, M.; Bazire, A.; Hémery, G.; Meylheuc, T.; Mollé, D.; Orange, N.; Fito-Boncompte, L.; Feuilloley, M.; Haras, D.; Dufour, A.; et al. Outer membrane Modifications ofPseudomonas fluorescensMF37 in Response to Hyperosmolarity. J. Proteome Res. 2008, 7, 1218–1225. [Google Scholar] [CrossRef]
- Kentache, T.; Ben Abdelkrim, A.; Jouenne, T.; Dé, E.; Hardouin, J. Global Dynamic Proteome Study of a Pellicle-forming Acinetobacter baumannii Strain. Mol. Cell. Proteom. 2016, 16, 100–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Hao, S.; Lan, R.; Wang, G.; Xiao, D.; Sun, H.; Xu, J. The Type VI Secretion System Modulates Flagellar Gene Expression and Secretion in Citrobacter freundii and Contributes to Adhesion and Cytotoxicity to Host Cells. Infect. Immun. 2015, 83, 2596–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pace, F.; De Paiva, J.B.; Nakazato, G.; Lancellotti, M.; Sircili, M.P.; Stehling, E.G.; Da Silveira, W.D.; Sperandio, V. Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology 2011, 157, 2954–2962. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, J.; Xu, J.; Zhang, H.; He, L.; Feng, J. TssB is essential for virulence and required for Type VI secretion system in Ralstonia solanacearum. Microb. Pathog. 2014, 74, 1–7. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef]
- Nakamura, S.; Minamino, T. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef] [Green Version]
- Redondo-Nieto, M.; Lloret, J.; Larenas, J.; Barahona, E.; Navazo, A.; Martiínez-Granero, F.; Capdevila, S.; Rivilla, R.; Martiín, M. Transcriptional Organization of the Region Encoding the Synthesis of the Flagellar Filament in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 4106–4109. [Google Scholar] [CrossRef] [Green Version]
- Schulz, S.; Eckweiler, D.; Bielecka, A.; Nicolai, T.; Franke, R.; Dötsch, A.; Hornischer, K.; Bruchmann, S.; Düvel, J.; Häussler, S. Elucidation of Sigma Factor-Associated Networks in Pseudomonas aeruginosa Reveals a Modular Architecture with Limited and Function-Specific Crosstalk. PLoS Pathog. 2015, 11, e1004744. [Google Scholar] [CrossRef] [Green Version]
- Postel, S.; Deredge, D.; Bonsor, D.A.; Yu, X.; Diederichs, K.; Helmsing, S.; Vromen, A.; Friedler, A.; Hust, M.; Egelman, E.H.; et al. Bacterial flagellar capping proteins adopt diverse oligomeric states. eLife 2016, 5, 213. [Google Scholar] [CrossRef]
- Herlihey, F.A.; Clarke, A. Controlling Autolysis During Flagella Insertion in Gram-Negative Bacteria. In Protein Reviews; Springer: Singapore, 2016; pp. 41–56. [Google Scholar]
- Arora, S.K.; Ritchings, B.W.; Almira, E.C.; Lory, S.; Ramphal, R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1997, 179, 5574–5581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herva, J.J.R.; Duque, E.; Molina-Henares, M.A.; Navarro-Avilés, G.; Van Dillewijn, P.; De La Torre, J.; Molina-Henares, A.J.; De La Campa, A.M.S.; Ran, F.A.; Segura, A.; et al. Physiological and transcriptomic characterization of a fliA mutant of Pseudomonas putida KT2440. Environ. Microbiol. Rep. 2009, 2, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Granero, F.; Navazo, A.; Barahona, E.; Redondo-Nieto, M.; De Heredia, E.G.; Baena, I.; Martín-Martín, I.; Rivilla, R.; Martín, M. Identification of flgZ as a Flagellar Gene Encoding a PilZ Domain Protein That Regulates Swimming Motility and Biofilm Formation in Pseudomonas. PLoS ONE 2014, 9, e87608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarter, L.L. Regulation of flagella. Curr. Opin. Microbiol. 