Association of Phytophthora with Declining Vegetation in an Urban Forest Environment
Abstract
1. Introduction
2. Materials and Methods
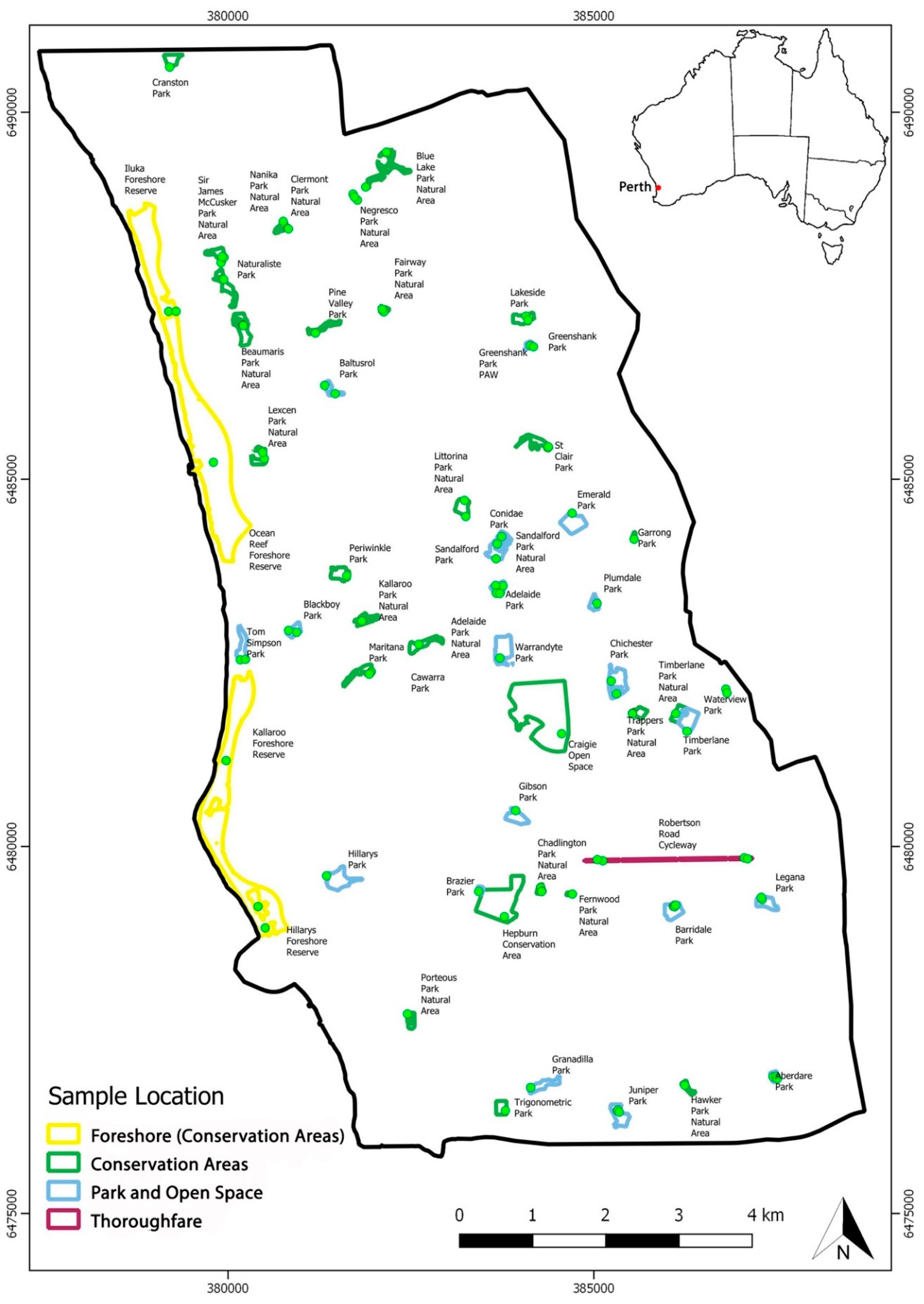
2.1. Study Area and Sample Collection
2.2. Baiting Technique
2.3. eDNA Extraction from Fine Roots and Metabarcoding
2.4. Data Analysis
3. Results
3.1. Isolation and Identification
3.2. Phytophthora Community
3.3. Distribution of Phytophthora cinnamomi and P. multivora
4. Discussion
4.1. Bridgehead Effect
4.2. Phytophthora Species Detected
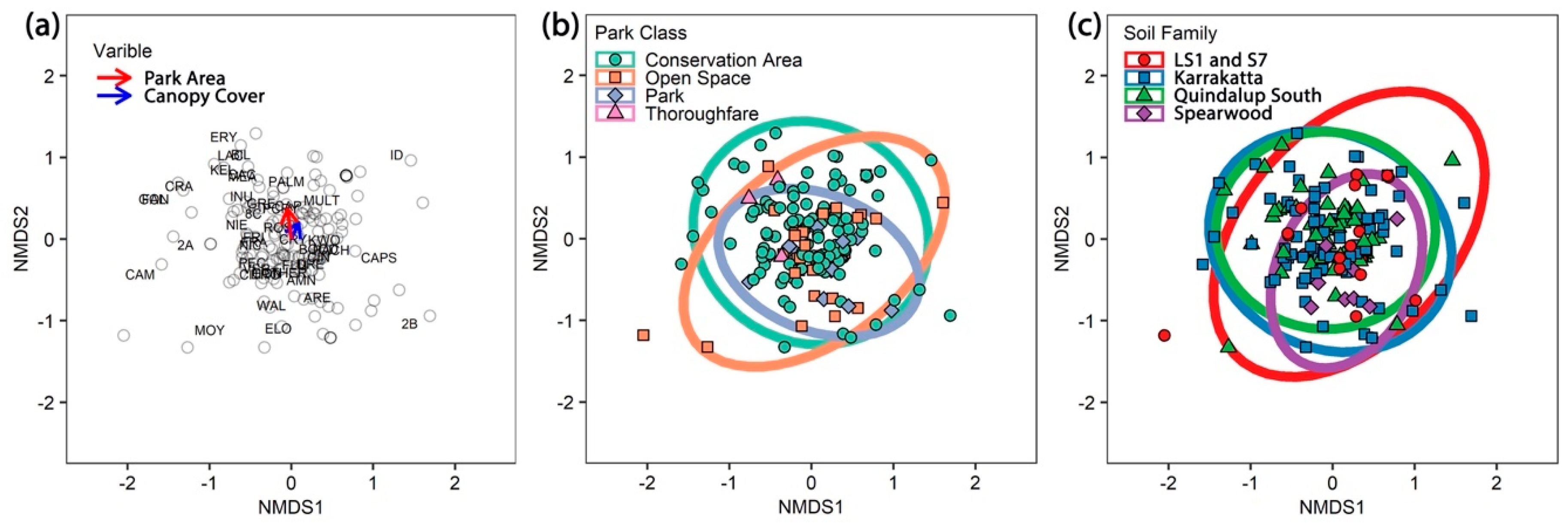
4.3. Impact of Parks Class and Area, Canopy Cover and Health on Phytophthora Community
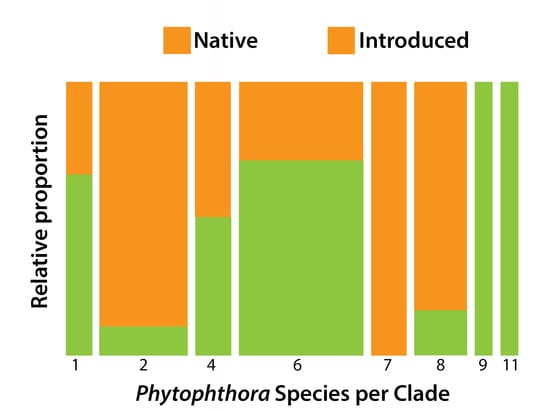
4.4. Relationship between Commonly Detected Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nowak, D.J.; Noble, M.H.; Sisinni, S.M.; Dwyer, J.F. People and trees: Assessing the US urban forest resource. J. Forestry Res. 2001, 99, 37–42. [Google Scholar]
- Konijnendijk, C.C.; Ricard, R.M.; Kenney, A.; Randrup, T.B. Defining urban forestry–A comparative perspective of North America and Europe. Urban For. Urban Gree. 2006, 4, 93–103. [Google Scholar] [CrossRef]
- Nowak, D.J.; Walton, J.T. Projected urban growth (2000–2050) and its estimated impact on the US forest resource. J. Forest. 2005, 103, 383–389. [Google Scholar]
- Colunga-Garcia, M.; Magarey, R.A.; Haack, R.A.; Gage, S.H.; Qi, J. Enhancing early detection of exotic pests in agricultural and forest ecosystems using an urban-gradient framework. Ecol. Appl. 2010, 20, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Simberloff, D. The role of propagule pressure in biological invasions. Ann. Rev. Ecol. Evol. Syst. 2009, 40, 81–102. [Google Scholar] [CrossRef]
- Pautasso, M.; Schlegel, M.; Holdenrieder, O. Forest health in a changing world. Microb. Ecol. 2015, 69, 826–842. [Google Scholar] [CrossRef]
- Paap, T.; Burgess, T.I.; Wingfield, M.J. Urban trees: Bridge-heads for forest pest invasions and sentinels for early detection. Biol. Invasions 2017, 19, 3515–3526. [Google Scholar] [CrossRef]
