1. Introduction
Toxoplasma gondii is an obligate intracellular apicomplexan protozoan parasite that is widely distributed throughout the world. The disease associated with
T. gondii infection, toxoplasmosis, can lead to severe central nervous system (CNS) pathologies [
1]. Although
T. gondii infection is usually asymptomatic in healthy individuals, infection can be fatal in immunocompromised patients and in congenitally infected fetuses [
2,
3]. Given the pathological damage and neuroinflammation that can be caused by
T. gondii infection of the brain [
4,
5], many epidemiological studies have shown an association between
T. gondii infection and a number of neuropsychiatric conditions, such as schizophrenia and bipolar disorder [
4,
5,
6]. Besides the lack of a vaccine, current treatment options for toxoplasmosis are limited and responses to commonly used medicines are often unsatisfactory [
7]. There is a clear need for new and more potent anti-
T. gondii therapeutics to reduce the clinical impact of toxoplasmosis. Ideal therapeutic agents should prevent parasite growth, without perturbing the proliferation or homeostasis of host cells. However, in general, although available therapeutic agents display potent anti-
T. gondii properties, many are cytotoxic to mammalian cells [
8].
The blood-brain barrier (BBB) is the initial site of interaction between neuro-pathogens such as
T. gondii and the mammalian brain [
9]. Breaching this protective biophysical barrier is a key mechanism by which
T. gondii tachyzoites invade and damage the CNS [
9]. During
T. gondii infection, different signalling pathways control the expression of a wide range of genes that orchestrate molecular and cellular events to eliminate the invading parasite and regulate the associated inflammation [
10]. One of these pathways is the Wnt pathway, which is considered an important regulatory axis in the immune system, where genes involved in this pathway were upregulated in
T. gondii-infected mice [
10]. Moreover, other intracellular pathogens have been shown to manipulate this pathway to facilitate their entry into the cell and establish infection, such as
Chlamydia trachomatis [
11] and the influenza virus [
12].
Small molecule inhibitors of the Wnt pathway are becoming more prominent in the pharmaceutical industry [
13], and are increasingly being used to target cancer cells and intracellular pathogens [
14]. Natural polyether ionophores, such as monensin, have been substances of great interest, because of their antimicrobial activities [
15]. Monensin has been extensively used in veterinary applications as a growth promoter [
16] and to treat coccidiosis and cryptosporidiosis [
15,
17]. In addition, monensin was proposed as an anticancer agent [
18,
19]. Monensin, secreted by the bacteria
Streptomyces cinnamonensis, has been shown to modulate Wnt signalling by antagonizing β-catenin and the lipoprotein receptor-related protein LRP [
18]. Investigation into how manipulation of the Wnt pathway by monensin can influence the growth of
T. gondii could be beneficial in the development of new anti-
T. gondii therapeutics.
In this study, we investigated whether inhibition of the Wnt signalling pathway by monensin can reduce the growth of T. gondii infecting human brain microvascular endothelial cells (hBMECs) or microglial cells, and whether suppression of the growth of T. gondii within hBMECs using monensin can restore the impairment of the BBB integrity. We also performed systems-level transcriptional analysis of hBMECs infected by T. gondii in the presence or absence of monensin to obtain a comprehensive insight into how monensin treatment alters gene expression of hBMECs during T. gondii infection.
2. Materials and Methods
2.1. Chemicals
Monensin was purchased as a sodium salt powder from Alfa Aesar (Ward Hill, MA, USA) and 1.73 mg was dissolved in 250 μL of absolute methanol. The 10 mM of stock solution was diluted with complete culture medium to prepare five working concentrations 0.01 μM, 0.1 μM, 1 μM, 10 μM and 100 μM, which were stored at 4 °C until use. Sulforhodamine B (SRB) was purchased from Sigma-Aldrich (SIGMA-ALDRICH®/Merck KGaA, Darmstadt, Germany) and a solution of 0.4% (w/v) SRB dissolved in 1% acetic acid (Fisher Scientific, Leicestershire, UK) was prepared and stored protected from light at 4 °C.
