Growth Performance and Adaptability of European Sea Bass (Dicentrarchus labrax) Gut Microbiota to Alternative Diets Free of Fish Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Feed Formulation and Feeding
2.3. Fish and Experimental Conditions
2.4. Digestibility Methods
2.5. Sampling of Gastrointestinal Contents
2.6. DNA Extraction and 16S rRNA Gene Sequencing and Analyses
2.7. Bioinformatic and Biostatistical Analyses of Microbiota Datasets
2.8. Statistical Analyses of Body Weight Measures
3. Results and Discussion
3.1. Changing Protein Sources in Sea Bass Diet Influences Growth Performance and Feed Efficiency
3.2. Changes of Protein Sources in Fish Diet do not Significantly Alter Gut Microbiota Diversity
3.3. Sea Bass Microbiota Composition Daisplays High Diet-independent Inter-individual Variability
3.4. Diet does not Influence Microbiota Compositional Diversity
3.5. Growth does not Correlate with Diet-dependent Changes in Microbiota Composition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020. Sustainability in Action; Food and Agriculture Organization of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Stickney, R.R. Aquaculture: An Introductory Text, 3rd ed.; CABI: Wallingford, UK; Oxfordshire, UK; Boston, MA, USA, 2017; Volume xiii, 337p. [Google Scholar]
- Gatlin, D.M., III; Barrows, F.T.; Brown, P.; Dabrowski, K.; Gaylord, T.G.; Hardy, R.W.; Herman, E.; Hu, G.; Krogdahl, Å.; Nelson, R.; et al. Expanding the utilization of sustainable plant products in aquafeeds: A review. Aquac. Res. 2007, 38, 551–579. [Google Scholar] [CrossRef]
- Parma, L.; Yúfera, M.; Navarro-Guillén, C.; Moyano, F.J.; Soverini, M.; D’Amico, F.; Candela, M.; Fontanillas, R.; Gatta, P.P.; Bonaldo, A. Effects of calcium carbonate inclusion in low fishmeal diets on growth, gastrointestinal pH, digestive enzyme activity and gut bacterial community of European sea bass (Dicentrarchus labrax L.) juveniles. Aquaculture 2019, 510, 283–292. [Google Scholar] [CrossRef]
- Naylor, R.L.; Hardy, R.W.; Bureau, D.P.; Chiu, A.; Elliott, M.; Farrell, A.P.; Forster, I.; Gatlin, D.M.; Goldburg, R.J.; Hua, K.; et al. Feeding aquaculture in an era of finite resources. Proc. Natl. Acad. Sci. USA 2009, 106, 15103–15110. [Google Scholar] [CrossRef] [PubMed]
- Little, D.C.; Newton, R.W.; Beveridge, M.C.M. Aquaculture: A rapidly growing and significant source of sustainable food? Status, transitions and potential. Proc. Nutr. Soc. 2016, 75, 274–286. [Google Scholar] [CrossRef]
- Caruso, G. Use of Plant Products as Candidate Fish Meal Substitutes: An Emerging Issue in Aquaculture Productions. Fish. Aquac. J. 2015, 6, e123. [Google Scholar]
- Pelletier, N.; Klinger, D.H.; Sims, N.A.; Yoshioka, J.-R.; Kittinger, J.N. Nutritional Attributes, Substitutability, Scalability, and Environmental Intensity of an Illustrative Subset of Current and Future Protein Sources for Aquaculture Feeds: Joint Consideration of Potential Synergies and Trade-offs. Environ. Sci. Technol. 2018, 52, 5532–5544. [Google Scholar] [CrossRef]
- Gasco, L.; Biasato, I.; Dabbou, S.; Schiavone, A.; Gai, F. Animals Fed Insect-Based Diets: State-of-the-Art on Digestibility, Performance and Product Quality. Animals 2019, 9, 170. [Google Scholar] [CrossRef]
- Ringø, E.; Zhou, Z.; Vecino, J.L.G.; Wadsworth, S.; Romero, J.; Krogdahl, Å.; Olsen, R.E.; Dimitroglou, A.; Foey, A.; Davies, S.; et al. Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac. Nutr. 2016, 22, 219–282. [Google Scholar] [CrossRef]
- Vandeputte, M.; Gagnaire, P.-A.; Allal, F. The European sea bass: A key marine fish model in the wild and in aquaculture. Anim. Genet. 2019, 50, 195–206. [Google Scholar] [CrossRef]
- Kaushik, S.J.; Covès, D.; Dutto, G.; Blanc, D. Almost total replacement of fish meal by plant protein sources in the diet of a marine teleost, the European seabass, Dicentrarchus labrax. Aquaculture 2004, 230, 391–404. [Google Scholar] [CrossRef]
