Isotopic Tracers Unveil Distinct Fates for Nitrogen Sources during Wine Fermentation with Two Non-Saccharomyces Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
2.2. Fermentations and Sampling
2.3. Quantification of Consumed and Proteinogenic Amino Acids
2.4. Isotopic Enrichment of Intracellular Amino Acid Analysis
2.5. Isotopic Enrichment of Volatile Compounds Analysis
2.6. Outline of Experiment Design and Data Analysis
3. Results
3.1. Incorporation of Nitrogen from Glutamine, Ammonium, and Arginine into Proteinogenic Amino Acids
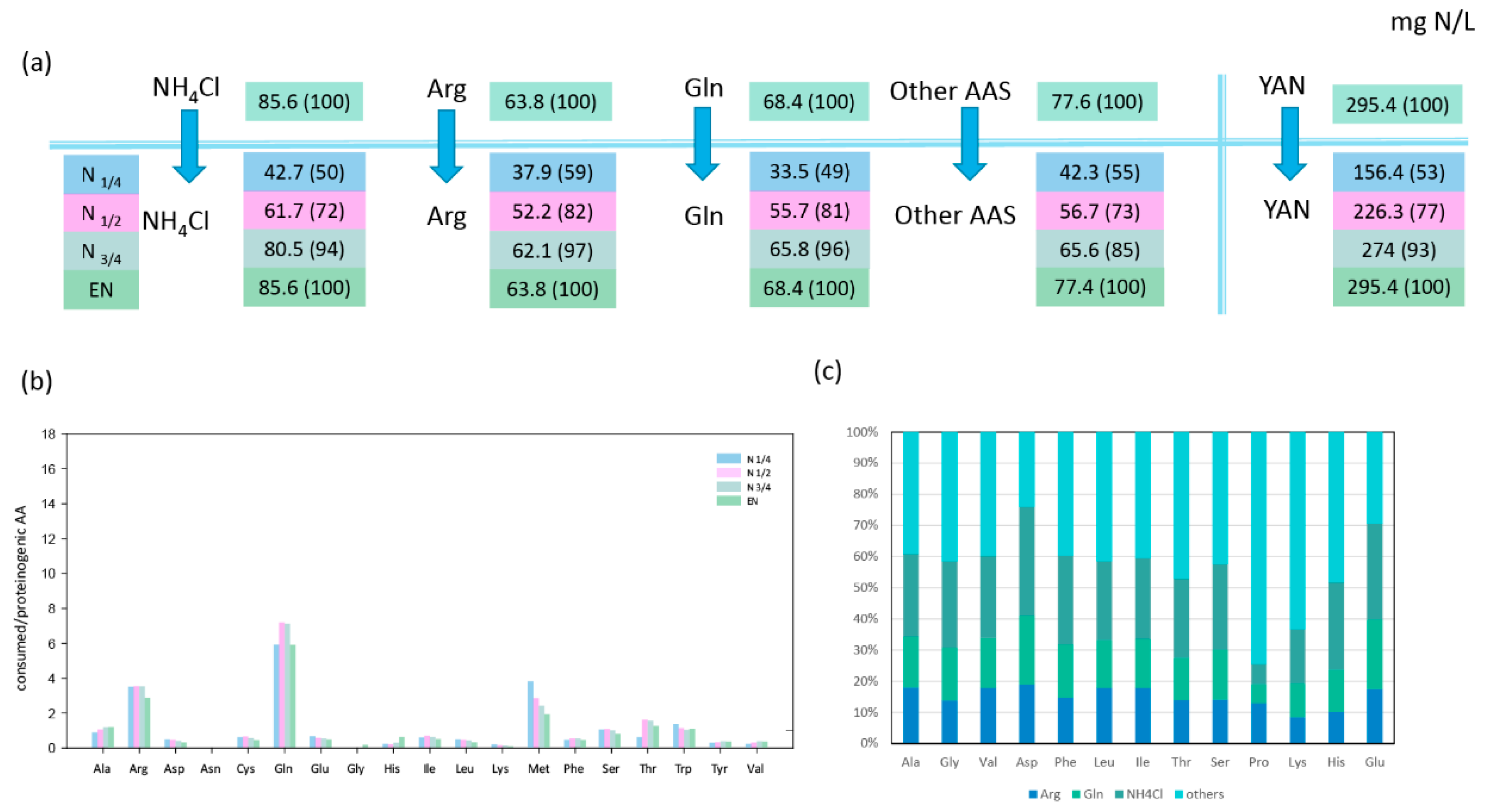
3.1.1. Consumption of Glutamine, Ammonium, and Arginine during the Growth Phase
3.1.2. Relationships between Consumption of Amino Acids and Anabolic Requirements in Proteinogenic Amino Acids
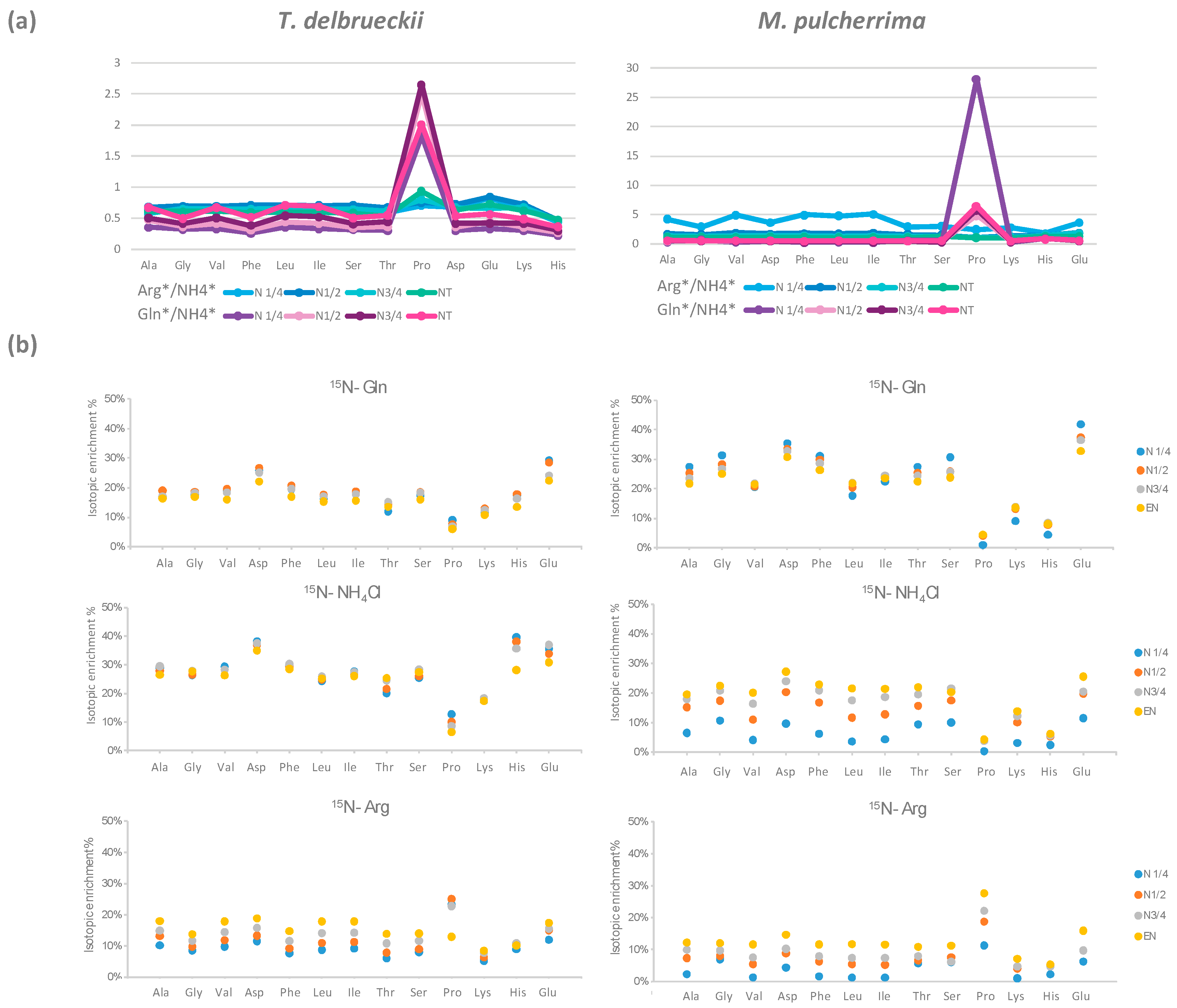
3.1.3. Redistribution of Glutamine, Ammonium, and Arginine for De Novo Synthesis of Other Amino Acids
3.1.4. Different Isotopic Enrichment Patterns of Proteinogenic Amino Acids
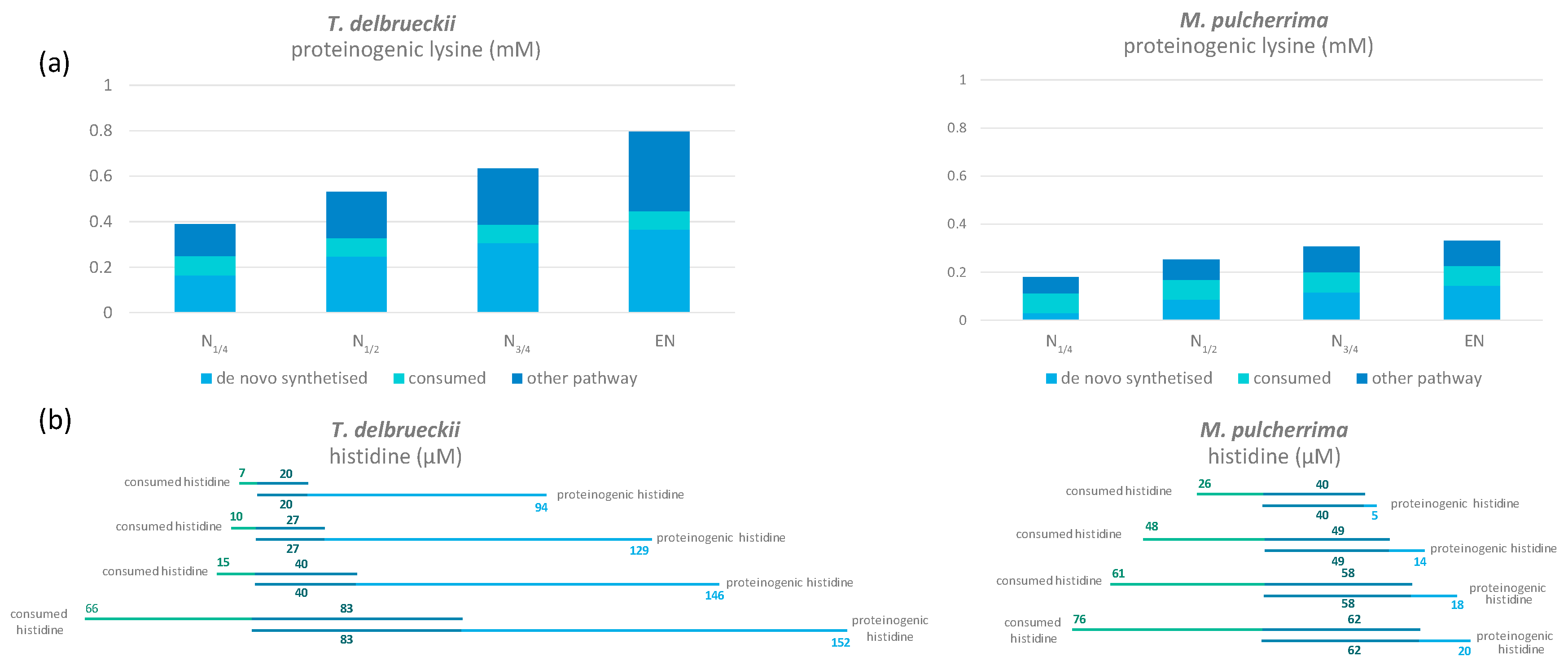
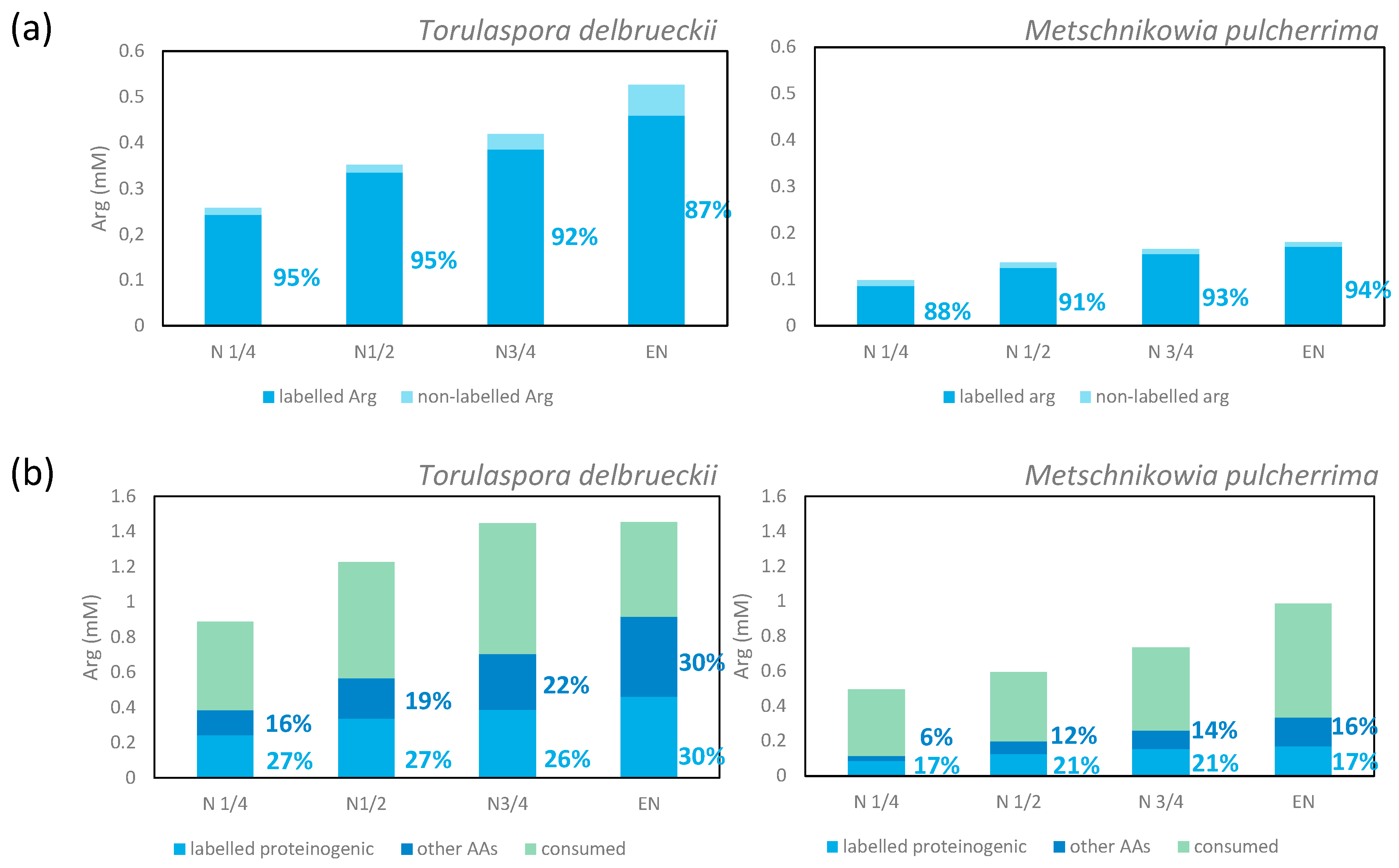
3.1.5. Metabolism of Arginine and Proline during Fermentation
3.2. Origin of the Carbon Backbone of Proteinogenic Aliphatic Amino Acids
3.2.1. Consumed Aliphatic Amino Acids Recovered in Proteinogenic Amino Acids
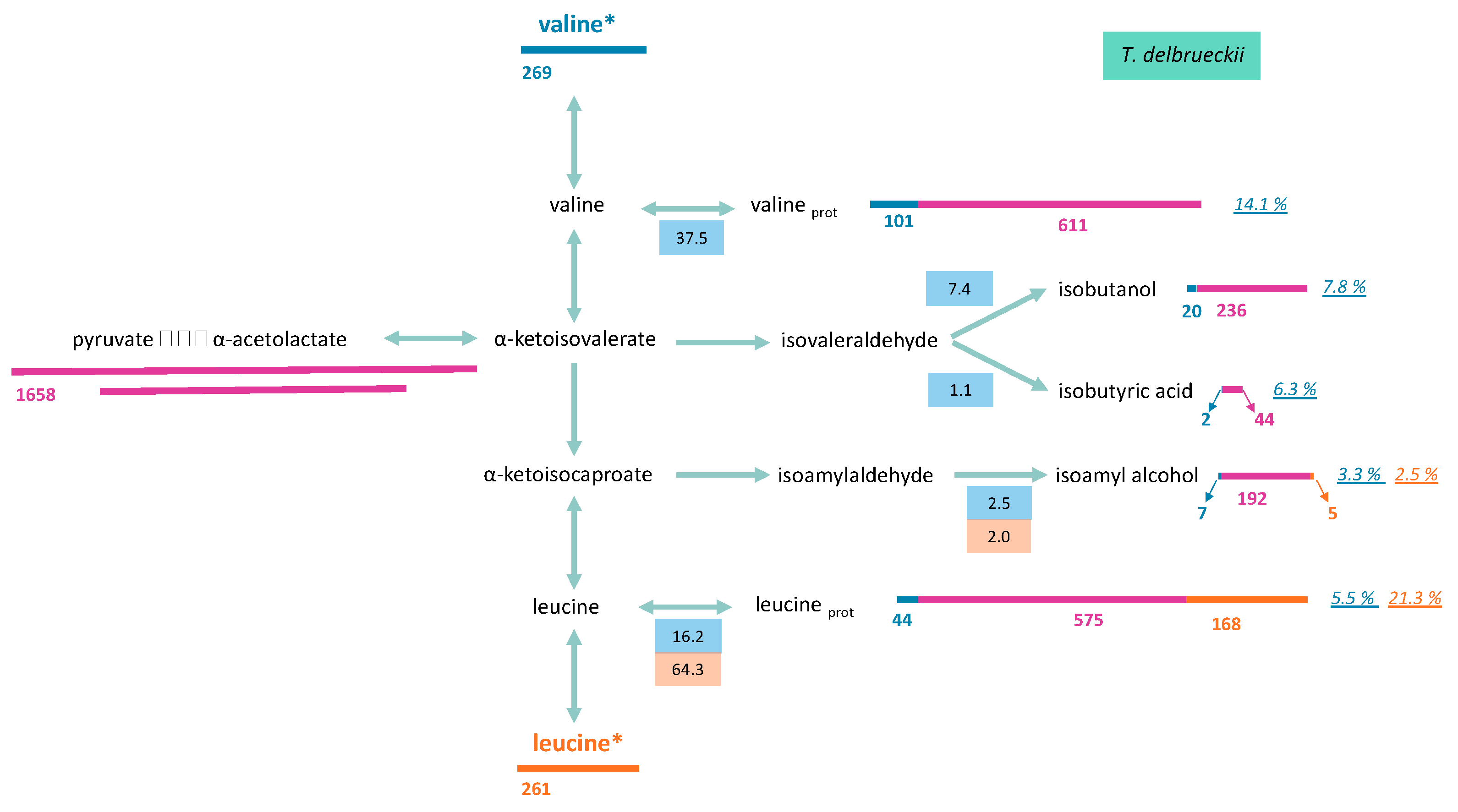
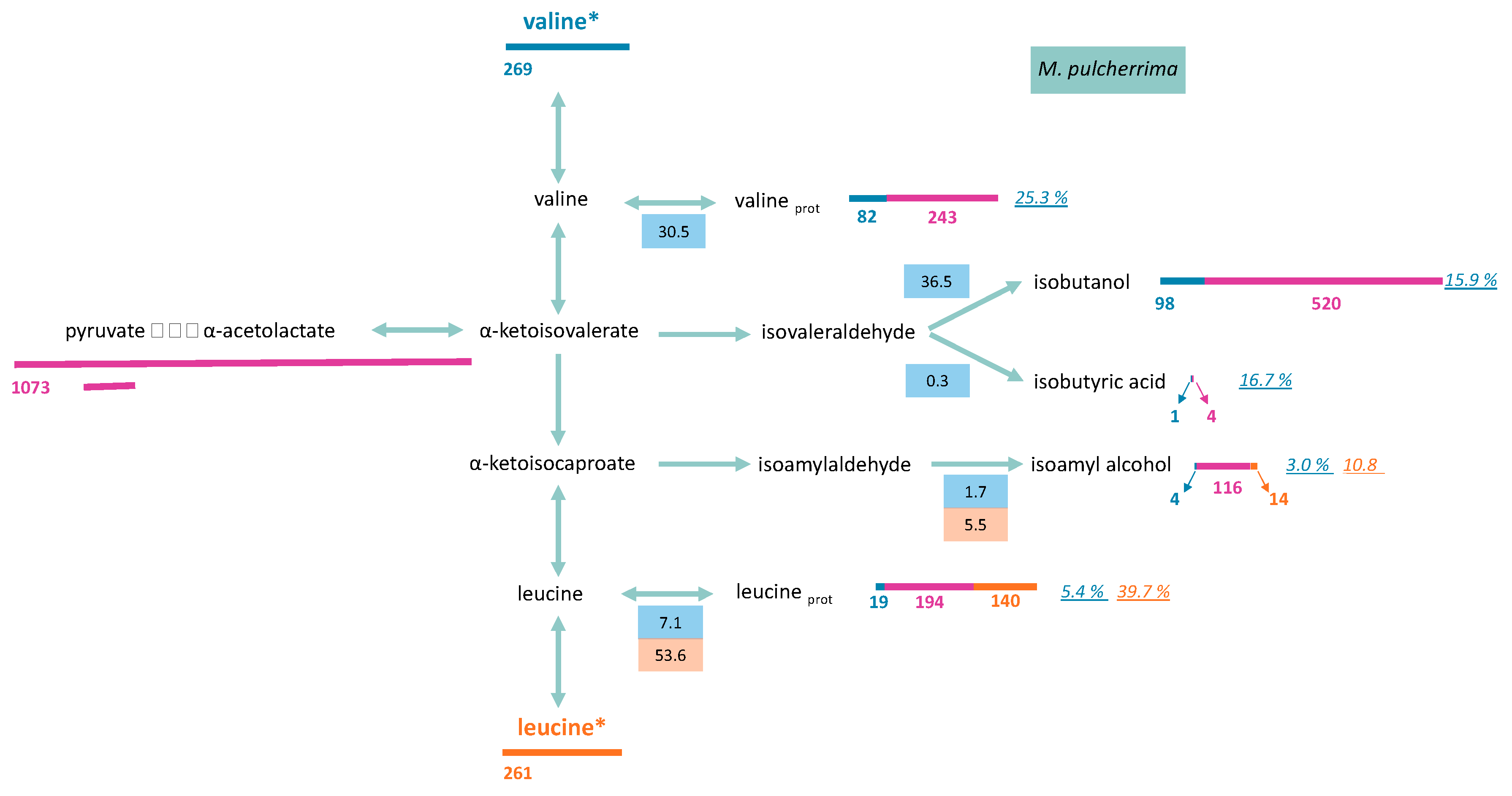
3.2.2. Formation of Volatile Compounds through Valine and Leucine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Hansen, E.C. Recherches sur la physiologie et la morphologie des ferments alcooliques V. Methodes pour obtenir des cultures pures de Saccharomyces et de mikroorganismes analogues. C. R. Trav. Lab. Carlsberg 1883, 2, 92–105. [Google Scholar]
- Bisson, L.F. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 1999, 50, 107–119. [Google Scholar]