2.2. Cell Lines
Primary human brain microvascular endothelial cells (hBMECs) kindly provided by Naveed Khan (American University of Sharjah, Sharjah, UAE) were maintained in modified Gibco® Roswell Park Memorial Institute (RPMI) 1640 media (Thermo Fisher Scientific, Waltham, MA, USA) containing 20% Gibco® Heat Inactivated Fetal Bovine Serum (HI-FBS), 1% Gibco® non-essential amino-acids (100×), 1% Gibco® sodium pyruvate (100 mM), 1% Gibco® MEM vitamins (100×), 1% L-glutamine (200 mM), and 1% Gibco® antibiotic-antimycotic solution 100× (10,000 units/mL of penicillin, 10,000 µg/mL of streptomycin, and 25 µg/mL of Amphotericin B). Human microglial cells (ATCC CRL-3304) originally obtained from the American Type Culture Collection were grown in modified Gibco® Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) containing 5% Gibco® HI-FBS and 1% Gibco® antibiotic-antimycotic solution 100X. Both cell lines were passaged twice a week and grown in T75 (75 cm2) NUNC™ tissue culture flasks (Fisher Scientific, Leicestershire, UK) for two weeks before being used in the experiments. All cultures were maintained in a cell culture incubator at 37 °C with 5% atmospheric CO2.
2.3. Parasite Strain and Culture Conditions
Tachyzoites of the T. gondii RH strain were cultured in NUNC™ T75 tissue culture flasks containing hBMECs using the same cell culture conditions described above. The tachyzoites were harvested from their feeder cell cultures when 70–80% of the hBMECs were lysed in about three days. Then, they were separated from host cell debris via centrifugation at 500 g for 5 min. Based on the cell line used in the subsequent experiments, the number of tachyzoites was adjusted with the respective medium (RPMI for hBMECs or DMEM for microglial cells) to achieve a multiplicity of infection (MOI) of 5 (5 tachyzoites: 1 cell) for each cell line.
2.4. Efficacy of Monensin on the In Vitro Growth of T. gondii
We used parasite counting assay in 24-well plates and the sulforhodamine B (SRB) assay in 96-well plates to investigate the effects of inhibition of the Wnt signalling pathway using monensin on the intracellular growth of T. gondii.
2.4.1. Parasite Counting Assay
Microglial and hBMEC cultures were seeded on Nunclon™ Delta Surface 24-well culture plates (Thermo Fisher Scientific, Waltham, MA, USA) at seeding densities of 5 × 104 cells and 105 cells per well, respectively. Cultures of both cell types were infected with T. gondii tachyzoites at a MOI of 5. Approximately 3 h post-infection, the culture medium in each well was discarded and replaced with 1 mL of fresh medium supplemented with one of the five working concentrations of monensin (0.01 μM, 0.1 μM, 1 μM, 10 μM and 100 μM). Growth of cell cultures was monitored daily by microscopic observation using a Zeiss Axiovert 25 inverted microscope (Leica Microsystems, Milton Keynes, UK) with a 10X objective lens. When the parasites had completed a few cycles of development and began to exit the cells in large numbers at ~3 days after infection of the untreated cells, the absolute number of tachyzoites in each well of treated and untreated cultures was counted. To achieve this, the remaining medium in the wells was topped up with phosphate buffered saline (PBS) with pipetting to produce a homogenous parasite suspension. A small amount (50 μL) of the suspension from each well was used to count tachyzoites using a standard hemocytometer counting slide.