- Bonvini, E.; Bonaldo, A.; Mandrioli, L.; Sirri, R.; Dondi, F.; Bianco, C.; Fontanillas, R.; Mongile, F.; Gatta, P.P.; Parma, L. Effects of feeding low fishmeal diets with increasing soybean meal levels on growth, gut histology and plasma biochemistry of sea bass. Animal 2018, 12, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Tulli, F.; Vachot, C.; Tibaldi, E.; Fournier, V.; Kaushik, S.J. Contribution of dietary arginine to nitrogen utilisation and excretion in juvenile sea bass (Dicentrarchus labrax) fed diets differing in protein source. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 147, 179–188. [Google Scholar] [CrossRef]
- Geay, F.; Ferraresso, S.; Zambonino-Infante, J.L.; Bargelloni, L.; Quentel, C.; Vandeputte, M.; Kaushik, S.; Cahu, C.; Mazurais, D. Effects of the total replacement of fish-based diet with plant-based diet on the hepatic transcriptome of two European sea bass (Dicentrarchus labrax) half-sibfamilies showing different growth rates with the plant-based diet. BMC Genom. 2011, 12, 522. [Google Scholar] [CrossRef]
- Torrecillas, S.; Mompel, D.; Caballero, M.J.; Montero, D.; Merrifield, D.; Rodiles, A.; Robaina, L.; Zamorano, M.; Karalazos, V.; Kaushik, S.; et al. Effect of fishmeal and fish oil replacement by vegetable meals and oils on gut health of European sea bass (Dicentrarchus labrax). Aquaculture 2017, 468, 386–398. [Google Scholar] [CrossRef]
- Bonaldo, A.; Roem, A.J.; Fagioli, P.; Pecchini, A.; Cipollini, I.; Gatta, P.P. Influence of dietary levels of soybean meal on the performance and gut histology of gilthead sea bream (Sparus aurata L.) and European sea bass (Dicentrarchus labrax L.). Aquac. Res. 2008, 39, 970–978. [Google Scholar] [CrossRef]
- Gomes, E.F.; Rema, P.; Kaushik, S.J. Replacement of fish meal by plant proteins in the diet of rainbow trout (Oncorhynchus mykiss): Digestibility and growth performance. Aquaculture 1995, 130, 177–186. [Google Scholar] [CrossRef]
- Gomes, E.F.; Corraze, G.; Kaushik, S. Effects of dietary incorporation of a co-extruded plant protein (rapeseed and peas) on growth, nutrient utilization and muscle fatty acid composition of rainbow trout (Oncorhynchus mykiss). Aquaculture 1993, 113, 339–353. [Google Scholar] [CrossRef]
- Francis, G.; Makkar, H.P.S.; Becker, K. Antinutritional factors present in plant-derived alternate fish feed ingredients and their effects in fish. Aquaculture 2001, 199, 197–227. [Google Scholar] [CrossRef]
- Oliva-Teles, A.; Gonçalves, P. Partial replacement of fishmeal by brewers yeast (Saccaromyces cerevisae) in diets for sea bass (Dicentrarchus labrax) juveniles. Aquaculture 2001, 202, 269–278. [Google Scholar] [CrossRef]
- Tibaldi, E.; Zittelli, G.C.; Parisi, G.; Bruno, M.; Giorgi, G.; Tulli, F.; Venturini, S.; Tredici, M.; Poli, B. Growth performance and quality traits of European sea bass (D. labrax) fed diets including increasing levels of freeze-dried Isochrysis sp. (T-ISO) biomass as a source of protein and n-3 long chain PUFA in partial substitution of fish derivatives. Aquaculture 2015, 440, 60–68. [Google Scholar] [CrossRef]
- Gasco, L.; Henry, M.; Piccolo, G.; Marono, S.; Gai, F.; Renna, M.; Lussiana, C.; Antonopoulou, E.; Mola, P.; Chatzifotis, S. Tenebrio molitor meal in diets for European sea bass (Dicentrarchus labrax L.) juveniles: Growth performance, whole body composition and in vivo apparent digestibility. Anim. Feed Sci. Technol. 2016, 220, 34–45. [Google Scholar] [CrossRef]
- Campos, I.; Matos, E.; Marques, A.; Valente, L.M.P. Hydrolyzed feather meal as a partial fishmeal replacement in diets for European seabass (Dicentrarchus labrax) juveniles. Aquaculture 2017, 476, 152–159. [Google Scholar] [CrossRef]
- Magalhães, R.; Sánchez-López, A.; Leal, R.S.; Martínez-Llorens, S.; Oliva-Teles, A.; Peres, H. Black soldier fly (Hermetia illucens) pre-pupae meal as a fish meal replacement in diets for European seabass (Dicentrarchus labrax). Aquaculture 2017, 476, 79–85. [Google Scholar] [CrossRef]