- Bauer, F.F.; Pretorius, I.S. Yeast stress response and fermentation efficiency: How to survive the making of wine—A review. S. Afr. J. Enol. Vitic. 2000, 21, 27–51. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine. In Food Microbiology Fundamentals and Frontiers, 2nd ed.; Doyle, M.P., Beuchat, L.R., Montville, T.J., Eds.; ASM Press: Washington, DC, USA, 2001; pp. 747–772. [Google Scholar]
- Belda, I.; Ruiz, J.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Benito, S.; Santos, A. Influence of Torulaspora delbrueckii in varietal thiol (3-SH and 4-MSP) release in wine sequential fermentations. Int. J. Food Microbiol. 2017, 257, 183–191. [Google Scholar] [CrossRef]
- Jolly, N.P.; Augustyn, O.P.H.; Pretorius, I.S. The Role and Use of Non-Saccharomyces Yeasts in Wine Production. S. Afr. J. Enol. Vitic. 2006, 27, 15–39. [Google Scholar] [CrossRef]
- Seguinot, P.; Bloem, A.; Brial, P.; Meudec, E.; Ortiz-Julien, A.; Camarasa, C. Analysing the impact of the nature of the nitrogen source on the formation of volatile compounds to unravel the aroma metabolism of two non-Saccharomyces strains. Int. J. Food Microbiol. 2019, 316, 108441. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Translational Regulation of Gcn4 and the General Amino Acid Control of Yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [Green Version]
- Magasanik, B.; Kaiser, C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef]
- Crépin, L.; Nidelet, T.; Sanchez, I.; Dequin, S.; Camarasa, C. Sequential use of nitrogen compounds by saccharomyces cerevisiae during wine fermentation: A model based on kinetic and regulation characteristics of nitrogen permeases. Appl. Environ. Microbiol. 2012, 78, 8102–8111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crépin, L.; Truong, N.M.; Bloem, A.; Sanchez, I.; Dequin, S.; Camarasa, C. Management of multiple nitrogen sources during wine fermentation by S. cerevisiae. Appl. Environ. Microbiol. 2017, 83, AEM.02617-16. [Google Scholar] [CrossRef] [Green Version]
