Bifidobacterium β-Glucosidase Activity and Fermentation of Dietary Plant Glucosides Is Species and Strain Specific
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
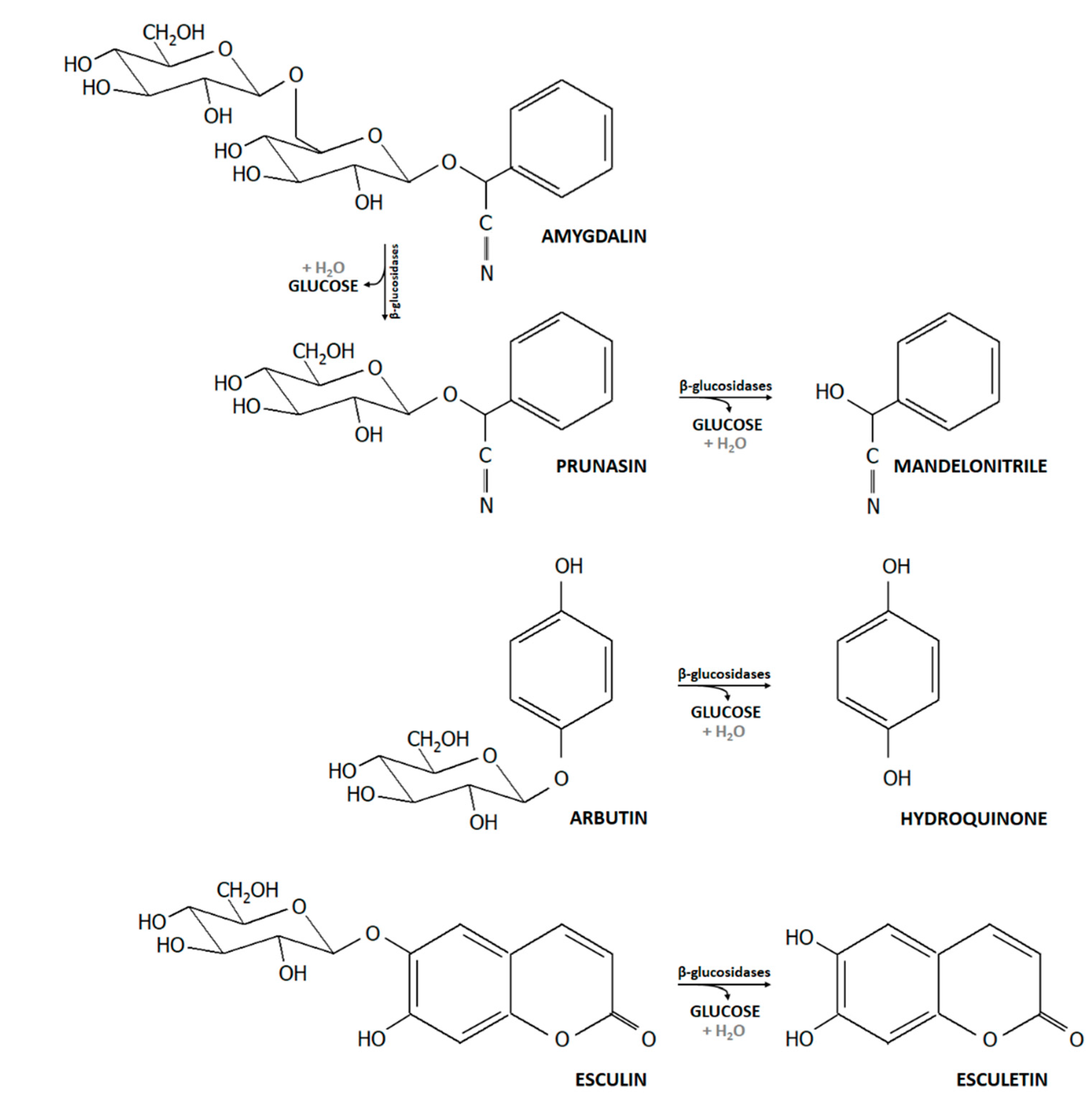
2.2. Utilization of Selected Dietary Plant Glucosides
2.3. Metabolite Formation Analysis Using Ion Chromatography with Suppressed Conductivity Detection
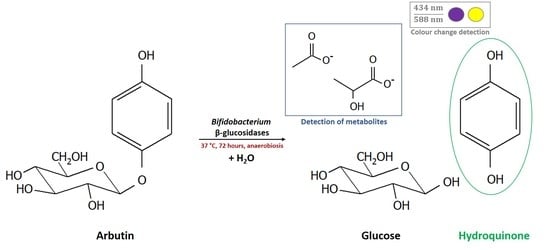
2.4. Determination of Whole Cell β-Glucosidase Activity, and Release of the Acylglycone Esculetin
2.5. Antibacterial Activity of Arbutin and Hydroquinone against Selected Bifidobacterium Strains
2.6. Identification and Comparison of β-Glucosidases Encoded by Representative Bifidobacterium spp. of the Species Investigated