- Valente, L.M.P.; Custódio, M.; Batista, S.; Fernandes, H.; Kiron, V. Defatted microalgae (Nannochloropsis sp.) from biorefinery as a potential feed protein source to replace fishmeal in European sea bass diets. Fish Physiol. Biochem. 2019, 45, 1067–1081. [Google Scholar] [CrossRef]
- Gatesoupe, F.J.; Huelvan, C.; Le Bayon, N.; Le Delliou, H.; Madec, L.; Mouchel, O.; Quazuguel, P.; Mazurais, D.; Zambonino-Infante, J.-L. The highly variable microbiota associated to intestinal mucosa correlates with growth and hypoxia resistance of sea bass, Dicentrarchus labrax, submitted to different nutritional histories. BMC Microbiol. 2016, 16, 266. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The Gut Microbiota of Marine Fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Llewellyn, M.S.; Boutin, S.; Hoseinifar, S.H.; Derome, N. Teleost microbiomes: The state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 2014, 5, 207. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A.M. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture 2017, 467, 149–157. [Google Scholar] [CrossRef]
- Walter, J.; Ley, R. The Human Gut Microbiome: Ecology and Recent Evolutionary Changes. Annu. Rev. Microbiol. 2011, 65, 411–429. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, A.; Aizpurua, O.; Bohmann, K.; Zepeda-Mendoza, M.L.; Gilbert, M.T.P. Do Vertebrate Gut Metagenomes Confer Rapid Ecological Adaptation? Trends Ecol. Evolut. 2016, 31, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.; Terova, G.; Ascione, C.; Giannico, R.; Brambilla, F. Next generation sequencing for gut microbiome characterization in rainbow trout (Oncorhynchus mykiss) fed animal by-product meals as an alternative to fishmeal protein sources. PLoS ONE 2018, 13, e0193652. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.K. Role of gastrointestinal microbiota in fish. Aquac. Res. 2010, 41, 1553–1573. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Snowberg, L.K.; Hirsch, P.E.; Lauber, C.L.; Knight, R.; Caporaso, J.G.; Svanbäck, R. Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 2014, 17, 979–987. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Fish and Shrimp; The National Academies Press: Washington, DC, USA, 2011; 392p. [Google Scholar]
- Mladineo, I.; Bočina, I.; Przybyla, C.; Fievet, J.; Blancheton, J.-P. Fish growth and health aspects of sea bass (Dicentrarchus labrax) reared in standard vs. high rate algal pond recirculation systems. Aquat. Living Resour. 2010, 23, 217–224. [Google Scholar] [CrossRef]
- Pichavant, K.; Person-Le-Ruyet, J.; Le Bayon, N.; Severe, A.; Le Roux, A.; Boeuf, G. Comparative effects of long-term hypoxia on growth, feeding and oxygen consumption in juvenile turbot and European sea bass. J. Fish Biol. 2001, 59, 875–883. [Google Scholar] [CrossRef]
- Przybyla, C.; Fievet, J.; Callier, M.; Blancheton, J.-P. Effect of dietary water content on European sea bass (Dicentrarchus labrax) growth and disease resistance. Aquat. Living Resour. 2014, 27, 73–81. [Google Scholar] [CrossRef]
- Parma, L.; Candela, M.; Soverini, M.; Turroni, S.; Consolandi, C.; Brigidi, P.; Mandrioli, L.; Sirri, R.; Fontanillas, R.; Gatta, P.P.; et al. Next-generation sequencing characterization of the gut bacterial community of gilthead sea bream (Sparus aurata, L.) fed low fishmeal based diets with increasing soybean meal levels. Anim. Feed Sci. Technol. 2016, 222, 204–216. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rideout, J.R.; He, Y.; Navas-Molina, J.A.; Walters, W.A.; Ursell, L.K.; Gibbons, S.M.; Chase, J.; McDonald, D.; Gonzalez, A.; Robbins-Pianka, A.; et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2014, 2, e545. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial; R Doc 43. 2015. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 2 September 2020).