2.4.2. Colorimetric SRB Assay
The SRB assay involves the measurement of cellular protein content using a plate reader to quantify the absorbance after cells have been fixed and stained with SRB dye. The SRB absorbance values represent the spectrophotometric quantification of protein concentration of the cells, which is directly proportional to the number of cells (i.e. increased absorbance correlates with increased protein content, which reflects an increase in the cells’ number). Hence, SRB absorbance values can be used as an indicator of host cell proliferation rates in
T. gondii-infected versus mock-infected (control) cultures to determine the level of
T. gondii growth indirectly. Based on this rationale, we can expect infected cells to produce less absorbance due to the disruption of cell proliferation caused by the damaging effects of the parasite growth on the host cell viability. Here, a microplate-based SRB staining assay was used as a simple, cost-effective, convenient approach to determine the inhibitory effect of monensin on
T. gondii growth in vitro. Briefly, cells were cultured in Corning
® Costar
® 96-well plates (Sigma-Aldrich/Merck KGaA, Darmstadt, Germany). Each well was seeded with 5 × 10
3 microglial cells or 10
4 hBMECs in 100 µL of the respective medium and then grown to confluency (~1 day). Tachyzoites used to infect each well at a MOI of 5 were able to invade the host cells within 3 h. Then, the medium was discarded to remove any extracellular tachyzoites and 100 µL of media containing one of the five concentrations of monensin (0.01 μM, 0.1 μM, 1 μM, 10 μM, and 100 µM) was added to each well. Wells that received medium without monensin (untreated) served as controls. After five days of incubation, the SRB assay was performed according to Vichai and Kirtikara [
20]. Ice-cold 100 µM of 10% trichloroacetic acid (TCA) solution (Fisher Scientific, Leicestershire, UK) was added to each well. The plates were kept at 4 °C for 1 h. Afterwards, the solution was discarded from the wells followed by thorough washing with distilled water. Next, 25 µL of SRB dye was added to each well and incubated for 15 min at ambient temperature. The plates were wrapped with foil to protect the dye from the light. Then, the wells were washed with 1% acetic acid to remove any unattached stain, followed by addition of 100 µL of 10 mM Tris base solution (Sigma-Aldrich/Merck KGaA, Darmstadt, Germany) to each well. Finally, the plates were gently agitated on a plate shaker at 35 rotations/min for 5 min to ensure a homogeneous solution. The absorbance was measured at 492 nm using a plate reader (LT-4000 Microplate Reader, LabTech International Ltd., Heathfield, UK).
2.5. Effect of Monensin on BBB Integrity
The hBMECs are fundamental constituents of the BBB and play important roles in the maintenance of barrier integrity. To determine if and to what extent integrity of hBMECs is affected by exposure to 0.1 µM monensin in the presence or absence of T. gondii infection, transendothelial electrical resistance (TEER) was measured. Briefly, 0.5 µM PET-inserts (24-well Millicell® Cell Culture Inserts, Millipore, Watford, UK) were fitted to wells of the 24-well culture plates. Then, 104 cells in 100 µL of RPMI medium were added to each insert and 100 μL of the culture medium was added to the bottom of the well. The plate was incubated for 1 h; then the medium was discarded and replaced with 1.3 mL fresh media in the bottom of each well. Also, 300 µL fresh media was added onto the cell monolayer inside the inserts. Three days after seeding the cells, the media in the inserts and wells were discarded and replaced with fresh media. Six days after seeding, one of the plates was infected with T. gondii by removing old media and adding tachyzoites at a MOI of 5, in fresh medium, to the inserts. The plate was incubated for 1 h before addition of monensin-supplemented RPMI or fresh RPMI (control) to hBMEC monolayer growing on the apical (top) side of insert. TEER was measured in each insert daily for 4 successive days. TEER was measured using an Evom voltohmmeter with the Endohm 12 and STX2 electrodes (World Precision Instruments, Inc., Sarasota, FL, USA). The electrodes were placed into the insert membrane and bottom of each well. TEER values were measured three times from each well, while changing the position of the electrodes slightly each time in order to obtain the overall average of the insert resistance. A background resistance was established by placing the electrodes into inserts with a blank medium only, without cells. This value was used to calculate the TEER value of each well using the equation (TEER (cells) = (TEER(total) − TEER(blank)) culture area). The culture area of the insert was 0.33 cm2. The TEER value was expected to increase with increased resistance of the hBMEC monolayer.
2.6. Microarray Analysis
The aim of this experiment was to identify the effect of 0.1 µM monensin on the overall expression of host cell genes, and in particular on the expression of genes involved in the Wnt signalling pathway. We used cDNA microarrays to detect changes in mRNA expression in four experimental groups. These included: uninfected, un-treated hBMECs (Control); hBMECs treated with 0.1 µM monensin for 24 h (Monensin_only); T. gondii infected-hBMECs for 24 h (TG_only); and T. gondii-infected hBMECs treated with 0.1µM monensin after 3 h of infection and incubated for further 24 h (TG_Monensin).