- Park, J.; Park, B.; Veeraraghavan, N.; Jung, K.; Lee, Y.-H.; Blair, J.E.; Geiser, D.M.; Isard, S.; Mansfield, M.A.; Nikolaeva, E. Phytophthora database: A forensic database supporting the identification and monitoring of Phytophthora. Plant Dis. 2008, 92, 966–972. [Google Scholar] [CrossRef]
- Freer-Smith, P.; El-Khatib, A.; Taylor, G. Capture of particulate pollution by trees: A comparison of species typical of semi-arid areas (Ficus nitida and Eucalyptus globulus) with European and North American species. Water Air Soil Poll. 2004, 155, 173–187. [Google Scholar] [CrossRef]
- Tubby, K.; Webber, J. Pests and diseases threatening urban trees under a changing climate. Forestry 2010, 83, 451–459. [Google Scholar] [CrossRef]
- Niinemets, Ü.; Valladares, F. Tolerance to shade, drought, and waterlogging of temperate Northern Hemisphere trees and shrubs. Ecol. Monog. 2006, 76, 521–547. [Google Scholar] [CrossRef]
- Lloret, F.; Keeling, E.G.; Sala, A. Components of tree resilience: Effects of successive low-growth episodes in old ponderosa pine forests. Oikos 2011, 120, 1909–1920. [Google Scholar] [CrossRef]
- Hulbert, J.M.; Agne, M.C.; Burgess, T.I.; Roets, F.; Wingfield, M.J. Urban environments provide opportunities for early detections of Phytophthora invasions. Biol. Invasions 2017, 19, 3629–3644. [Google Scholar] [CrossRef]
- Riddell, C.E.; Frederickson-Matika, D.; Armstrong, A.C.; Elliot, M.; Forster, J.; Hedley, P.E.; Morris, J.; Thorpe, P.; Cooke, D.E.L.; Pritchard, L.; et al. Metabarcoding reveals a high diversity of woody host-associated Phytophthora spp. in soils at public gardens and amenity woodlands in Britain. PeerJ 2019, 7, e6931. [Google Scholar] [CrossRef]
- Yang, X.; Tyler, B.M.; Hong, C. An expanded phylogeny for the genus Phytophthora. IMA Fungus 2017, 8, 355–384. [Google Scholar] [CrossRef]
- Khaliq, I.; Hardy, G.E.S.J.; White, D.; Burgess, T.I. eDNA from roots: A robust tool for determining Phytophthora communities in natural ecosystems. FEMS Microb. Ecol. 2018, 94, fiy048. [Google Scholar] [CrossRef]