3. Results
3.1. Distribution of (Putative) β-Glucosidases Encoding Genes in Genomes of Representative Bifidobacterium spp.
3.2. Beta-Glucosidase Activity of Bifidobacterium spp.
3.3. Growth in the Presence of β-Glucosides
3.4. Release of the Acylglycone Esculetin
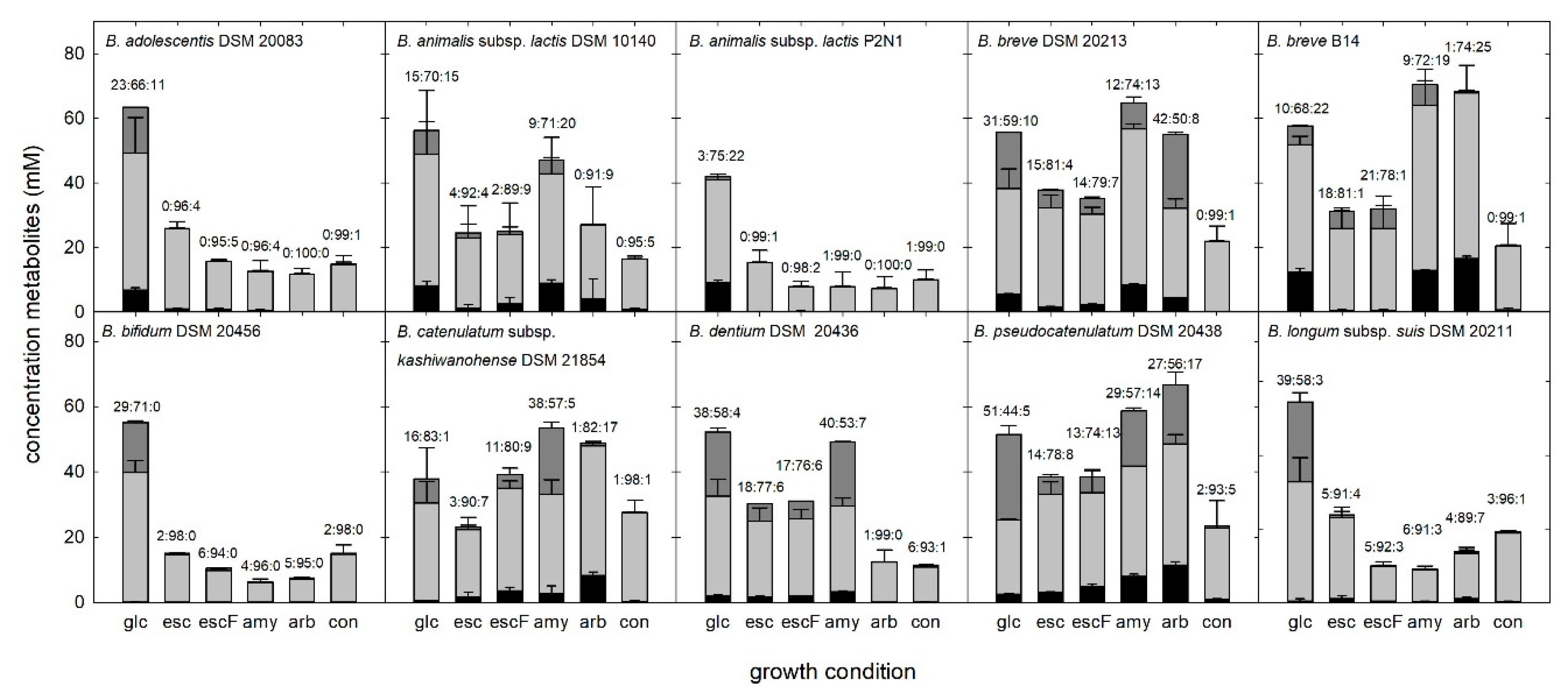
3.5. Metabolite Formation during Growth of Bifidobacteria in the Presence of Plant Glucosides
3.6. Antibacterial Activity of Hydroquinone and Arbutin and Impact on Growth Kinetics
4. Discussion
4.1. Genomic Potential for β-Glucosidase Activity Partly Predicts Activity
4.2. Bifidobacterium β-Glucosidase Increases Bioactivity and Bioavailability of Plant Glucosidases and Acylglycones
4.3. Niche Adaption of Bifidobacteria Seems Related to β-Glucosidase Activity
4.4. Substrate Source Impacted Fermentation Profiles
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Biernat, K.A.; Li, B.; Redinbo, M.R. Microbial unmasking of plant glycosides. MBio 2018, 9, e02433-17. [Google Scholar] [CrossRef]
- De Arriba, S.G.; Naser, B.; Nolte, K.-U. Risk assessment of free hydroquinone derived from Arctostaphylos Uva-ursi folium herbal preparations. Int. J. Toxicol. 2013, 32, 442–453. [Google Scholar] [CrossRef]
- Theilmann, M.C.; Goh, Y.J.; Nielsen, K.F.; Klaenhammer, T.R.; Barrangou, R.; Abou Hachem, M. Lactobacillus acidophilus metabolizes dietary plant glucosides and externalizes their bioactive phytochemicals. MBio 2017, 8. [Google Scholar] [CrossRef]
- Jurica, K.; Gobin, I.; Kremer, D.; Čepo, D.V.; Grubešić, R.J.; Karačonji, I.B.; Kosalec, I. Arbutin and its metabolite hydroquinone as the main factors in the antimicrobial effect of strawberry tree (Arbutus unedo L.) leaves. J. Herb. Med. 2017, 8, 17–23. [Google Scholar] [CrossRef]
- Dabek, M.; McCrae, S.I.; Stevens, V.J.; Duncan, S.H.; Louis, P. Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008, 66, 487–495. [Google Scholar] [CrossRef]
- Ventura, M.; Turroni, F.; Motherway, M.O.; MacSharry, J.; van Sinderen, D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012, 20, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.A.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. Metabolic activities and probiotic potential of bifidobacteria. Int. J. Food Microbiol. 2011, 149, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Ruscheweyh, H.J.; Bunesova, V.; Pham, V.T.; Beerenwinkel, N.; Lacroix, C. Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Mahony, J.; van Sinderen, D.; Ventura, M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018, 26, 339–350. [Google Scholar] [CrossRef]
- Milani, C.; Turroni, F.; Duranti, S.; Lugli, G.A.; Mancabelli, L.; Ferrario, C.; Van Sinderen, D.; Ventura, M. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 2016, 82, 980–991. [Google Scholar] [CrossRef]