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Manchester, UK) and eluted with nuclease-free water. The extracted RNA was stored at −80 °C for one week prior to microarray analysis. All subsequent sample handling, labelling, and GeneChip (GeneChip Human Gene 1.0 ST arrays, Affymetrix, Santa Clara, CA, USA) processing were performed at the transcriptomics service at Nottingham Arabidopsis Stock Centre (NASC). The RNA concentration and quality were assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA, USA) and the RNA 6000 Nano kit (Caliper Life Sciences, Mountain View, CA, USA), respectively, prior to the microarray analysis. Only RNA samples with a minimum RNA concentration of 100 ng/µL and RNA integrity number (RIN) value > 8 were used for microarray analysis.
Single-stranded complimentary DNA was prepared from 200 ng of total RNA as per the Ambion
® WT Expression Kit’s instructions (Thermo Fisher Scientific, Paisley, UK) for Affymetrix GeneChip Whole Transcript WT Expression Arrays (Affymetrix, Wycombe, UK). Total RNA was first converted to cDNA, followed by transcription to make cDNA. Single-stranded cDNA was synthesized, end labelled, and hybridized for 16 h at 45 °C to Human Gene 1.0 ST arrays (Affymetrix, Wycombe, UK). All liquid handling steps were performed by a GeneChip Fluidics Station 450 and GeneChips were scanned with the GeneChip Scanner 3000 7G (Affymetrix, Wycombe, UK) using Command Console v3.2.4. The microarray data have been deposited in the ArrayExpress database at EMBL-EBI (Available online:
www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-8817.
The R package software (R Studio Inc., Boston, MA, USA) was used to process the data. Briefly, a data file, called ‘pdatamon’ was created to link each microarray data file with ‘.ga.cel’ ending to one of the four experimental groups (i.e. Control, Monensin_only, TG_only, TG_Monensin). The libraries of modules (oligo, gplots, RColorBrewer, pvclust, colourspace, limma, gene filter, hugene21sttranscriptcluster.db) needed for quality control, differential gene expression and transcript cluster ID annotation were downloaded and uploaded into the workspace. Next, Affymetrix microarray data files were uploaded together with ‘pdatamon’ file. Quality control analysis and normalization were performed using the robust multi-array average (RMA). RMA normalization is sensitive for low expression values, and provides consistent fold-change estimates, and excellent background adjustment compared to MAS 5.0 and dChip normalization methods [
21,
22]. Only genes exhibiting a fold-change FC ≤ −1 (downregulated) or FC ≥ 1 (upregulated) relative to control and an adjusted
p-value < 0.05 were considered as differentially expressed genes (DEGs). The ranked list of DEGs were included in subsequent analyses. Comparison was made between control (untreated + uninfected) hBMECs and each of the other three groups, including hBMECs treated with monensin (Monensin_only),
T. gondii-infected hBMECs treated with monensin (TG_Monensin), and hBMECs infected with
T. gondii only (TG_only).
2.7. Gene Ontology (GO) Analysis
Functional annotations of the valid DEGs and statistical overrepresentation of the GO terms in the categories of biological processes, molecular functions, and cell components [
23], were retrieved in accordance with instructions [
24,
25], using the PANTHER online database (Available online:
www.pantherdb.org). The PANTHER library of annotated genes (
Homo sapiens) was used as reference list for calculating statistical overrepresentation with a false discovery rate (FDR) < 0.05 considered significantly enriched.
2.8. Pathway Enrichment Analysis
The DEGs genes were functionally annotated and positioned within the respective biological pathways using the Reactome pathway database (Available online:
https://reactome.org) and the Reactome Analysis Tool version v65 (Available online:
https://reactome.org/PathwayBrowser/#TOOL=AT). Significance was estimated using hypergeometric testing and FDR was controlled for using the Benjamini–Hochberg procedure. Pathways with FDR < 0.01 were considered significantly enriched.