- Rollero, S.; Mouret, J.R.; Bloem, A.; Sanchez, I.; Ortiz-Julien, A.; Sablayrolles, J.M.; Dequin, S.; Camarasa, C. Quantitative13C-isotope labelling-based analysis to elucidate the influence of environmental parameters on the production of fermentative aromas during wine fermentation. Microb. Biotechnol. 2017, 10, 1649–1662. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Tourdot-maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexndre, H. Non-Saccharomyces Yeasts Nitrogen Source Preferences: Impact on Sequential Fermentation and Wine Volatile Compounds Profile. Front. Microbiol. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemsawasd, V.; Viana, T.; Ardö, Y.; Arneborg, N. Influence of nitrogen sources on growth and fermentation performance of different wine yeast species during alcoholic fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 10191–10207. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Seguinot, P.; Sanchez, I.; Ortiz-Julien, A.; Heras, J.M.; Querol, A.; Camarasa, C.; Guillamón, J.M. Nitrogen sources preferences of non-Saccharomyces yeasts to sustain growth and fermentation under winemaking conditions. Food Microbiol. 2020, 85. [Google Scholar] [CrossRef]
- Lleixà, J.; Martín, V.; Portillo, M.D.C.; Carrau, F.; Beltran, G.; Mas, A. Comparison of fermentation and wines produced by inoculation of Hanseniaspora vineae and Saccharomyces cerevisiae. Front. Microbiol. 2016, 7, 338. [Google Scholar] [CrossRef] [Green Version]
