Antibiofilm Activity of a Trichoderma Metabolite against Xanthomonas campestris pv. campestris, Alone and in Association with a Phage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Growth of Xcc Phages
2.2. Complex φHA6PP
2.3. Isolation of 6-Pentyl-α-pyrone
2.4. Antibiofilm Activity of 6-Pentyl-α-pyrone
2.5. CLSM Analysis for Static Biofilm Evaluation
2.6. CLSM Analysis for Dynamic Biofilm Evaluation
2.7. RNA Extraction and Expression Profiling by qPCR
3. Results
3.1. Antibiofilm Activity of 6-Pentyl-α-pyrone
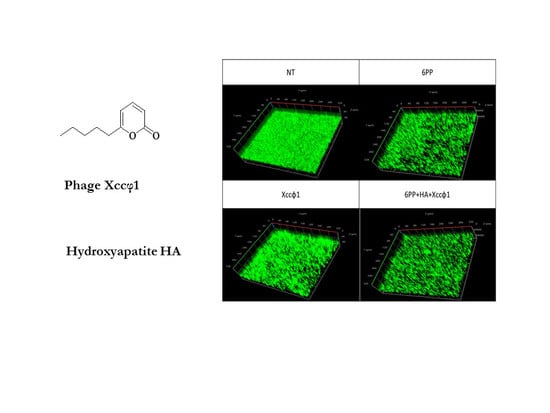
3.2. CLSM Analysis for Static Biofilm Evaluation
3.3. CLSM Analysis for Dynamic Biofilm Evaluation
3.4. RNA Extraction and Expression Profiling by qPCR
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Qian, W.; Jia, Y.; Ren, S.X.; He, Y.Q.; Feng, J.X.; Lu, L.F.; Wu, W. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 2005, 15, 757–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, C.T.; Chiang, Y.C.; Hsiao, Y.M. Functional characterization and proteomic analysis of lolA in Xanthomonas campestris pv. campestris. BMC Microbiol. 2019, 19, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dow, J.M.; Daniels, M.J. Pathogenicity determinants and global regulation of pathogenicity of Xanthomonas campestris pv. campestris. Curr. Top. Microbiol. Immunol. 1994, 192, 29–41. [Google Scholar] [PubMed]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Windler, M.; Leinweber, K.; Bartulos, C.R.; Philipp, B.; Kroth, P.G. Biofilm and capsule formation of the diatom Achnanthidium minutissimum are affected by a bacterium. J. Phycol. 2015, 51, 343–355. [Google Scholar] [CrossRef] [Green Version]
- Cepas, V.; López, Y.; Munoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Donlan, R.M. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009, 17, 66–72. [Google Scholar] [CrossRef]
- Stewart, P.S.; William Costerton, J. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef]
- Harper, D.R.; Parracho, H.M.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Morales, S. Bacteriophages and biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Vandenheuvel, D.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono-and dual-species staphylococcal biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, D.P.; Melo, L.D.R.; Vilas Boas, D.; Sillankorva, S.; Azeredo, J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 2017, 39, 48–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Barbetti, M.J.; Li, H.; Woo, S.L.; Lorito, M. A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 2008, 72, 80–86. [Google Scholar] [CrossRef]
- Marra, R.; Ambrosino, P.; Carbone, V.; Vinale, F.; Woo, S.L.; Ruocco, M.; Gigante, S. Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr. Genet. 2006, 50, 307–321. [Google Scholar] [CrossRef]
- Pascale, A.; Vinale, F.; Manganiello, G.; Nigro, M.; Lanzuise, S.; Ruocco, M.; Lorito, M. Trichoderma and its secondary metabolites improve yield and quality of grapes. Crop Prot. 2017, 92, S11–S12. [Google Scholar] [CrossRef] [Green Version]
- Fulgione, A.; Ianniello, F.; Papaianni, M.; Contaldi, F.; Sgamma, T.; Giannini, C.; Lelli, M. Biomimetic hydroxyapatite nanocrystals are an active carrier for Salmonella bacteriophages. Int. J. Nanomed. 2019, 14, 2219–2232. [Google Scholar] [CrossRef] [Green Version]
- Nocerino, N.; Fulgione, A.; Iannaccone, M.; Tomasetta, L.; Ianniello, F.; Martora, F.; Capparelli, R. Biological activity of lactoferrin-functionalized biomimetic hydroxyapatite nanocrystals. Int. J. Nanomed. 2014, 9, 1175–1184. [Google Scholar]
- Fulgione, A.; Nocerino, N.; Iannaccone, M.; Roperto, S.; Capuano, F.; Roveri, N.; Capparelli, R. Lactoferrin adsorbed onto biomimetic hydroxyapatite nanocrystals controlling-in vivo-the Helicobacter pylori infection. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Cross, T.; Schoff, C.; Chudoff, D.; Graves, L.; Broomell, H.; Terry, K.; Dunbar, D. An optimized enrichment technique for the isolation of Arthrobacter bacteriophage species from soil sample isolates. J. Vis. Exp. 2015. [Google Scholar] [CrossRef] [Green Version]
- Papaianni, M.; Contaldi, F.; Fulgione, A.; Woo, S.L.; Casillo, A.; Corsaro, M.M.; Garonzi, M. Role of phage ϕ1 in two strains of Salmonella Rissen, sensitive and resistant to phage ϕ1. BMC Microbiol. 2018, 18, 208. [Google Scholar] [CrossRef]