2.9. Statistical Analysis
Statistical analysis was performed using a Student’s t-test (two independent groups), one-way ANOVA (multiple groups, one independent factor) or two-way ANOVA (multiple groups, two independent factors) followed by a Bonferroni–Holm post hoc test using GraphPad Prism 7 software for Windows (GraphPad Software, San Diego, CA, USA). The mean of at least three independent experiments is presented, with error bars in the graphs showing the standard error of the mean (SEM). Differences were considered statistically significant at a p value of < 0.05. The level of significance was reported as: * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. The drug concentration that caused 50% inhibition of host cell growth was expressed as 50% cytotoxic concentration (CC50). The CC50 of monensin on microglia and hBMECs were calculated by plotting dose-response curves followed by performing simple linear regression analysis using Graph Pad Prism 7 software. The calculation of the half-maximal inhibitory concentration (IC50), which is the concentration of monensin that caused a 50% decrease of T. gondii growth compared to the control in both cell lines, were calculated using Graph Pad Prism 7’s nonlinear regression (curve fit) built-in analysis tool of dose-response inhibition (three parameters). The selectivity index (SI), which represents the ratio of the CC50 for host cells to the IC50 for T. gondii, was calculated by comparing the cytotoxicity of monensin for host cells to that of T. gondii.
4. Discussion
In this study, we utilized microarray analysis to evaluate the transcriptome profile of
T. gondii-infected hBMECs treated with the Wnt inhibitor monensin. First, we investigated the effect of Wnt inhibition by monensin on the in vitro growth of
T. gondii inside hBMECs and microglia cells.
T. gondii growth inhibition was evidenced by the significant reduction in the number of tachyzoites (
Figure 1) and the relative similarity in cell density levels (
Figure 2) between infected and non-infected cultures. Our data showed that parasite inhibition increased as the concentration of monensin increased (i.e. dose-dependent) and that monensin at 0.1 µM concentration significantly decreased the growth of
T. gondii tachyzoites. Consistent with this, it has been reported that monensin reduced
T. gondii growth in human foreskin fibroblasts [
26,
27]. Our result also fits within the effective range of monensin concentrations (0.01 and 0.1 µM) reported against human cytomegalovirus (HCMV) [
14]. Interestingly, monensin was more effective in inhibiting
T. gondii growth in hBMECs compared to microglia cells. Moreover, in the presence of monensin treatment, infected and noninfected hBMECs exhibited significant reduction in their growth rates. However, the differential variation in the magnitude of cell growth reduction between the two cell lines suggests that monensin had more cytotoxic effect on hBMECs compared to microglia cells. These findings were the basis for choosing hBMECs to determine the effects of monensin on host gene transcription in the context of
T. gondii infection.
Next, we determined the effect of
T. gondii with or without monensin treatment on the integrity of cerebrovascular endothelial cell monolayer. Our findings showed that
T. gondii infection can compromise the integrity of BBB endothelial cells compared to uninfected cells (
p < 0.05;
Figure 3). Consistent with our finding,
T. gondii was reported to compromise the cell integrity to facilitate a paracellular migration route across the BBB [
28]. Cellular barrier function is mediated through intercellular tight junction (TJ) proteins, which can be dysregulated by
T. gondii infection to increase barrier permeability and parasite crossing [
29]. Alteration in the TJ function may occur either through a direct effect of
T. gondii infection on the host cell cytoskeleton [
30] or via the indirect action of
T. gondii-induced proinflammatory cytokines, which can promote the opening of TJ that would in turn allow leukocytes to migrate across the endothelial barrier into cerebral tissue [
31,
32].
T. gondii-infected and treated hBMECs seemed to maintain their resistance, presumably due to the inhibitory effect of monensin on
T. gondii growth, thereby protecting cells from cytoskeletal rearrangement associated with parasite invasion and growth, which compromises TJ and BBB integrity. Given the ability of this parasite to invade and use leukocytes as a Trojan-horse to migrate across the BBB [