- Rollero, S.; Bloem, A.; Ortiz-Julien, A.; Bauer, F.F.; Camarasa, C.; Divol, B. A comparison of the nitrogen metabolic networks of Kluyveromyces marxianus and Saccharomyces cerevisiae. Environ. Microbiol. 2019, 21. [Google Scholar] [CrossRef]
- Rollero, S.; Bloem, A.; Ortiz-Julien, A.; Camarasa, C.; Divol, B. Altered fermentation performances, growth, and metabolic footprints reveal competition for nutrients between yeast species inoculated in synthetic grape juice-like medium. Front. Microbiol. 2018, 9, 196. [Google Scholar] [CrossRef]
- Azzolini, M.; Tosi, E.; Lorenzini, M.; Finato, F.; Zapparoli, G. Contribution to the aroma of white wines by controlled Torulaspora delbrueckii cultures in association with Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2015, 31, 277–293. [Google Scholar] [CrossRef]
- González-Royo, E.; Pascual, O.; Kontoudakis, N.; Esteruelas, M.; Esteve-Zarzoso, B.; Mas, A.; Canals, J.M.; Zamora, F. Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 2015, 240, 999–1012. [Google Scholar] [CrossRef]
- Varela, C.; Barker, A.; Tran, T.; Borneman, A.; Curtin, C. Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. Int. J. Food Microbiol. 2017, 252, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bely, M.; Sablayrolles, J.M.; Barre, P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 1990, 70, 246–252. [Google Scholar] [CrossRef]
- Su, Y.; Gamero, A.; Eugenia, M.; Ariel, C. Interspecific hybridisation among diverse Saccharomyces species: A combined biotechnological solution for low-temperature and nitrogen-limited wine fermentations. Int. J. Food Microbiol. 2019, 310, 108331. [Google Scholar] [CrossRef] [PubMed]