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw. 2017, 82, 26. [Google Scholar] [CrossRef]
- Lenth, R.V. Least-Squares Means: The R Package lsmeans. J. Stat. Softw. 2016, 69, 33. [Google Scholar] [CrossRef]
- Boujard, T.; Houlihan, D.; Jobling, M. Food Intake in Fish; Blackwell Science: Oxford, MA, USA, 2001. [Google Scholar]
- Huisman, E.A. Food conversion efficiencies at maintenance and production levels for carp, Cyprinus carpio L., and rainbow trout, Salmo gairdneri Richardson. Aquaculture 1976, 9, 259–273. [Google Scholar] [CrossRef]
- Brett, J.R. 10-Environmental Factors and Growth. In Fish Physiology 8; Hoar, W.S., Randall, D.J., Brett, J.R., Eds.; Academic Press: Cambridge, MA, USA, 1979; pp. 599–675. [Google Scholar]
- Olvera-Novoa, M.A.; Dominguez-Cen, L.J.; Olivera-Castillo, L.; Martinez-Palacios, C.A. Effect of the use of the microalga Spirulina maxima as fish meal replacement in diets for tilapia, Oreochromis mossambicus (Peters), fry. Aquac. Res. 1998, 29, 709–715. [Google Scholar] [CrossRef]
- Richmond, A. Spirulina. In Micro-Algual Biotechnology; Borowitzka, M.A., Borowitzka, L.J., Eds.; Cambridge University Press: Cambridge, UK, 1998; pp. 85–121. [Google Scholar]
- Lanari, D.; D’Agaro, E. Alternative plant protein sources in sea bass diets. Ital. J. Anim. Sci. 2005, 4, 365–374. [Google Scholar] [CrossRef]
- Krogdahl, Å.; Penn, M.; Thorsen, J.; Refstie, S.; Bakke, A.M. Important antinutrients in plant feedstuffs for aquaculture: An update on recent findings regarding responses in salmonids. Aquac. Res. 2010, 41, 333–344. [Google Scholar] [CrossRef]
- Nikouli, E.; Meziti, A.; Antonopoulou, E.; Mente, E.; Kormas, K.A. Gut Bacterial Communities in Geographically Distant Populations of Farmed Sea Bream (Sparus aurata) and Sea Bass (Dicentrarchus labrax). Microorganisms 2018, 6, 92. [Google Scholar] [CrossRef]
- Carda-Diéguez, M.; Mira, A.; Fouz, B. Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. FEMS Microbiol. Ecol. 2014, 87, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Kokou, F.; Sasson, G.; Friedman, J.; Eyal, S.; Ovadia, O.; Harpaz, S.; Cnaani, A.; Mizrahi, I. Core gut microbial communities are maintained by beneficial interactions and strain variability in fish. Nat. Microbiol. 2019, 4, 2456–2465. [Google Scholar] [CrossRef]