33], further studies should investigate how monensin may influence the effect of
T. gondii on leukocytes’ ability to cross the BBB. If monensin is effective in inhibiting
T. gondii growth inside leukocytes, then this may thwart the parasite’s ability to disseminate to the CNS tissue.
The exact mechanism of anti-
T. gondii activity of monensin is unknown, however this compound seems to exert its action by directly interfering with one or more functions in the parasite. For example, osmotic swelling of the parasite [
34], presumably due to its inability to regulate ionic homeostasis as a result of Na
+ influx [
35], interference with vesicular trafficking [
36], altering the parasite’s cell cycle [
26], or inducing nutrient stress and TgMSH-1-mediated autophagy [
27], have all been suggested to contribute to
T. gondii death. Interestingly, pre-treatment of hBMECs with monensin inhibited the susceptibility of hBMEC to gram-negative bacteria
Citrobacter freundii [
37] and
Brucella abortus [
38] invasion or survival in a dose-dependent manner. Monensin exhibits potent anticancer activity on several types of cancer, by inhibiting cell proliferation, cell cycle progression, and cell migration and by inducing apoptosis [
39]. Monensin may also accomplish its anticancer effect by targeting multiple signaling pathways, particularly the epidermal growth factor receptor (EGFR) and Wnt signaling pathways [
39,
40]. Given these pervasive effects of monensin, it is possible that monensin exerts its anti-
T. gondii effect, by altering the host response to infection in addition to its direct antiparasitic effect.
Therefore, we investigated whether the secondary effects of monensin on the host cells may contribute to parasite clearance. We performed microarray analysis to test the hypothesis that treatment with 0.1 µM monensin modulates signalling pathways, such as the Wnt pathway, which in turn inhibits
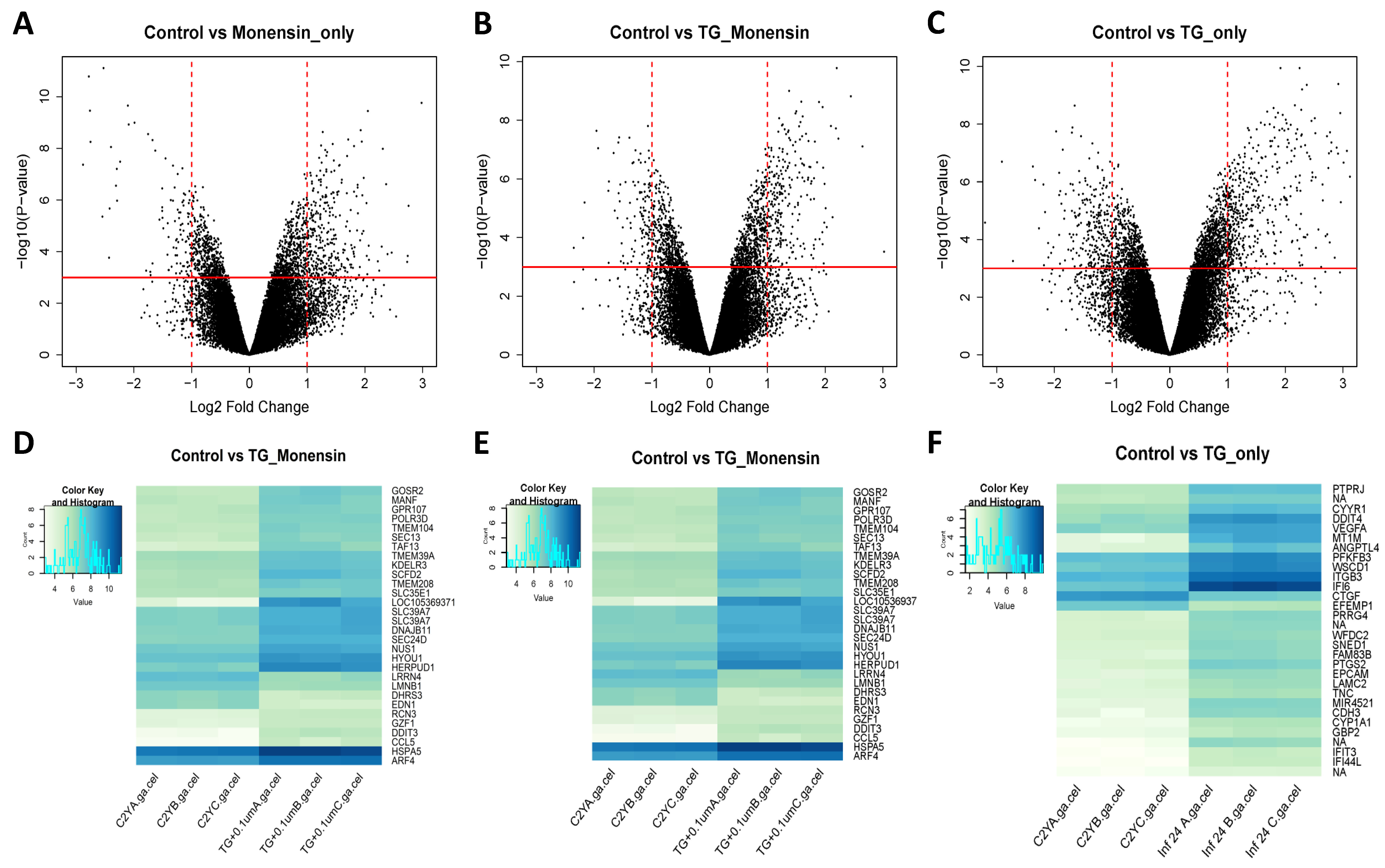
T. gondii infection and enhances hBMECs survival. The microarray quality control checks indicated that the datasets had acceptable quality. Variations within samples were minimal (
Figure 4) and variations between samples were made comparable by RMA normalization (
Figure S1). Our analysis revealed 480 DEGs in hBMECs treated with monensin compared to control untreated hBMECs. Since monensin induced significant changes in the gene transcription of treated cells, we wanted to investigate whether monensin also can cause the same changes in the presence of
T. gondii infection. Interestingly, 424 DEGs were detected in
T. gondii-infected hBMECs treated with monensin compared to control hBMECs. When we compared DEGs in monensin-treated uninfected cells to DEGs in monensin-treated
T. gondii-infected cells, different levels of expression of specific genes were detected between the two groups, showing the differential response of treated cells in the presence or absence of infection. This finding is anticipated because a subset of these DEGs can be expected to be mechanistically involved in the infection process and thus their expression is unlikely to change in response to treatment per se. Interestingly, the TG_only group expression profile was distinct from the expression profiles of the other three groups (TG_Monensin group, Monensin_only group and Control_only group) as shown in the dendrogram (