- Burgess, T.I.; White, D.; McDougall, K.L.; Garnas, J.; Dunstan, W.A.; Català, S.; Carnegie, A.J.; Worboys, S.; Cahill, D.; Vettraino, A.-M.; et al. Distribution and diversity of Phytophthora across Australia. Pacific Conserv. Biol. 2017, 23, 150–162. [Google Scholar] [CrossRef]
- Català, S.; Pérez-Sierra, A.; Abad-Campos, P. The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in northern Spain. PLoS ONE 2015, 10, e0119311. [Google Scholar]
- Bose, T.; Wingfield, M.J.; Roux, J.; Vivas, M.; Burgess, T.I. Community composition and distribution of Phytophthora species across adjacent native and non-native forests of South Africa. Fungal Ecol. 2018, 36, 17–25. [Google Scholar] [CrossRef]
- Hopper, S.D.; Gioia, P. The southwest Australian floristic region: Evolution and conservation of a global hot spot of biodiversity. Ann. Rev. Ecol. Evol. Syst. 2004, 35, 623–650. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Barber, P.A.; Harper, R.; Linh, T.V.K.; Dell, B. Vegetation trends associated with urban development: The role of golf courses. PLoS ONE 2020, 15, e0228090. [Google Scholar] [CrossRef]
- Gillis, T. Use of Remotely Sensed Imagery to Map Sudden Oak Death (Phytophthora ramorum) in the Santa Cruz Mountains. Master’s Thesis, University of Southern California, Los Angeles, CA, USA, 2014; p. 125. [Google Scholar]
- Cardillo, E.; Acedo, A.; Abad, E. Topographic effects on dispersal patterns of Phytophthora cinnamomi at a stand scale in a Spanish heathland. PLoS ONE 2018, 13, e0195060. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Wilson, B.; Rookes, J.; Cahill, D. Use of high resolution digital multi-spectral imagery to assess the distribution of disease caused by Phytophthora cinnamomi on heathland at Anglesea, Victoria. Australas. Plant Pathol. 2009, 38, 110–119. [Google Scholar] [CrossRef]
- Rowe, D.; Tille, P.; Kuswardiyanto, K. Land Capability Assessment for Expanding Irrigated Horticulture around Myalup; Department of Primary Industries and Regional Development: Perth, Australia, 2017. Available online: https://researchlibrary.agric.wa.gov.au/rmtr/389/ (accessed on 28 June 2020).