- Bloem, A.; Rollero, S.; Seguinot, P.; Crépin, L.; Perez, M.; Picou, C.; Camarasa, C. Workflow Based on the Combination of Isotopic Tracer Experiments to Investigate Microbial Metabolism of Multiple Nutrient Sources. J. Vis. Exp. 2018, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Millard, P.; Letisse, F.; Sokol, S.; Portais, J.C. IsoCor: Correcting MS data in isotope labeling experiments. Bioinformatics 2012, 28, 1294–1296. [Google Scholar] [CrossRef] [Green Version]
- Rollero, S.; Bloem, A.; Camarasa, C.; Sanchez, I.; Ortiz-Julien, A.; Sablayrolles, J.M.; Dequin, S.; Mouret, J.R. Combined effects of nutrients and temperature on the production of fermentative aromas by Saccharomyces cerevisiae during wine fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 2291–2304. [Google Scholar] [CrossRef]
- Roca-mesa, H.; Sendra, S.; Mas, A.; Beltran, G.; Torija, M.J. Nitrogen preferences during alcoholic fermentation of different non-saccharomyces yeasts of oenological interest. Microorganisms 2020, 8, 157. [Google Scholar] [CrossRef] [Green Version]
- Prior, K.J.; Bauer, F.F.; Divol, B. The utilisation of nitrogenous compounds by commercial non-Saccharomyces yeasts associated with wine. J. Food Microbiol. 2019, 79, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Marini, A.M.; Vissers, S.; Urrestarazu, A.; André, B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 1994, 13, 3456–3463. [Google Scholar] [CrossRef]
- Marini, A.M.; Soussi-Boudekou, S.; Vissers, S.; Andre, B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell Biol. 1997, 17, 4282–4293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proux-Wéra, E.; Armisén, D.; Byrne, K.P.; Wolfe, K.H. A pipeline for automated annotation of yeast genome sequences by a conserved-synteny approach. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, K.; Abadjieva, A.; Hilven, P.; Stankiewicz, A.; Crabeel, M. The N-acetylglutamate synthase/N-acetylglutamate kinase metabolon of Saccharomyces cerevisiae allows co-ordinated feedback regulation of the first two steps in arginine biosynthesis. Eur. J. Biochem. 