- Star, B.; Haverkamp, T.H.A.; Jentoft, S.; Jakobsen, K.S. Next generation sequencing shows high variation of the intestinal microbial species composition in Atlantic cod caught at a single location. BMC Microbiol. 2013, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, J.W.; Peterson, B.C.; Swanson, K.S.; Small, B.C. Ontogenetic Characterization of the Intestinal Microbiota of Channel Catfish through 16S rRNA Gene Sequencing Reveals Insights on Temporal Shifts and the Influence of Environmental Microbes. PLoS ONE 2016, 11, e0166379. [Google Scholar] [CrossRef]
- Apper, E.; Weissman, D.; Respondek, F.; Guyonvarch, A.; Baron, F.; Boisot, P.; Rodiles, A.; Merrifield, D. Hydrolysed wheat gluten as part of a diet based on animal and plant proteins supports good growth performance of Asian seabass (Lates calcarifer), without impairing intestinal morphology or microbiota. Aquaculture 2016, 453, 40–48. [Google Scholar] [CrossRef]
- Xiong, J.B.; Nie, L.; Chen, J. Current understanding on the roles of gut microbiota in fish disease and immunity. Zool. Res. 2019, 40, 70–76. [Google Scholar] [CrossRef]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Lokesh, J.; Kiron, V.; Sipkema, D.; Fernandes, J.M.O.; Moum, T. Succession of embryonic and the intestinal bacterial communities of Atlantic salmon (Salmo salar) reveals stage-specific microbial signatures. MicrobiologyOpen 2019, 8, e00672. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Fečkaninová, A.; Lokesh, J.; Koščová, J.; Sørensen, M.; Fernandes, J.; Kiron, V. Lactobacillus Dominate in the Intestine of Atlantic Salmon Fed Dietary Probiotics. Front. Microbiol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Józefiak, A.; Engberg, R.M. Insect proteins as a potential source of antimicrobial peptides in livestock production. A review. J. Anim. Feed Sci. 2017, 26, 87–99. [Google Scholar] [CrossRef]
- Nawaz, A.; Javaid, A.B.; Irshad, S.; Hoseinifar, S.H.; Xiong, H. The functionality of prebiotics as immunostimulant: Evidences from trials on terrestrial and aquatic animals. Fish Shellfish Immunol. 2018, 76, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Carnevali, O.; de Vivo, L.; Sulpizio, R.; Gioacchini, G.; Olivotto, I.; Silvi, S.; Cresci, A. Growth improvement by probiotic in European sea bass juveniles (Dicentrarchus labrax, L.), with particular attention to IGF-1, myostatin and cortisol gene expression. Aquaculture 2006, 258, 430–438. [Google Scholar] [CrossRef]