- Bessell-Browne, J. Kings Park Soil Survey; Department of Agriculture and Food: Kensington, Australia, 1990. Available online: https://researchlibrary.agric.wa.gov.au/rmtr/93/ (accessed on 28 June 2020).
- Evans, B.; Lyons, T.J.; Barber, P.A.; Stone, C.; Hardy, G.E.S.J. Dieback classification modelling using high resolution digital multi spectral imagery and in situ assessments of crown condition. Remote Sens. Lett. 2012, 3, 541–550. [Google Scholar] [CrossRef]
- Evans, B.; Lyons, T.J.; Barber, P.A.; Stone, C.; Hardy, G.E.S.J. Enhancing a eucalypt crown condition indicator driven by high spatial and spectral resolution remote sensing imagery. J. Appl. Remote Sens. 2012, 6, 063605. [Google Scholar] [CrossRef]
- Simamora, A.; Paap, T.; Howard, K.; Stukely, M.J.C.; Hardy, G.E.S.; Burgess, T.I. Phytophthora contamination in a nursery and its potential dispersal into the natural environment. Plant Dis. 2018, 102, 132–139. [Google Scholar] [CrossRef]
- Jeffers, S.; Aldwinckle, H. Enhancing detection of Phytophthora catorum in naturally infested soil. Phytopathology 1987, 77, 1475–1482. [Google Scholar] [CrossRef]
- Community Ecology Package. Version 2.5–6. Available online: https://rdrr.io/cran/vegan/ (accessed on 28 June 2020).
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. cooccur: Probabilistic species co-occurrence analysis in R. J. Stat. Softw. 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Brasier, C.M. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008, 57, 792–808. [Google Scholar] [CrossRef]
- Lombaert, E.; Guillemaud, T.; Cornuet, J.-M.; Malausa, T.; Facon, B.; Estoup, A. Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS ONE 2010, 5, e9743. [Google Scholar] [CrossRef]
- Blackburn, T.M.; Pyšek, P.; Bacher, S.; Carlton, J.T.; Duncan, R.P.; Jarošík, V.; Wilson, J.R.; Richardson, D.M. A proposed unified framework for biological invasions. Trends Ecol. Evol. 2011, 26, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Prigigallo, M.I.; Mosca, S.; Cacciola, S.O.; Cooke, D.E.L.; Schena, L. Molecular analysis of Phytophthora diversity in nursery-grown ornamental and fruit plants. Plant Pathol. 2015, 64, 1308–1319. [Google Scholar] [CrossRef]
- Burgess, T.I.; Webster, J.L.; Ciampini, J.A.; White, D.; Hardy, G.E.S.; Stukely, M.J.C. Re-evaluation of Phytophthora species isolated during 30 years of vegetation health surveys in Western Australia using molecular techniques. Plant Dis. 2009, 93, 215–223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scott, P.M.; Burgess, T.I.; Barber, P.A.; Shearer, B.L.; Stukely, M.J.C.; Hardy, G.E.S.; Jung, T. Phytophthora multivora sp. nov., a new species recovered from declining Eucalyptus, Banksia, Agonis and other plant species in Western Australia. Persoonia 2009, 22, 1–13. [Google Scholar] [CrossRef]
- Rea, A.; Jung, T.; Burgess, T.I.; Stukely, M.J.C.; Hardy, G.E.S. Phytophthora elongata sp. nov. a novel pathogen from the Eucalyptus marginata forest of Western Australia. Australas. Plant Pathol. 2010, 39, 477–491. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z.; Shivas, R.G.; Edwards, J.; Seifert, K.A.; Alfenas, A.C.; Alfenas, R.F.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.S.; et al. Fungal Planet description sheets: 69–91. Persoonia 2011, 26, 108–156. [Google Scholar] [CrossRef]