- Bunesova, V.; Killer, J.; Javurkova, B.; Vlkova, E.; Tejnecky, V.; Musilova, S.; Rada, V. Diversity of the subspecies Bifidobacterium animalis subsp. lactis. Anaerobe 2017, 44, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Bottacini, F.; Morrissey, R.; Esteban-Torres, M.; James, K.; Van Breen, J.; Dikareva, E.; Egan, M.; Lambert, J.; Van Limpt, K.; Knol, J.; et al. Comparative genomics and genotype-phenotype associations in Bifidobacterium breve. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Hylckama Vlieg, J.E.T. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Bunešová, V.; Joch, M.; Musilová, S.; Rada, V. Bifidobacteria, lactobacilli, and short chain fatty acids of vegetarians and omnivores. Sci. Agric. Bohem. 2017, 48, 47–54. [Google Scholar] [CrossRef]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Kubota, H.; Gawad, A.; Sakai, T.; Oishi, K.; Martin, R.; Ben-Amor, K.; Knol, J. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS ONE 2013, 8, e78331. [Google Scholar] [CrossRef]
- Nouioui, I.; Carro, L.; García-López, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Pukall, R.; Klenk, H.-P.; Goodfellow, M.; Göker, M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef]
- Sharma, V.; Mobeen, F.; Prakash, T. Exploration of survival traits, probiotic determinants, host interactions, and functional evolution of bifidobacterial genomes using comparative genomics. Genes 2018, 9, 477. [Google Scholar] [CrossRef]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Milani, C.; James, K.; Mancabelli, L.; Turroni, F.; Alessandri, G.; Mangifesta, M.; Mancino, W.; Ossiprandi, M.C.; et al. Bifidobacterium bifidum and the infant gut microbiota: An intriguing case of microbe-host co-evolution. Environ. Microbiol. 2019, 21, 3683–3695. [Google Scholar] [CrossRef]
- Masco, L.; Ventura, M.; Zink, R.; Huys, G.; Swings, J. Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: Reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1137–1143. [Google Scholar] [CrossRef]
- Henne, K.; Rheinberg, A.; Melzer-Krick, B.; Conrads, G. Aciduric microbial taxa including Scardovia wiggsiae and Bifidobacterium spp. in caries and caries free subjects. Anaerobe 2015, 35, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Neves, B.G.; Stipp, R.N.; Bezerra, D.D.S.; Guedes, S.F.D.F.; Rodrigues, L.K.A. Quantitative analysis of biofilm bacteria according to different stages of early childhood caries. Arch. Oral. Biol. 2018, 96, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Modrackova, N.; Makovska, M.; Mekadim, C.; Vlkova, E.; Tejnecky, V.; Bolechova, P.; Bunesova, V. Prebiotic potential of natural gums and starch for bifidobacteria of variable origins. Bioact. Carbohydr. Diet. Fibre 2019, 20. [Google Scholar] [CrossRef]
- Bunesova, V.; Domig, K.J.; Killer, J.; Vlkova, E.; Kopecny, J.; Mrazek, J.; Rockova, S.; Rada, V. Characterization of bifidobacteria suitable for probiotic use in calves. Anaerobe 2012, 18, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Bunesova, V.; Vlkova, E.; Killer, J.; Rada, V.; Rockova, S. Identification of Bifidobacterium strains from faeces of lambs. Small Rumin. Res. 2012, 105, 355–360. [Google Scholar] [CrossRef]