- Frouël, S.; Le Bihan, E.; Serpentini, A.; Lebel, J.M.; Koueta, N.; Nicolas, J.L. Preliminary Study of the Effects of Commercial Lactobacilli Preparations on Digestive Metabolism of Juvenile Sea Bass (Dicentrarchus labrax). J. Mol. Microbiol. Biotechnol. 2008, 14, 100–106. [Google Scholar] [CrossRef]
- Abelli, L.; Randelli, E.; Carnevali, O.; Picchietti, S. Stimulation of Gut Immune System by Early Administration of Probiotic Strains in Dicentrarchus labrax and Sparus aurata. Ann. N. Y. Acad. Sci. 2009, 1163, 340–342. [Google Scholar] [CrossRef]
- Piccolo, G.; Bovera, F.; Lombardi, P.; Mastellone, V.; Nizza, S.; Di Meo, C.; Marono, S.; Nizza, A. Effect of Lactobacillus plantarum on growth performance and hematological traits of European sea bass (Dicentrarchus labrax). Aquac. Int. 2015, 23, 1025–1032. [Google Scholar] [CrossRef]
- Gatesoupe, F.J. Updating the Importance of Lactic Acid Bacteria in Fish Farming: Natural Occurrence and Probiotic Treatments. J. Mol. Microbiol. Biotechnol. 2008, 14, 107–114. [Google Scholar] [CrossRef]
- Ringø, E.; Hoseinifar, S.H.; Ghosh, K.; Doan, H.V.; Beck, B.R.; Song, S.K. Lactic Acid Bacteria in Finfish—An Update. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Meena, D.K.; Das, P.; Kumar, S.; Mandal, S.C.; Prusty, A.K.; Singh, S.K.; Akhtar, M.S.; Behera, B.K.; Kumar, K.; Pal, A.K.; et al. Beta-glucan: An ideal immunostimulant in aquaculture (a review). Fish Physiol. Biochem. 2013, 39, 431–457. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish, 4th ed.; Springer: Dordrecht, The Netherlands; Chichester, UK, 2007; Volume xxvi, 552p. [Google Scholar]

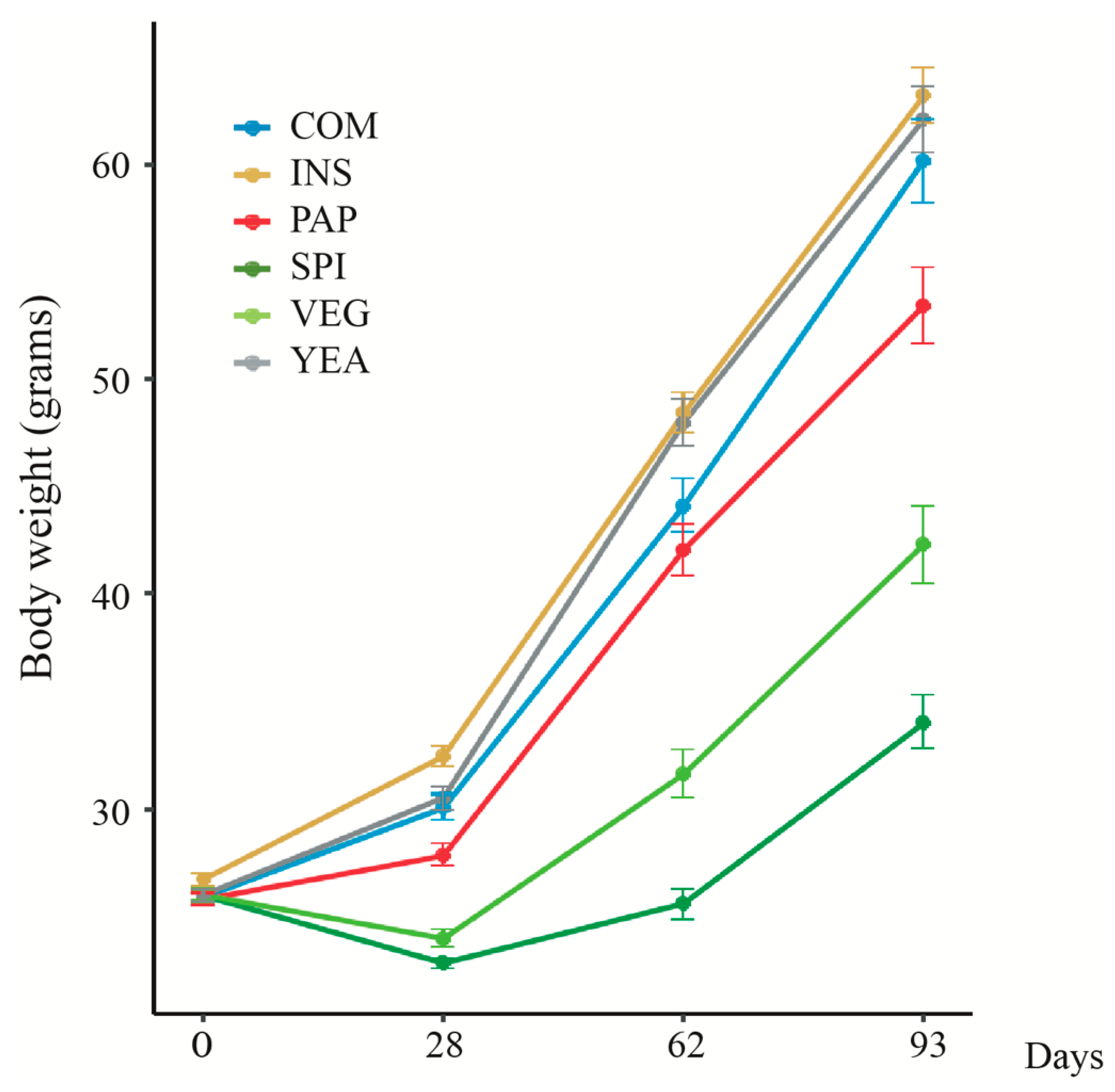


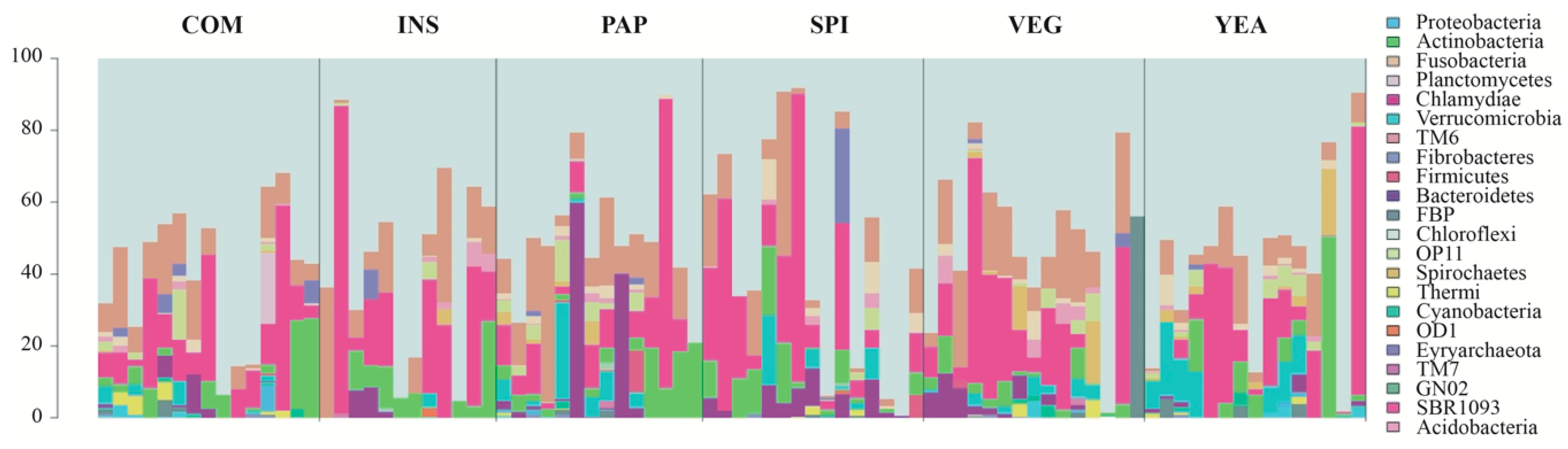
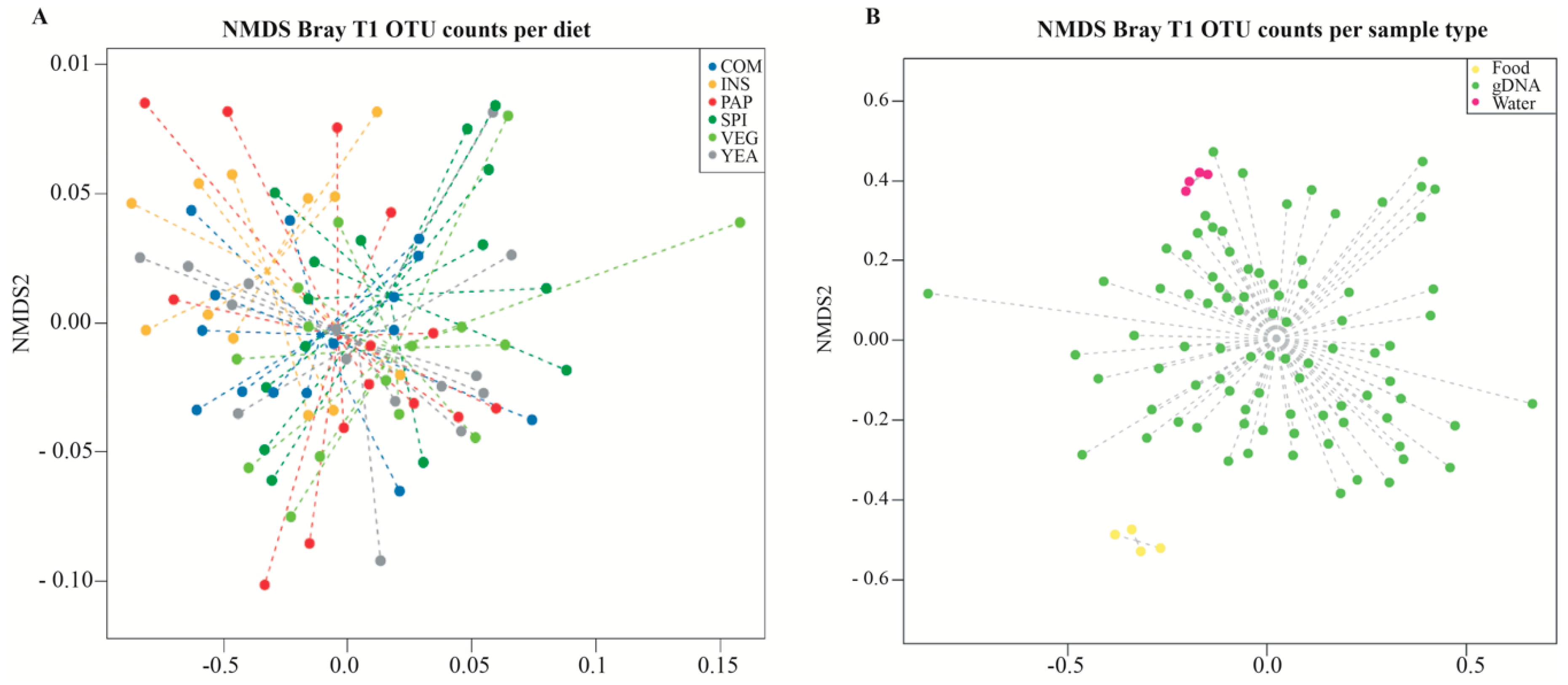
| COM * | VEG | INS | YEA | PAP | SPI | |
|---|---|---|---|---|---|---|
| ADC dry matter (%) | 81.9 | 70.4 ± 1.1 b | 76.9 ± 0.4 a | 73.2 ± 2.0 a,b | 72.5 ± 3.3 a,b | 68.2 ± 2.8 b |
| ADC protein (%) | 93.6 | 92.9 ± 0.9 a | 93.1 ± 0.4 a | 92.6 ± 0.9 a | 90.3 ± 2.6 a | 82.6 ± 1.7 b |
| ADC lipid (%) | 94.8 | 90.4 ± 0.5 a | 91.4 ± 0.4 a | 91.3 ± 0.3 a | 90.6 ± 2.6 a | 87.9 ± 1.5 b |
| ADC energy (%) | 89.4 | 81.1 ± 0.5 b,c | 85.2 ± 0.6 a | 84.6 ± 0.6 a,b | 84 ± 1.7 a,b | 79.7 ± 0.7 c |
| Phylum | Class | Order | Family | INS * | PAP | SPI | YEA | COM |
|---|---|---|---|---|---|---|---|---|