- Jung, T.; Stukely, M.J.C.; Hardy, G.E.S.; White, D.; Paap, T.; Dunstan, W.A.; Burgess, T.I. Multiple new Phytophthora species from ITS clade 6 associated with natural ecosystems in Australia: Evolutionary and ecological implications. Persoonia 2011, 26, 13–39. [Google Scholar] [CrossRef]
- Rea, A.; Burgess, T.I.; Hardy, G.E.S.; Stukely, M.J.C.; Jung, T. Two novel species of Phytophthora associated with episodic dieback of kwongan vegetation of south-west Western Australia. Plant Pathol. 2011, 60, 1055–1068. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Shivas, R.G.; Burgess, T.I.; Decock, C.A.; Dreyer, L.L.; Granke, L.L.; Guest, D.I.; Hardy, G.E.S.; Hausbeck, M.K.; et al. Fungal Planet description sheets: 107–127. Persoonia 2012, 28, 138–182. [Google Scholar] [CrossRef]
- Aghighi, S.; Fontanini, L.; Yeoh, P.; Hardy, G.E.S.; Burgess, T.I.; Scott, J.K. A conceptual model to describe the decline of European blackberry (Rubus anglocandicans), a weed of national significance in Australia. Plant Dis. 2014, 98, 580–589. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Schumacher, R.K.; Summerell, B.A.; Giraldo, A.; Gené, J.; Guarro, J.; Wanasinghe, D.N.; Hyde, K.D.; Camporesi, E.; et al. Fungal Planet Description Sheets: 281–319. Persoonia 2014, 33, 212–289. [Google Scholar] [CrossRef] [PubMed]
- Safaiefarahani, B.; Mostowfizadeh-Ghalamfarsa, R.; Hardy, G.E.S.; Burgess, T.I. Re-evaluation of the Phytophthora cryptogea species complex and the description of a new species, Phytophthora pseudocryptogea sp. nov. Mycol. Prog. 2015, 14, 1–12. [Google Scholar] [CrossRef]
- Simamora, A.; Stukely, M.J.C.; Hardy, G.E.S.; Burgess, T.I. Phytophthora boodjera sp. nov., a damping-off pathogen in production nurseries and from urban and natural landscapes, with an update on the status of P. alticola. IMA Fungus 2015, 6, 319–335. [Google Scholar] [CrossRef]
- Paap, T.; Croeser, L.; White, D.; Aghighi, S.; Barber, P.A.; Hardy, G.E.S.; Burgess, T.I. Phytophthora versiformis sp nov., a new species from Australia related to P. quercina. Australas. Plant Pathol. 2017, 46, 369–378. [Google Scholar] [CrossRef]
- Burgess, T.I.; Simamora, A.; White, D.; Williams, B.; Schwager, M.; Stukely, M.J.C.; Hardy, G.E.S.J. New species from Phytophthora Clade 6a: Evidence for recent radiation. Persoonia 2018, 41, 1–17. [Google Scholar] [CrossRef]
- Davison, E.M.; Drenth, A.; Kumar, S.; Mack, S.; Mackie, A.E.; McKirdy, S. Pathogens associated with nursery plants imported into Western Australia. Australas. Plant Pathol. 2006, 35, 473–475. [Google Scholar] [CrossRef]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; APS Press: St. Paul, MN, USA, 1996; p. 562. [Google Scholar]
- Barber, P.A.; Paap, T.; Burgess, T.I.; Dunstan, W.A.; Hardy, G.E.S. A diverse range of Phytophthora species are associated with dying urban trees in an Australian capital city. Urban For. Urban Gree. 2013, 12, 569–575. [Google Scholar] [CrossRef]