- Pei, K.; Xiong, Y.; Li, X.; Jiang, H.; Xiong, Y. Colorimetric ELISA with an acid–base indicator for sensitive detection of ochratoxin A in corn samples. Anal. Methods 2018, 10, 30–36. [Google Scholar] [CrossRef]
- Youn, S.Y.; Park, M.S.; Ji, G.E. Identification of the β-glucosidase gene from Bifidobacterium animalis subsp. lactis and its expression in B. bifidum BGN4. J. Microbiol. Biotechnol. 2012, 22, 1714–1723. [Google Scholar] [CrossRef]
- Guadamuro, L.; Flórez, A.B.; Alegría, Á.; Vázquez, L.; Mayo, B. Characterization of four β-glucosidases acting on isoflavone-glycosides from Bifidobacterium pseudocatenulatum IPLA 36007. Food Res. Int. 2017, 100, 522–528. [Google Scholar] [CrossRef][Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9. [Google Scholar] [CrossRef]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. DbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef]
- Jung, I.H.; Lee, J.H.; Hyun, Y.J.; Kim, D.H. Metabolism of ginsenoside Rb1 by human intestinal microflora and cloning of its metabolizing β-D-glucosidase from Bifidobacterium longum H-1. Biol. Pharm. Bull. 2012, 35, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Florindo, R.N.; Souza, V.P.; Manzine, L.R.; Camilo, C.M.; Marana, S.R.; Polikarpov, I.; Nascimento, A.S. Structural and biochemical characterization of a GH3 β-glucosidase from the probiotic bacteria Bifidobacterium adolescentis. Biochimie 2018, 148, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ding, W.; Xu, Y.; Wu, D.; Li, S.; Chen, J.; Guo, B. New insights into the antibacterial activity of hydroxycoumarins against Ralstonia solanacearum. Molecules 2016, 21, 468. [Google Scholar] [CrossRef] [PubMed]
- Rúa, J.; Fernández-Álvarez, L.; De Castro, C.; Del Valle, P.; De Arriaga, D.; García-Armesto, M.R. Antibacterial activity against foodborne Staphylococcus aureus and antioxidant capacity of various pure phenolic compounds. Foodborne Path Dis. 2011, 8, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; He, N.; Zhao, Y.; Xia, D.; Wei, J.; Kang, W. Antimicrobial mechanism of hydroquinone. Appl. Biochem. Biotechnol. 2019, 189, 1291–1303. [Google Scholar] [CrossRef]
- Byeon, S.E.; Yi, Y.S.; Lee, J.; Yang, W.S.; Kim, J.H.; Kim, J.; Hong, S.; Kim, J.-H.; Cho, J.Y. Hydroquinone exhibits in vitro and in vivo anti-cancer activity in cancer cells and mice. Int. J. Mol. Sci. 2018, 19, 903. [Google Scholar] [CrossRef] [PubMed]
- Horn, C.M.; Aucamp, J.; Smit, F.J.; Seldon, R.; Jordaan, A.; Warner, D.F.; N’Da, D.D. Synthesis and in vitro antimycobacterial and antileishmanial activities of hydroquinone-triazole hybrids. Med. Chem. Res. 2020. [Google Scholar] [CrossRef]
- Mykkänen, H.; Tikka, J.; Pitkänen, T.; Hänninen, O. Fecal bacterial enzyme activities in infants increase with age and adoption of adult-type diet. J. Pediatr. Gastroenterol. Nutr. 1997, 25, 312–316. [Google Scholar] [CrossRef]
- De Vries, W.; Stouthamer, A. Pathway of glucose fermentation in relation to the taxonomy of bifidobacteria. J. Bacteriol. 1967, 93, 574–576. [Google Scholar] [CrossRef] [PubMed]