2006, 9, 180–186. [Google Scholar] [CrossRef]
- Bernal, P.; Allsopp, L.; Filloux, A.; Llamas, M.A. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017, 11, 972–987. [Google Scholar] [CrossRef] [Green Version]




| Strain or Plasmid | Relevant Characteristics | Reference/Source |
|---|---|---|
| Pseudomonas fluorescens | ||
| MFE01 | Air isolate, RifR | [33] |
| MFE01+pPSV35 | MFE01 with pPSV35 empty vector, GmR | [33] |
| MFE01+fliA | MFE01 with pPSV35 carrying wild-type fliA gene, GmR | This study |
| MFE01ΔtssC | MFE01 with a in frame central deletion in tssC gene | [9] |
| MFE01ΔtssC+pPSV35 | MFE01ΔtssC with pPSV35 empty vector, GmR | [34] |
| MFE01ΔtssC+tssC | MFE01 with pPSV35 carrying wild-type tssC gene, GmR | [34] |
| MFE01ΔtssC-R | MFE01ΔtssC with chromosomal introduction of wild-type tssC gene | This study |
| MFE01ΔtssC+fliA | MFE01ΔtssC with pPSV35 carrying wild-type fliA gene, GmR | This study |
| MFE01Δhcp1 | MFE01 with hcp1 gene disruption, TcR | [9] |
| MFE01Δhcp1+pPSV35 | MFE01Δhcp1 with pPSV35 empty vector, TcR, GmR | [9] |
| MFE01Δhcp1+hcp1 | MFE01Δhcp1 with pPSV35 carrying wild-type hcp1 gene, TcR, GmR | [9] |
| MFE01Δhcp1+fliA | MFE01Δhcp1 with pPSV35 carrying wild-type fliA gene, TcR, GmR | This study |
| MFE01Δhcp2 | MFE01 with early stop codon in hcp2 gene | [33] |
| MFE01Δhcp2+pPSV35 | MFE01Δhcp2 with pPSV35 empty vector, GmR | This study |
| MFE01Δhcp3 | MFE01 with in frame deletion in hcp3 gene | [34] |
| MFE01Δhcp3+pPSV35 | MFE01Δhcp3 with pPSV35 empty vector, GmR | [34] |
| MFE01ΔfliA | MFE01 with in frame central deletion in fliA gene | This study |
| MFE01ΔfliA+pPSV35 | MFE01ΔfliA with pPSV35 empty vector, GmR | This study |
| MFE01ΔfliA+fliA | MFE01ΔfliA with pPSV35 carrying wild-type fliA gene, GmR | This study |
| MFE01+pJN105 | MFE01 with pJN105 empty vector, GmR | This study |
| MFE01+flgM | MFE01 with pJN105 carrying wild-type flgM gene, GmR | This study |
| MFE01-flgM::flag | MFE01 with 3’ flgM::flag transcriptional fusion | This study |
| MFE01Δhcp1-flgM::flag | MFE01Δhcp1 with 3’ flgM::flag transcriptional fusion | This study |
| MFN1032 | Clinical isolate | [35] |
| MFN1032+pJN105 | MFN1032 with pJN105 empty vector, GmR | This study |
| MFN1032+flgM | MFN1032 with pJN105 carrying wild-type flgM gene, GmR | This study |
| Escherichia coli | ||
| S17.1 | RP4-2-Tc::Mu, aph::Tn7, recA, SmR, donor strain for conjugation | [36] |
| Top10® | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG | ThermoFischer Scientific |
| Plasmids | ||
| pPSV35 | Pseudomonas aeruginosa oriV, lacIq mob+, PlacUV5, pUC18MCS, expression vector, GmR | [37] |