- Hüberli, D.; Hardy, G.E.S.; White, D.; Williams, N.; Burgess, T.I. Fishing for Phytophthora from Western Australia’s waterways: A distribution and diversity survey. Australas. Plant Pathol. 2013, 42, 251–260. [Google Scholar] [CrossRef]
- Hickman, C.J. The red core root disease of the strawberry caused by Phytophthora fragariae n. sp. J. Pomol. Hort. Sci. 1941, 18, 89–118. [Google Scholar]
- Themann, K.; Werres, S.; Lüttmann, R.; Diener, H.-A. Observations of Phytophthora spp. in water recirculation systems in commercial hardy ornamental nursery stock. Eur. J. Plant Pathol. 2002, 108, 337–343. [Google Scholar] [CrossRef]
- Jung, T.; Chang, T.T.; Bakonyi, J.; Seress, D.; Pérez-Sierra, A.; Yang, X.; Hong, C.; Scanu, B.; Fu, C.H.; Hsueh, K.L.; et al. Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms. Plant Pathol. 2016, 66, 194–211. [Google Scholar] [CrossRef]
- Eschen, R.; Britton, K.; Brockerhoff, E.; Burgess, T.; Dalley, V.; Epanchin-Niell, R.S.; Gupta, K.; Hardy, G.E.S.; Huang, Y.; Kenis, M.; et al. International variation in phytosanitary legislation and regulations governing importation of live plants. Environ. Sci. Policy 2015, 51, 228–237. [Google Scholar] [CrossRef]
- ArborCarbon. Pathogen Sampling & Mapping Project; Final Report; Prepared for City of Joondalup, June 2016; ArborCarbon: Murdoch, Australia, 2016; p. 83. [Google Scholar]
- Burgess, T.I.; McDougall, K.L.; Scott, P.; Hardy, G.E.S.J.; Garnas, J. Predictors of Phytophthora diversity and distribution in natural areas across diverse Australian ecoregions. Ecography 2019, 42, 594–607. [Google Scholar] [CrossRef]
- Burgess, T.I. Molecular characterization of natural hybrids formed between five related indigenous clade 6 Phytophthora species. PLoS ONE 2015, 10, e0134225. [Google Scholar] [CrossRef]
- Oh, E.; Gryzenhout, M.; Wingfield, B.D.; Wingfield, M.J.; Burgess, T.I. Surveys of soil and water reveal a goldmine of Phytophthora diversity in South African natural ecosystems. IMA Fungus 2013, 4, 123–131. [Google Scholar] [CrossRef]
- Panabieres, F.; Ali, G.S.; Allagui, M.B.; Dalio, R.J.; Gudmestad, N.C.; Kuhn, M.L.; Guha Roy, S.; Schena, L.; Zampounis, A. Phytophthora nicotianae diseases worldwide: New knowledge of a long-recognised pathogen. Phytopathol. Mediterr. 2016, 55, 20–40. [Google Scholar]
- Lowe, S.; Browne, M.; Boudjelas, S.; De Poortner, M. One-hundred of the world’s worst invasive alien species. In A Selection from the Global Invasive Species Database; The Invasive Species Specialist Group, International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2000. [Google Scholar]
- Burgess, T.I.; Scott, J.K.; McDougall, K.L.; Stukely, M.J.C.; Crane, C.; Dunstan, W.A.; Brigg, F.; Andjic, V.; White, D.; Rudman, T.; et al. Current and projected global distribution of Phytophthora cinnamomi, one of the world’s worst plant pathogens. Glob. Chang. Biol. 2017, 23, 1661–1674. [Google Scholar] [CrossRef] [PubMed]
- Migliorini, D.; Khdiar, M.Y.; Rodríguez Padrón, C.; Vivas, M.; Barber, P.A.; Hardy, G.E.S.; Burgess, T.I. Extending the host range of Phytophthora multivora, a pathogen of woody plants in horticulture, nurseries, urban environments and natural ecosystems. Urban For. Urban Gree. 2019, 46, e12646. [Google Scholar] [CrossRef]