- Palframan, R.J.; Gibson, G.R.; Rastall, R.A. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr. Issues Intest. Microbiol. 2003, 4, 71–75. [Google Scholar]
- Van der Meulen, R.; Adriany, T.; Verbrugghe, K.; De Vuyst, L. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl. Environ. Microbiol. 2006, 72, 5204–5210. [Google Scholar] [CrossRef] [PubMed]

| Species/Subspecies | Strain | Origin | GLU | ESC | AMYG | ARB | API | β-GLU | ESN rel. |
|---|---|---|---|---|---|---|---|---|---|
| B. adolescentis | DSM 20083 | Intestine of adult | +++ | - | - | - | - | + | + |
| B34 | Stool of infant | +++ | - | +++ | - | - | + | + | |
| B35 | Stool of infant | +++ | ++ | +++ | + | - | + | + | |
| B36 | Stool of infant | +++ | - | - | - | - | +R | + | |
| B38 | Stool of infant | +++ | + | - | - | - | + | + | |
| B2 | Stool of adult | ++ | - | ++ | - | - | +R | + | |
| B9 | Stool of adult | ++ | - | ++ | - | - | +R | + | |
| B30 | Stool of adult | +++ | - | +++ | - | - | +R | + | |
| B39 | Stool of adult | +++ | ++ | +++ | +++ | - | + | + | |
| B41 | Stool of adult | +++ | - | +++ | - | - | + | + | |
| B56 | Stool of adult | ++ | - | - | - | - | + | + | |
| PEG038 | Stool of adult | +++ | - | +++ | - | - | + | + | |
| 10/6d | Dog feces | +++ | - | +++ | - | - | + | + | |
| B. animalis subsp. animalis | DSM 20104 | Rat feces | +++ | - | - | - | - | + | + |
| 805P4 | Calf feces | +++ | +++ | - | - | - | +R | + | |
| 012II1 | Calf feces | +++ | + | - | - | - | +R | + | |
| 023II | Calf feces | +++ | - | - | - | - | +R | + | |
| J1 (L1) | Lamb feces | ++ | - | - | - | - | +R | + | |
| J5 (L4) | Lamb feces | +++ | + | - | - | - | +R | + | |
| J6 (L3) | Lamb feces | +++ | ++ | - | - | - | + | + | |
| B. animalis subsp. lactis | DSM 10140 | Yoghurt | +++ | - | +++ | - | - | + | + |
| BB12 | Probiotic product | ++ | ++ | ++ | - | - | + | + | |
| Dan | Probiotic product | ++ | + | +++ | - | - | + | + | |
| Nestlé | Infant nutrition | +++ | ++ | ++ | - | - | + | + | |
| S7 | Ovine cheese | ++ | ++ | ++ | - | - | + | + | |
| B22 | Stool of infant | +++ | ++ | +++ | - | - | + | + | |
| B25 | Stool of infant | +++ | + | +++ | - | - | + | + | |
| PEG042 | Stool of adult | +++ | - | +++ | - | - | + | + | |
| PEG084 | Stool of adult | ++ | + | ++ | +++ | - | + | + | |
| P2N1 | Dog feces | +++ | - | - | - | - | + | + | |
| 43/7nb | Dog feces | +++ | - | - | - | - | + | + | |
| 11/6a | Dog feces | ++ | - | - | - | - | + | + | |
| ZDK1 | Cameroon sheep feces | +++ | ++ | +++ | - | - | + | + | |
| ZDK4 | Barbary sheep feces | ++ | ++ | ++ | - | - | + | + | |
| ZDK7 | Okapi feces | +++ | ++ | +++ | - | - | + | + | |
| B. bifidum | DSM 20456 | Stool of infant | +++ | - | - | - | - | - | - |
| DSM 20239 | Stool of infant | +++ | - | - | - | - | - | - | |
| B6 | Stool of infant | +++ | - | - | - | - | -R | - | |
| B33 | Stool of infant | +++ | - | - | - | - | -R | - | |
| B10 | Stool of adult | +++ | - | - | - | - | -R | - | |