Figure 4A). This clustering is supported by the detection of 902 DEGs in hBMECs infected by
T. gondii without monensin treatment (TG_only group), compared to 424 DEGs detected in infected and treated cells (TG_Monensin group). Such a significantly large number of DEGs in the TG_only group indicates that monensin must have attenuated the adverse impact of
T. gondii infection on the treated cells, probably by interfering with signalling pathways essential for the parasite growth or via direct inhibition of the parasite proliferation, providing further support to the antiparasitic effect of monensin observed in
Figure 1 and
Figure 2.
PANTHER GO enrichment analysis, used to functionally annotate all DEGs in each group, revealed similar terms in the three GO categories between the examined groups. Reactome pathway enrichment analysis of the DEGs of hBMECs treated with monensin revealed that monensin treatment downregulated pathways related to steroids, cholesterol and protein biosynthesis and their transport between endoplasmic reticulum (ER) and Golgi apparatus. Transcriptional analysis of
T. gondii-infected hBMECs treated with monensin revealed a similar transcriptional pattern to the Monensin_only group. These results suggest that parasite inhibition may have been achieved through dysregulation of the biosynthesis of key nutrients in the host cells and/or inhibition of nutrient transport between cell organelles, limiting their transport into parasitophorous vacuole. This finding is consistent with a previous transcriptomic study of monensin-treated porcine kidney (PK)-15 cells, which revealed an overrepresentation of the downregulated genes involved in the biosynthesis of spliceosome, ribosome, and protein processing in ER, suggesting that monensin, via down-regulation of protein biosynthesis, can limit the parasite growth and proliferation [
41].
Our data also showed that monensin administration modulated pathways related to DNA replication in the nucleus and cell cycle. In our study, we have identified 39 significantly upregulated pathways related to DNA replication, cell cycle and cell division in the TG_Monensin group compared to only three significantly upregulated pathways from the Monensin_only transcriptome data. The significantly increased number of these upregulated pathways in the TG_Monensin group is probably attributed to the dual effects of monensin and parasite infection on the host cell transcriptome. The effects of monensin on DNA synthesis, and repair of the parasite and the host cells, can be also inferred from the downregulated pathways identified in both the Monensin_only and the TG_Monensin groups. Some of the downregulated pathways were related to DNA repair mechanisms, such as resolution of D-loop structures through Holliday junction intermediates (R-HSA-5693568), resolution of D-loop structures (R-HSA-5693537), resolution of D-loop structures through synthesis-dependent strand annealing (R-HSA-5693554), and unwinding of DNA (R-HSA-176974). In addition, key DNA repair mechanisms related to molecular functions, such as DNA helicase activity (GO:0003678), helicase activity (GO:0004386), kinase regulator activity (GO:0019207), nucleic acid binding (GO:0003676) and binding (GO:0005488), were also identified as overrepresented (PANTHER Overrepresentation Test) from downregulated genes of the Monensin_only group.