2003, 270, 1014–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crépin, L.; Sanchez, I.; Nidelet, T.; Dequin, S.; Camarasa, C. Efficient ammonium uptake and mobilization of vacuolar arginine by Saccharomyces cerevisiae wine strains during wine fermentation. Microb. Cell Fact. 2014, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, A.; Sancho, M.; Beltran, G.; Guillamón, J.M.; Warringer, J. Replenishment and mobilization of intracellular nitrogen pools decouples wine yeast nitrogen uptake from growth. Appl. Microbiol. Biotechnol. 2016, 100, 3255–3265. [Google Scholar] [CrossRef]
- Cooper, T.G. Nitrogen metabolism in Saccharomyces cerevisiae. In The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression; Strathern, J.N., Jones, E.W., Broach, J.R., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1982; pp. 39–99. [Google Scholar]
- Brunke, S.; Seider, K.; Richter, M.E.; Bremer-Streck, S.; Ramachandra, S.; Kiehntopf, M.; Brock, M.; Hube, B. Histidine Degradation via an Aminotransferase Increases the Nutritional Flexibility of Candida glabrata. Eukaryot. Cell 2014, 13, 758–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Z.H.; Garrad, R.C.; Winston, M.K.; Bhattacharjee, J.K. Use of α-aminoadipate and lysine as sole nitrogen source by Schizosacharomyces pombe and selected pathogenic fungi. J. Basic Microbiol. 1991, 31, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Zabriskie, T.M.; Jackson, M.D. Lysine biosynthesis and metabolism in fungi. Nat. Prod. Rep. 2000, 17, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Fani, R.; Lio, P.; Lazcano, A.; Bazzicalupo, M.; Carlomagno, M.S.; Bruni, C.B. Histidine biosynthetic pathway and genes: Structure, regulation and evolution. Microbiol. Rev. 1996, 60, 44–69. [Google Scholar] [CrossRef]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Bell, S.J.; Henschke, P.A. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust. J. Grape Wine Res. 2005, 11, 242–295. [Google Scholar] [CrossRef]
- Seguinot, P.; Ortiz-Julien, A.; Camarasa, C. Impact of Nutrient Availability on the Fermentation and Production of Aroma Compounds Under Sequential Inoculation with M. pulcherrima and S. cerevisiae. Front. Microbiol. 2020, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Mouret, J.R.; Camarasa, C.; Angenieux, M.; Agosin, E.; Perez, M.; Farines, V.; Sablayrolles, J.M. Kinetic analysis and gas—liquid balances of the production of fermentative aromas during winemaking fermentations: Effect of assimilable nitrogen and temperature. Food Res. Int. 2014, 62, 1–10. [Google Scholar] [CrossRef]
- Buhaescu, I.; Izzedine, H. Mevalonate pathway: A review of clinical and therapeutical implications. Clin. Biochem. 2007, 40, 575–584. [Google Scholar] [CrossRef] [PubMed]
- VanNice, J.C.; Skaff, D.A.; Keightley, A.; Addo, J.K.; Wyckoff, G.J.; Miziorko, H.M. Identification in Haloferax volcanii of Phosphomevalonate Decarboxylase and Isopentenyl Phosphate Kinase as Catalysts of the Terminal Enzyme Reactions in an Archaeal Alternate Mevalonate Pathway. J. Bacteriol. 2014, 196, 1055–1063. [Google Scholar] [CrossRef]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009, 48, 4688–4716. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, L.; Liu, H.W.; Sherman, D.H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: Architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 1998, 95, 12111–12116. [Google Scholar] [CrossRef] [Green Version]