| B29 | Stool of adult | +++ | - | - | - | - | -R | - | |
| B40 | Stool of adult | +++ | - | - | - | - | - | - | |
| B55 | Stool of adult | +++ | - | - | - | - | - | - | |
| B. breve | DSM 20213 | Intestine of infant | +++ | ++ | +++ | +++ | - | + | + |
| BR03 | Probiotic product | ++ | ++ | +++ | - | - | + | + | |
| B13 | Stool of infant | +++ | +++ | +++ | ++ | - | +R | + | |
| B14 | Stool of infant | +++ | ++ | +++ | +++ | - | +R | + | |
| B37 | Stool of infant | +++ | +++ | +++ | - | - | + | + | |
| B42 | Stool of infant | ++ | ++ | ++ | +++ | - | + | + | |
| B43 | Stool of infant | +++ | - | ++ | - | - | + | + | |
| B50 | Stool of infant | +++ | +++ | +++ | +++ | - | + | + | |
| B57 | Stool of infant | +++ | +++ | +++ | - | - | + | + | |
| PEG010 | Stool of adult | ++ | ++ | +++ | - | - | + | + | |
| PEG064 | Stool of adult | +++ | +++ | +++ | - | - | + | + | |
| PEG071 | Stool of adult | +++ | ++ | +++ | - | - | + | + | |
| PEG074 | Stool of adult | +++ | +++ | +++ | ++ | - | + | + | |
| B. catenulatum B. catenulatum subsp. kashiwanohense B. pseudocatenulatum B. catenulatum/ pseudocatenulatum | DSM 16992 | Human feces | +++ | +++ | +++ | +++ | - | + | + |
| DSM 21854 | Stool of infant | +++ | - | +++ | + | - | + | + | |
| DSM 20438 | Stool of infant | +++ | +++ | +++ | +++ | - | + | + | |
| B12 | Stool of infant | +++ | - | +++ | - | - | +R | + | |
| B46 | Stool of infant | +++ | ++ | +++ | +++ | - | + | + | |
| B48 | Stool of infant | +++ | +++ | - | - | - | + | + | |
| B23 | Stool of adult | +++ | ++ | +++ | - | - | +R | + | |
| B32 | Stool of adult | +++ | ++ | +++ | - | - | +R | + | |
| B51 | Stool of adult | ++ | - | +++ | - | - | + | + | |
| B52 | Stool of adult | +++ | + | - | +++ | - | + | + | |
| B53 | Stool of adult | +++ | - | +++ | - | - | + | + | |
| 22/4nb | Dog feces | +++ | ++ | +++ | - | - | + | + | |
| B. dentium | DSM 20436 | Dental caries | +++ | +++ | +++ | - | - | + | + |
| FD1 | Stool of infant | +++ | ++ | +++ | +++ | - | + A | + | |
| TH1 | Stool of infant | +++ | +++ | +++ | - | - | + A | + | |
| VOK II | Stool of infant | +++ | ++ | +++ | - | - | + A | + | |
| PEG020 | Stool of adult | +++ | ++ | +++ | +++ | - | + | + | |
| A1/5A | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N12 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N21 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N23 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N26 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N77 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N79 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N105 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N109 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N110 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N111 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| N112 | Monkey feces | +++ | +++ | +++ | +++ | - | + A | + | |
| B. longum subsp. infantis B. longum subsp. longum B. longum subsp. suillum B. longum subsp. suis B. longum | DSM 20088 | Stool of infant | +++ | - | - | - | - | - | - |