Interestingly, both the Monensin_only and the TG_Monensin groups downregulated cell cycle-related pathways (e.g. cell cycle (R-HSA-1640170); G0 and early G1 (R-HSA-1538133) and transcription of E2F targets under negative control by p107/RBL1 and p130/RBL2 in complex with HDAC1 [R-HSA-1362300]) and upregulated PERK regulates gene expression (R-HSA-381042), which leads to cell cycle arrest in host cells. Monensin has been shown to arrest the parasite cell cycle process [
26]. Thus, we can infer that monensin can inhibit both parasite and host cell cycle processes that ultimately lead to growth inhibition of
T. gondii. This finding is consistent with the upregulation of the DNA damage-inducible transcript 3 (
DDIT3), a key gene in the upregulated pathway, which encodes a stress-inducible transcription factor that induces cell cycle arrest and apoptosis in mammalian cells. Our finding is consistent with a previous study that showed that monensin induced
DDIT3, a key inhibitor of the Wnt signalling pathway [
42], in prostate cancer cells [
43]. The key genes identified in those downregulated pathways are
CCNA2, that controls both the G1/S and the G2/M transition phases of the cell cycle, and
MYBL2, that functions as a physiological regulator of cell cycle progression. While there are no published data that show the inhibitory effect of monensin on
CCNA2, there are reports on monensin as a potent inhibitor of the
MYB gene family that includes
MYBL1 and
MYBL2 [
44,
45,
46].
The link between
T. gondii and the Wnt signalling pathway has been already established. In one study, the Wnt pathway was found to be upregulated during
T. gondii infection in brain cells [
47]. In another study, the virulence factor ROP18 of
T. gondii was found to inhibit the Wnt signalling pathway in neural stem cells [
48]. In the present study, Wnt signalling and Wnt-related pathways were not significantly affected by treatment with 0.1 µM monensin, rejecting the hypothesis that inhibition of
T. gondii growth is largely mediated by the aberrant Wnt signalling in the host cells due to monensin treatment. All Wnt-related genes, such as Frizzled receptors, beta-catenin and protein kinase C (PKC) had modest expression after treatment. The same also applies to the transcriptome of TG_only and TG_Monensin groups. This finding suggests that monensin treatment did not affect the Wnt signalling pathway in hBMECs and that inhibition of
T. gondii infection in hBMECs following monensin treatment was not achieved through direct blocking of the Wnt signalling pathway. Our study did however identify changes in the expression of genes related to other signal pathways involved in cell cycle control, apoptosis, and DNA synthesis and repair. These data further support the pleiotropic effects of monensin on multiple biological processes.
We were also interested in comparing host gene expression in uninfected to
T. gondii-infected hBMECs in the absence of monensin treatment to better understand the types of host responses induced by infection. The transcriptome analysis of hBMECs infected with
T. gondii for 24 h (TG_only) revealed significantly increased interferon-alpha/beta and gamma signalling-related pathways. This finding is consistent with previous transcriptome studies in murine macrophages [
49] and human fibroblast cells [
50], reaffirming the deregulation of the immune response following
T. gondii infection of hBMECs. The increased expression of the
MT1M gene was also reported previously [
51], where
MT1M upregulation has been suggested to contribute to the cellular homeostasis of transition metals, particularly zinc, during
T. gondii infection. The down-regulation of microRNA 29a has also been reported in a mouse brain infected by the cystogenic strain (type II) [
52], which is less virulent than the RH (type I) strain used in our study, suggesting that inhibition of microRNA 29a is common in
T. gondii infection, independent of the parasite virulence.