- Nesbitt, H.J.; Malajczuk, N.; Glenn, A.R. Effect of organic matteron the survival of Phytophthora cinnamomi Rands in soil. Soil Biol. Biochem. 1979, 11, 133–136. [Google Scholar] [CrossRef]
- McCarren, K.L.; McComb, J.A.; Shearer, B.L.; Hardy, G.E.S. The role of chlamydospores of Phytophthora cinnamomi—A review. Australas. Plant Pathol. 2005, 34, 333–338. [Google Scholar] [CrossRef]
- Redondo, M.A.; Boberg, J.; Stenlid, J.; Oliva, J. Functional traits associated with the establishment of introduced Phytophthora spp. in Swedish forests. J. Appl. Ecol. 2018, 55, 1538–1552. [Google Scholar] [CrossRef]
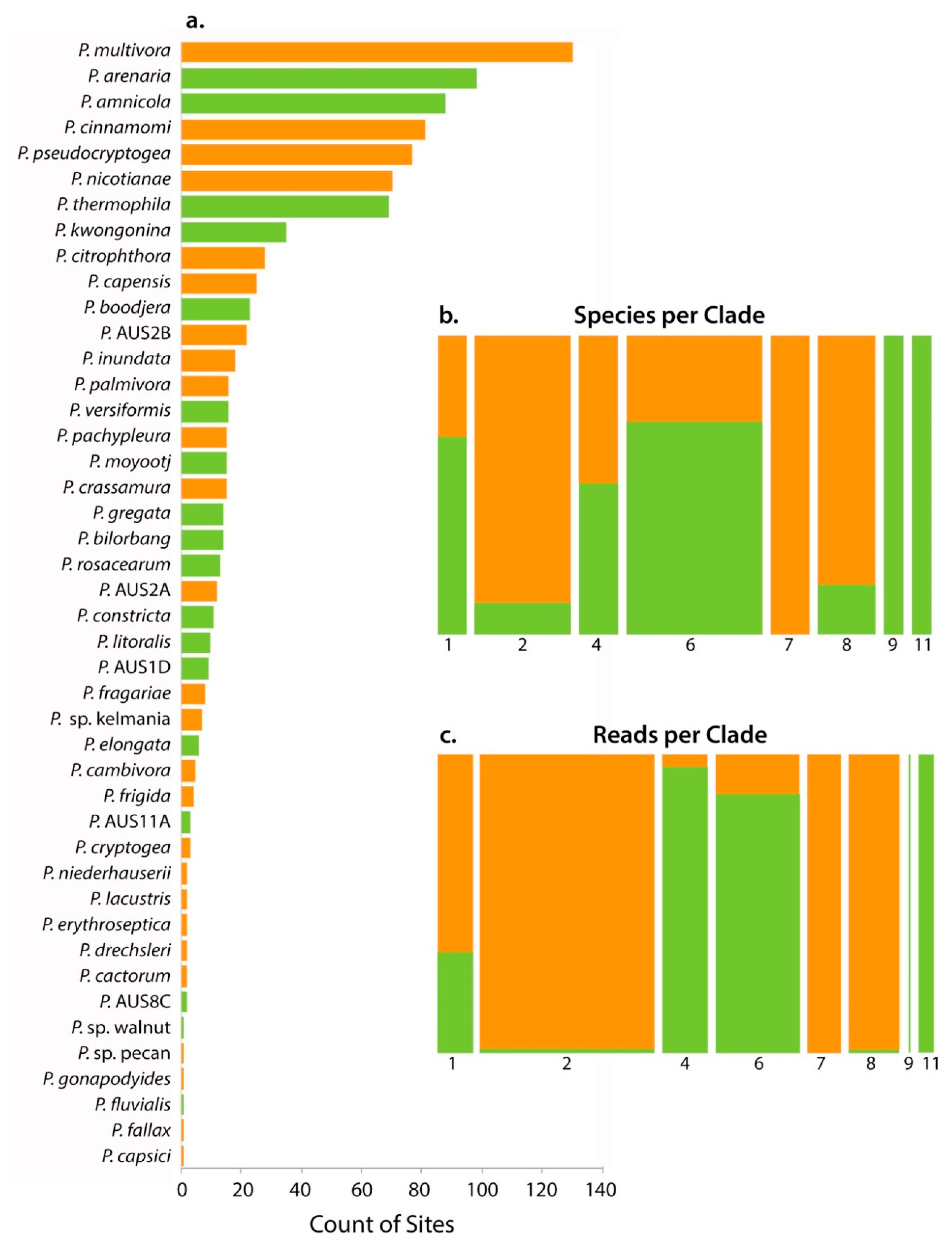

| Factor | Source | Description |
|---|---|---|
| Park Class | City of Joondalup | GIS layer units; open space—mainly sport parks (25), conservation areas (117), parks (32) and thoroughfares (4). See Figure 1. |
| Soil Family | Department of Primary Industries and Regional Development | DAFWA-033 Soil Landscape Mapping WA—best available; LS1 and S7 (24), Quindalup (46), Karrakatta (88) and Spearwood (10) soils. |
| Park Area | City of Joondalup | Internal GIS layers. Area calculated from polygon layers. Ranged in area from 0.5 to 108 ha. Large park area was deemed to be those sites with area >0.5 SD above the mean, medium ≤0.5 SD from the mean, and small at >0.5 SD below the mean. |
| Canopy cover | digital multispectral imagery | Standard deviation (SD) of plant cell density (PCD = IR/Red) from pixels within 5 m of the sample point, ranged from 0.005 to 170 ha. High canopy cover was deemed to be those sites with canopy cover >0.5 SD above the mean, medium ≤0.5 SD from the mean, and small at >0.5 SD below the mean. |
| Canopy health | digital multispectral imagery | SD of Red Edge Extrema Index (REEI = NIR/Red) from pixels of the sample point. The images were acquired in 2012 and 2015, ranging from −17.4 to + 19.5. A -ve value represents a decrease in canopy health. |
| Phytophthora Species | Clade | No. of Parks | No. of Samples | Reads | Rarefied Read No. | Baiting 1 | First Record 2 | Status 3 |
|---|---|---|---|---|---|---|---|---|
| P. cactorum | 1 | 2 | 2 | 165 | 13 | 2014 | I | |
| P. nicotianae | 1 | 42 | 70 | 8262 | 550 | 5 (4) | 2004 | I |
| P. AUS1D 4 | 1 | 7 | 9 | 7667 | 287 | N | ||
| P. AUS2A 4,5 | 2 | 11 | 12 | 620 | 33 | I | ||
| P. AUS2B 4,5 | 2 | 16 | 22 | 8912 | 303 | I | ||
| P. capensis4 | 2 | 14 | 25 | 6359 | 251 | I | ||
| P. capsici4 | 2 | 1 | 1 | 92 | 12 | I | ||
| P. citrophthora | 2 | 18 | 28 | 3113 | 169 | 2015 | I | |
| P. elongata | 2 | 6 | 6 | 105 | 2 | N | ||
| P. frigida4 | 2 | 4 | 4 | 43 | 10 | I | ||
| P. multivora | 2 | 52 | 130 | 63,649 | 3251 | 15 (11) | 1985 | I |
| P. pachypleura4 | 2 | 13 | 15 | 2268 | 83 | I | ||
| P. arenaria | 4 | 54 | 98 | 13,221 | 783 | 3 (2) | N | |
| P. boodjera | 4 | 19 | 23 | 4049 | 263 | 1 (1) | 2011 | N |
| P. palmivora | 4 | 14 | 16 | 461 | 41 | 2011 | I | |
| P. sp. pecan 4,5 | 4 | 1 | 1 | 2 | 0 | I | ||
| P. amnicola | 6 | 46 | 88 | 12,384 | 750 | N | ||
| P. bilorbang | 6 | 11 | 14 | 962 | 81 | N | ||
| P. crassamura | 6 | 11 | 15 | 6225 | 224 | I | ||
| P. fluvialis | 6 | 1 | 1 | 68 | 5 | N | ||
| P. gonapodyides4,5 | 6 | 1 | 1 | 9 | 1 | I | ||