| pAKE604 | Conjugative suicide vector, oriT, lacZ, sacB, ApR, KmR | [38] |
| pJN105 | Arabinose-inducible expression plasmid, GmR | [39] |
| Mutagenesis Primers | Primer Sequence (5’--> 3’) |
| Muta1-fliA-F | ACACTGGCCGACGTTATC |
| Muta2-fliA-EcoRI-R | TAATAAGAATTCGTAAAGATTCATGCCACTGG |
| Muta3-fliA-EcoRI-F | TAATAAGAATTCCTGTTCAGTTTCGACGAC |
| Muta4-fliA-R | CTTCAGCAGTCACCATCAA |
| Muta1-3’flag-flgM | TGATCAGGTCATCACACTG |
| Muta2-3’flag-flgM | CTTGTCATCGTCATCTTTATAATCGCGCTGGGCTTCGAAGTTG |
| Muta3-3’flag-flgM | GATTATAAAGATGACGATGACAAGTAGGCTTTTGCCGGCGCCAG |
| Muta4-3’flag-flgM | TTCATGGAAGGTGATGATCA |
| Muta1-tssC-F | CTGAGACTCCAGTAGCCAAG |
| Muta4-tssC-R | ATGTCATTGAGATCGGGCAA |
| Surexpression primers | Primer sequence (5’--> 3’) |
| fliA-EcoRI-F | TAATAAGAATTCGGCATCTGGAATTTTTCGT |
| fliA-XbaI-R | TAATAATCTAGATCCCCACACTGCCTTCA |
| flgM-EcoRI-F | TAATAAGAATTCTCCAAATTCCCAGAGGTTTT |
| flgM-XbaI-R | TAATAATCTAGAGTCGTTGATCAGTTGCAATA |
| qPCR primers | Primer sequence (5’--> 3’) |
| qRT-PCR-RecA-F | AAGGGTGCCGTAATGCGTAT |
| qRT-PCR-RecA-R | ATATCCAGACCCAGAGAGCCAGTA |
| qRT-PCR-FliA-F | CTGGTGTTGGCGCTGTACTAC |
| qRT-PCR-FliA-R | GCCAAGGACTTCACCGATTT |
| qRT-PCR-FlgM-F | GTACCAGCAACGCCAAGGAA |
| qRT-PCR-FlgM-R | TGTACCGACTCCCCGCTTT |
| qRT-PCR-FleQ-F | CATCGCGAACCCAATCTGT |
| qRT-PCR-FleQ-R | GGCCACTTGCTGCATCATCT |
| qRT-PCR-RpoN-F | ACTGGTCGCAGCGGAAAAT |
| qRT-PCR-RpoN-R | ATGCCTTGTGCCTCCAGTAAA |
| qRT-PCR-FliS-F | GATGTTAGCCCTTCGGCAGTAC |
| qRT-PCR-FliS-R | CACCTTCCATCAACATTTGCA |
| qRT-PCR-FlaA-F | ACACCCAGGCCATCCAGAA |
| qRT-PCR-FlaA-R | TGCAGGATGTCGGTCGAA |
| qRT-PCR-MotA-F | GCGTTCGTCTGCGATTACCT |
| qRT-PCR-MotA-R | CGTGCGGAGCCATGTTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouteiller, M.; Gallique, M.; Bourigault, Y.; Kosta, A.; Hardouin, J.; Massier, S.; Konto-Ghiorghi, Y.; Barbey, C.; Latour, X.; Chane, A.; et al. Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the Pseudomonas fluorescens MFE01 Strain. Microorganisms 2020, 8, 622. https://doi.org/10.3390/microorganisms8050622
Bouteiller M, Gallique M, Bourigault Y, Kosta A, Hardouin J, Massier S, Konto-Ghiorghi Y, Barbey C, Latour X, Chane A, et al. Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the Pseudomonas fluorescens MFE01 Strain. Microorganisms. 2020; 8(5):622. https://doi.org/10.3390/microorganisms8050622
Chicago/Turabian StyleBouteiller, Mathilde, Mathias Gallique, Yvann Bourigault, Artemis Kosta, Julie Hardouin, Sebastien Massier, Yoan Konto-Ghiorghi, Corinne Barbey, Xavier Latour, Andréa Chane, and et al. 2020. "Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the Pseudomonas fluorescens MFE01 Strain" Microorganisms 8, no. 5: 622. https://doi.org/10.3390/microorganisms8050622
APA StyleBouteiller, M., Gallique, M., Bourigault, Y., Kosta, A., Hardouin, J., Massier, S., Konto-Ghiorghi, Y., Barbey, C., Latour, X., Chane, A., Feuilloley, M., & Merieau, A. (2020). Crosstalk between the Type VI Secretion System and the Expression of Class IV Flagellar Genes in the Pseudomonas fluorescens MFE01 Strain. Microorganisms, 8(5), 622. https://doi.org/10.3390/microorganisms8050622