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | Rhodospirillaceae | −8359 | - | −8275 | - | - |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Hyphomicrobiaceae | −8302 | - | - | - | - |
| Planctomycetes | Planctomycetia | Pirellulales | Pirellulaceae | −5084 | - | - | - | - |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | −7244 | −4738 | −5618 | −3665 | - |
| TM6 | SJA-4 | −6680 | - | −8252 | - | - | ||
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | −5239 | - | −3614 | −3442 | - |
| Proteobacteria | Gammaproteobacteria | Legionellales | −5547 | - | −3171 | - | - | |
| Proteobacteria | Gammaproteobacteria | Legionellales | Legionellaceae | −5194 | - | - | - | - |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | −5904 | - | - | - | - |
| OP11 | WCHB1–64 | d153 | −5139 | - | - | - | - | |
| Bacteroidetes | Flavobacteriia | Flavobacteriales | Weeksellaceae | −4257 | - | - | - | - |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | −3516 | - | - | - | - |
| Proteobacteria | Alphaproteobacteria | −3722 | - | - | - | - | ||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | - | 6551 | 7300 | 3892 | - |
| Proteobacteria | Betaproteobacteria | - | 5635 | - | - | - | ||
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | - | - | 4820 | - | - |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | - | - | −3272 | - | - |
| Bacteroidetes | Chitinophagia | Chitinophagales | Chitinophagaceae | - | - | 3831 | 4628 | - |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | - | - | −3652 | - | - |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | - | - | - | −5510 | - |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | - | - | - | −4561 | - |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | - | - | - | −3922 | - |
| Phylum | Class | Order | Family | Genus | INS * | PAP | SPI | YEA | COM |
|---|---|---|---|---|---|---|---|---|---|
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | Rhodospirillaceae | −8298 | - | −8279 | - | - | |
| Proteobacteria | Gammaproteobacteria | Legionellales | Legionellaceae | −7147 | - | −4141 | - | - | |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Sulfitobacter | −7449 | - | −6105 | −3795 | - |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | −6970 | −4782 | −5822 | −3921 | - | |
| TM6 | SJA-4 | −6540 | - | −8506 | - | −4498 | |||
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | −4824 | - | −3505 | −3825 | - | |