| P. gregata | 6 | 11 | 14 | 300 | 31 | 2015 | N | |
| P. inundata | 6 | 12 | 18 | 405 | 30 | 2011 | I | |
| P. kwongonina | 6 | 25 | 35 | 4006 | 188 | 2010 | N | |
| P. lacustris | 6 | 2 | 2 | 19 | 3 | 1995 | I | |
| P. litoralis | 6 | 6 | 10 | 852 | 35 | 2011 | N | |
| P. moyootj | 6 | 11 | 15 | 2602 | 132 | N | ||
| P. rosacearum | 6 | 9 | 13 | 1332 | 67 | 2015 | N? | |
| P. sp. walnut 4,5 | 6 | 1 | 1 | 32 | 2 | N? | ||
| P. thermophila | 6 | 39 | 69 | 6667 | 432 | 1995 | N | |
| P. cambivora | 7 | 5 | 5 | 1329 | 67 | I | ||
| P. cinnamomi | 7 | 40 | 81 | 11,623 | 707 | ∝1980 | I | |
| P. fragariae4,5 | 7 | 6 | 8 | 342 | 14 | I | ||
| P. niederhauserii | 7 | 2 | 2 | 276 | 20 | 2012 | I | |
| P. cryptogea | 8 | 3 | 3 | 189 | 12 | 2015 | I | |
| P. drechsleri4 | 8 | 2 | 2 | 27 | 0 | I | ||
| P. erythroseptica | 8 | 2 | 2 | 19 | 2 | I | ||
| P. pseudocryptogea | 8 | 39 | 77 | 14,329 | 924 | 2016 | I | |
| P. sp. kelmania | 8 | 5 | 7 | 1832 | 232 | 2016 | I | |
| P. AUS8C 4,5 | 8 | 1 | 2 | 162 | 10 | N? | ||
| P. constricta | 9 | 7 | 11 | 500 | 20 | N | ||
| P. fallax4 | 9 | 1 | 1 | 26 | 3 | N? | ||
| P. versiformis | 11 | 15 | 16 | 6119 | 298 | 2011 | N | |
| P. AUS11A 4 | 11 | 3 | 3 | 2126 | 65 | N | ||
| Total | 193,747 | 10,416 | 24 (18) |
| Presence | Abundance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor | df | SS | MS | F-Value | R2 | P-Value | SS | MS | F-Value | R2 | P-Value |
| Global | |||||||||||
| Park Area | 1 | 1.64 | 1.64 | 5.51 | 0.03 | 0.0001 | 1.52 | 1.52 | 4.71 | 0.03 | 0.0001 |
| Park Class | 3 | 1.06 | 0.35 | 1.19 | 0.02 | 0.1809 | 0.94 | 0.31 | 0.97 | 0.02 | 0.5009 |
| Canopy Cover | 1 | 0.90 | 0.90 | 3.01 | 0.02 | 0.0001 | 1.51 | 1.51 | 4.68 | 0.03 | 0.0001 |
| Canopy Health | 1 | 0.35 | 0.35 | 1.16 | 0.01 | 0.2890 | 0.40 | 0.40 | 1.25 | 0.01 | 0.2256 |
| Soil Family | 3 | 1.33 | 0.44 | 1.48 | 0.03 | 0.0204 | 1.35 | 0.45 | 1.39 | 0.02 | 0.0532 |
| Residuals | 158 | 47.04 | 0.30 | 0.89 | 51.08 | 0.89 | |||||
| Total | 167 | 52.31 | 1.00 | 56.81 | 1.00 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khdiar, M.Y.; Barber, P.A.; Hardy, G.E.S.; Shaw, C.; Steel, E.J.; McMains, C.; Burgess, T.I. Association of Phytophthora with Declining Vegetation in an Urban Forest Environment. Microorganisms 2020, 8, 973. https://doi.org/10.3390/microorganisms8070973
Khdiar MY, Barber PA, Hardy GES, Shaw C, Steel EJ, McMains C, Burgess TI. Association of Phytophthora with Declining Vegetation in an Urban Forest Environment. Microorganisms. 2020; 8(7):973. https://doi.org/10.3390/microorganisms8070973
Chicago/Turabian StyleKhdiar, Mohammed Y., Paul A. Barber, Giles E. StJ. Hardy, Chris Shaw, Emma J. Steel, Cameron McMains, and Treena I. Burgess. 2020. "Association of Phytophthora with Declining Vegetation in an Urban Forest Environment" Microorganisms 8, no. 7: 973. https://doi.org/10.3390/microorganisms8070973
APA StyleKhdiar, M. Y., Barber, P. A., Hardy, G. E. S., Shaw, C., Steel, E. J., McMains, C., & Burgess, T. I. (2020). Association of Phytophthora with Declining Vegetation in an Urban Forest Environment. Microorganisms, 8(7), 973. https://doi.org/10.3390/microorganisms8070973