| DSM 20219 | Intestine of adult | +++ | - | - | - | - | - | - | |
| DSM 28597 | Feces of piglets | +++ | - | - | - | - | -A | - | |
| DSM 20211 | Pig feces | +++ | - | +++ | - | - | + | - | |
| 5/9 | Calf feces | +++ | - | - | - | - | - | - | |
| INFNUT | Probiotic product | ++ | - | - | - | - | - | - | |
| B3 | Stool of infant | +++ | - | - | - | - | -R | - | |
| B4 | Stool of infant | ++ | - | - | - | - | -R | - | |
| B7 | Stool of infant | +++ | - | - | - | - | - | - | |
| B8 | Stool of infant | +++ | - | - | - | - | - | - | |
| B11 | Stool of infant | +++ | - | - | - | - | - | - | |
| B16 | Stool of infant | +++ | - | - | - | - | - | - | |
| B17 | Stool of infant | +++ | - | - | - | - | - R | - | |
| B19 | Stool of infant | +++ | - | - | - | - | -R | - | |
| B20 | Stool of infant | ++ | - | - | - | - | -R | - | |
| B27 | Stool of infant | ++ | - | - | - | - | - | - | |
| B28 | Stool of infant | +++ | - | - | - | - | - | - | |
| B44 | Stool of infant | +++ | - | - | - | - | -A | - | |
| B49 | Stool of infant | +++ | - | - | - | - | - | - | |
| B1 | Stool of adult | +++ | - | - | - | - | + | - | |
| B26 | Stool of adult | +++ | - | - | - | - | -R | - | |
| PEG057 | Stool of adult | +++ | - | - | - | - | - | - | |
| PEG059 | Stool of adult | +++ | - | - | - | - | - | - | |
| PEG080 | Stool of adult | ++ | - | ++ | - | - | + | + | |
| PEG104 | Stool of adult | +++ | - | - | - | - | - | - | |
| 022II | Calf feces | +++ | - | - | - | - | -R | - | |
| 10/6b | Dog feces | +++ | - | - | - | - | + | + | |
| 32/3na | Dog feces | +++ | - | - | - | - | + | - | |
| 33/5nb | Dog feces | ++ | - | - | - | - | + | - | |
| 33/4nc | Dog feces | ++ | - | - | - | - | + | - |
| Species or Subspecies | Strain | Minimal Inhibitory Concentration (mM) | |
|---|---|---|---|
| Hydroquinone | Arbutin | ||
| B. adolescentis | DSM 20083 | 0.05–0.10 | >25.5 |
| B. animalis subsp. animalis | DSM 20104 | ≤0.05 | >25.5 |
| B. animalis subsp. lactis | DSM 10140 | 0.10–0.20 | >25.5 |
| B. bifidum | DSM 20456 | 0.10–0.20 | >25.5 |
| B. breve | DSM 20213 | 0.10–0.20 | >25.5 |
| B. longum subsp. suis | DSM 20211 | ≤0.05 | >25.5 |
| B. longum subsp. longum | DSM 20219 | ≤0.05 | >25.5 |
| B. longum subsp. infantis | DSM 20088 | ≤0.05 | >25.5 |
| Species | Strain | Characterized Beta-Glucosidase | Not Yet Characterized β-Glucosidases | ||||
|---|---|---|---|---|---|---|---|
| GH Family 1 | GH family 3 | ||||||
| Bbg572 (JX274651) 461 AA | r-β-gluE (AW18_08090, KEF28001.1) 787 AA | r-β-gluB (AW18_09810, KEF27912.1) 809 AA | r-β-gluD (AW18_08145, KEF28010.1) 748 AA | r-β-gluA (AW18_01575, KEF29323.1) 964 AA | |||
| B. breve | DSM 20213 | - | EFE88733.1A 93% I, 90% P in 774 AA | EFE90113.1 80% I, 97% P in 833 AA | EFE88739.1 82% I, 90% P in 757 AA | EFE90117.1 70% I, 81% P in 811 AA | |
| B. adolescentis | DSM 20083 | - | BAF39978.1 96% I, 98% P in 780 AA | BAF40379.1 90% I, 94% P in 811 AA | BAF39975.1 B 85% I, 92% P in 748 AA | BAF40392.1 97% I, 93% P in 962 AA | BAF40391.1 |