- Lachance, M.A. Metschnikowia: Half tetrads, a regicide and the fountain of youth. Yeast 2016, 33, 563–574. [Google Scholar] [CrossRef]
- MacDonald, J.C. Biosynthesis of pulcherriminic acid. Biochem. J. 1965, 96, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Krause, D.J.; Kominek, J.; Opulente, D.A.; Shen, X.X.; Zhou, X.; Langdon, Q.K.; DeVirgilio, J.; Hulfachor, A.B.; Kurtzman, C.P.; Rokas, A.; et al. Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl. Acad. Sci. USA 2018, 115, 11030–11035. [Google Scholar] [CrossRef] [Green Version]

| Time (min) | 0.0 | 3.0 | 5.0 | 11.0 | 12.5 | 14.0 | 18.0 | 21.0 | 23.0 | 25.0 | 26.0 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phase A (%) | 95.0 | 94.0 | 92.0 | 90.0 | 88.0 | 82.0 | 80.0 | 70.0 | 60.0 | 25.0 | 20.0 |
| Phase B (%) | 5.0 | 6.0 | 8.0 | 10.0 | 12.0 | 18.0 | 20.0 | 30.0 | 40.0 | 75.0 | 80.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.; Seguinot, P.; Bloem, A.; Ortiz-Julien, A.; Heras, J.M.; Guillamón, J.M.; Camarasa, C. Isotopic Tracers Unveil Distinct Fates for Nitrogen Sources during Wine Fermentation with Two Non-Saccharomyces Strains. Microorganisms 2020, 8, 904. https://doi.org/10.3390/microorganisms8060904
Su Y, Seguinot P, Bloem A, Ortiz-Julien A, Heras JM, Guillamón JM, Camarasa C. Isotopic Tracers Unveil Distinct Fates for Nitrogen Sources during Wine Fermentation with Two Non-Saccharomyces Strains. Microorganisms. 2020; 8(6):904. https://doi.org/10.3390/microorganisms8060904
Chicago/Turabian StyleSu, Ying, Pauline Seguinot, Audrey Bloem, Anne Ortiz-Julien, José María Heras, José Manuel Guillamón, and Carole Camarasa. 2020. "Isotopic Tracers Unveil Distinct Fates for Nitrogen Sources during Wine Fermentation with Two Non-Saccharomyces Strains" Microorganisms 8, no. 6: 904. https://doi.org/10.3390/microorganisms8060904