- Casillo, A.; Papa, R.; Ricciardelli, A.; Sannino, F.; Ziaco, M.; Tilotta, M.; Artini, M. Anti-Biofilm activity of a long-chain fatty aldehyde from antarctic Pseudoalteromonas haloplanktis TAC125 against Staphylococcus epidermidis biofilm. Front. Cell. Infect. Microbiol. 2017, 7, 46. [Google Scholar] [CrossRef] [Green Version]
- Casillo, A.; Ziaco, M.; Lindner, B.; Parrilli, E.; Schwudke, D.; Holgado, A.; Tutino, M.L. Unusual Lipid A from a cold-adapted Bacterium: Detailed structural characterization. ChemBioChem 2017, 18, 1845–1854. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Allam, A.F.; Farag, H.F.; Zaki, A.; Kader, O.A.; Abdul-Ghani, R.; Shehab, A.Y. Detection of low-intensity Schistosoma mansoni infection by Percoll sedimentation and real-time PCR techniques in a low-endemicity Egyptian setting. Trop. Med. Int. Heal. 2015, 20, 658–664. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dow, J.M.; Crossman, L.; Findlay, K.; He, Y.Q.; Feng, J.X.; Tang, J.L. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 2003, 100, 10995–11000. [Google Scholar] [CrossRef] [Green Version]
- Ryan, R.P.; Dow, J.M. Communication with a growing family: Diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011, 19, 145–152. [Google Scholar] [CrossRef]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef]
- Harman, G.E. Multifunctional fungal plant symbionts: New tools to enhance plant growth and productivity. New Phytol. 2011, 189, 647–649. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species-Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Horwitz, B.A.; Herrera-Estrella, A.; Schmoll, M.; Kenerley, C.M. Trichoderma research in the genome era. Annu. Rev. Phytopathol. 2013, 51, 105–129. [Google Scholar] [CrossRef]
- Ozkan, A.; Erdogan, A. A comparative study of the antioxidant/prooxidant effects of carvacrol and thymol at various concentrations on membrane and DNA of parental and drug resistant H1299 cells. Nat. Prod. Commun. 2012, 7, 1557–1560. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [Green Version]
- Torres, P.S.; Malamud, F.; Rigano, L.A.; Russo, D.M.; Marano, M.R.; Castagnaro, A.P.; Vojnov, A.A. Controlled synthesis of the DSF cell-cell signal is required for biofilm formation and virulence in Xanthomonas campestris. Environ. Microbiol. 2007, 9, 2101–2109. [Google Scholar] [CrossRef] [Green Version]
- Papaianni, M.; Paris, D.; Woo, S.L.; Fulgione, A.; Rigano, M.M.; Parrilli, E.; Limone, A. Plant dynamic metabolic response to bacteriophage treatment after Xanthomonas campestris pv. campestris infection. Front. Microbiol. 2020, 11, 732. [Google Scholar] [CrossRef]
- Papaianni, M.; Cuomo, P.; Fulgione, A.; Albanese, D.; Gallo, M.; Paris, D.; Motta, A.; Iannelli, D.; Capparelli, R. Bacteriophages promote metabolic changes in bacteria biofilm. Microorganisms 2020, 8, 480. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaianni, M.; Ricciardelli, A.; Fulgione, A.; d’Errico, G.; Zoina, A.; Lorito, M.; Woo, S.L.; Vinale, F.; Capparelli, R. Antibiofilm Activity of a Trichoderma Metabolite against Xanthomonas campestris pv. campestris, Alone and in Association with a Phage. Microorganisms 2020, 8, 620. https://doi.org/10.3390/microorganisms8050620
Papaianni M, Ricciardelli A, Fulgione A, d’Errico G, Zoina A, Lorito M, Woo SL, Vinale F, Capparelli R. Antibiofilm Activity of a Trichoderma Metabolite against Xanthomonas campestris pv. campestris, Alone and in Association with a Phage. Microorganisms. 2020; 8(5):620. https://doi.org/10.3390/microorganisms8050620
Chicago/Turabian StylePapaianni, Marina, Annarita Ricciardelli, Andrea Fulgione, Giada d’Errico, Astolfo Zoina, Matteo Lorito, Sheridan L. Woo, Francesco Vinale, and Rosanna Capparelli. 2020. "Antibiofilm Activity of a Trichoderma Metabolite against Xanthomonas campestris pv. campestris, Alone and in Association with a Phage" Microorganisms 8, no. 5: 620. https://doi.org/10.3390/microorganisms8050620
APA StylePapaianni, M., Ricciardelli, A., Fulgione, A., d’Errico, G., Zoina, A., Lorito, M., Woo, S. L., Vinale, F., & Capparelli, R. (2020). Antibiofilm Activity of a Trichoderma Metabolite against Xanthomonas campestris pv. campestris, Alone and in Association with a Phage. Microorganisms, 8(5), 620. https://doi.org/10.3390/microorganisms8050620