| B. longum subsp. longum | DSM 20019 | - | BAJ67169.1 C 82% I, 89% P in 776 AA | - | BAJ67164.1 78% I, 87% P in 507 AA D | - | |
| B. longum subsp. suis | DSM 20211 | - | KFI73778.1 E 82% I, 90% P in 775 AA | - | KFI73782.1 83% I, 91% P in 752 AA | - | KFI73422.1 |
| B. longum subsp. infantis | DSM 20088 | ACJ52977.1 69% I, 81% P in 417 AA | - | - | ACJ51732.1 82% I, 90% P in 756 AA | - | |
| B. animalis subsp. animalis | DSM 20104 | AFI62379.1 96% I, 98% P in 460 AA | AFI63691.1 73% I, 84% P in 776 AA | - | - | - | |
| B. animalis subsp. lactis | DSM 10140 | ACS47112.1 100% I, 100% P in 476 AA | ACS48458.1 73% I, 84% P in 771 AA | - | - | - | |
| B. animalis subsp. lactis | BB12 | ADC85172.1 100% I, 100% P in 460 AA | ADC84934.1 73% I, 84% P in 771 AA | - | - | - | ADC84934.1 |
| B. bifidum | DSM 20456 | BAQ97280.1 47% I, 63% P in 437 AA | - | - | - | - | |
| B. catenulatum subsp. kashiwanohense | DSM 21854 | KFI63440.1 71% I, 82% P in 458 AA | KFI67404.1 97% I, 98% P in 780 AA | KFI63941.1 95% I, 97% P in 728 AA | KFI67400.1 98% I, 99% P in 748 AA | KFI63834.1 92% I, 99% P in 299 AA* | |
| B. catenulatum | DSM 16992 | - | EEB22212.1 96% I, 98% P in 780 AA | EEB21148.1 95% I, 97% P in 809 AA | EEB22216.1 98% I, 99% P in 748 AA | EEB22373.1 93% I, 96% P in 696 AA | EEB22212.1 |
| B. pseudocatenulatum | DSM 20438 | - | EEG71163.1 96% I, 98% P in 780 AA | EEG71238.1 99% I, 99% P in 809 AA | EEG71159.1 98% I, 99% P in 748 AA | EEG70226.1 99% I, 99% P in 964 AA | |
| B. dentium | DSM 20436 | SEC02936.1 69% I, 80% P in 457 AA | SEC47920.1 99% I, 95% P in 774 AA | SEC11609.1 87% I, 93% P in 809 AA | SEC48734.1 94% I, 98% P in 748 AA | SEC11364.1 90% I, 95% P in 962 AA SEC11543.1 61% I, 76% P in 962 AA | SEC18208.1 SEB97687.1 SEB79266.1 SEC47658.1 SEC14043.1 SEB96905.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Modrackova, N.; Vlkova, E.; Tejnecky, V.; Schwab, C.; Neuzil-Bunesova, V. Bifidobacterium β-Glucosidase Activity and Fermentation of Dietary Plant Glucosides Is Species and Strain Specific. Microorganisms 2020, 8, 839. https://doi.org/10.3390/microorganisms8060839
Modrackova N, Vlkova E, Tejnecky V, Schwab C, Neuzil-Bunesova V. Bifidobacterium β-Glucosidase Activity and Fermentation of Dietary Plant Glucosides Is Species and Strain Specific. Microorganisms. 2020; 8(6):839. https://doi.org/10.3390/microorganisms8060839
Chicago/Turabian StyleModrackova, Nikol, Eva Vlkova, Vaclav Tejnecky, Clarissa Schwab, and Vera Neuzil-Bunesova. 2020. "Bifidobacterium β-Glucosidase Activity and Fermentation of Dietary Plant Glucosides Is Species and Strain Specific" Microorganisms 8, no. 6: 839. https://doi.org/10.3390/microorganisms8060839
APA StyleModrackova, N., Vlkova, E., Tejnecky, V., Schwab, C., & Neuzil-Bunesova, V. (2020). Bifidobacterium β-Glucosidase Activity and Fermentation of Dietary Plant Glucosides Is Species and Strain Specific. Microorganisms, 8(6), 839. https://doi.org/10.3390/microorganisms8060839