| Planctomycetes | Planctomycetia | Pirellulales | Pirellulaceae | −4518 | - | - | - | - | |
| Proteobacteria | Gammaproteobacteria | Legionellales | −4592 | - | - | - | - | ||
| OP11 | WCHB1–64 | d153 | −5099 | - | - | - | - | ||
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Marivita | −5114 | - | - | - | - |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Paracoccus | 4100 | 4611 | - | - | 4942 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobiaceae | Persicirhabdus | - | 3829 | 5959 | 5601 | 6226 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | - | 4085 | 7628 | 3756 | 4152 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | - | 5097 | - | - | −4518 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter | - | - | 3217 | - | 2616 |
| Bacteroidetes | Chitinophagia | Chitinophagales | Chitinophagaceae | Sediminibacterium | - | - | 3647 | 4625 | - |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | - | - | - | −5295 | - |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | - | - | - | - | 7736 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Pascual, D.; Estellé, J.; Dutto, G.; Rodde, C.; Bernardet, J.-F.; Marchand, Y.; Duchaud, E.; Przybyla, C.; Ghigo, J.-M. Growth Performance and Adaptability of European Sea Bass (Dicentrarchus labrax) Gut Microbiota to Alternative Diets Free of Fish Products. Microorganisms 2020, 8, 1346. https://doi.org/10.3390/microorganisms8091346
Pérez-Pascual D, Estellé J, Dutto G, Rodde C, Bernardet J-F, Marchand Y, Duchaud E, Przybyla C, Ghigo J-M. Growth Performance and Adaptability of European Sea Bass (Dicentrarchus labrax) Gut Microbiota to Alternative Diets Free of Fish Products. Microorganisms. 2020; 8(9):1346. https://doi.org/10.3390/microorganisms8091346
Chicago/Turabian StylePérez-Pascual, David, Jordi Estellé, Gilbert Dutto, Charles Rodde, Jean-François Bernardet, Yann Marchand, Eric Duchaud, Cyrille Przybyla, and Jean-Marc Ghigo. 2020. "Growth Performance and Adaptability of European Sea Bass (Dicentrarchus labrax) Gut Microbiota to Alternative Diets Free of Fish Products" Microorganisms 8, no. 9: 1346. https://doi.org/10.3390/microorganisms8091346
APA StylePérez-Pascual, D., Estellé, J., Dutto, G., Rodde, C., Bernardet, J.-F., Marchand, Y., Duchaud, E., Przybyla, C., & Ghigo, J.-M. (2020). Growth Performance and Adaptability of European Sea Bass (Dicentrarchus labrax) Gut Microbiota to Alternative Diets Free of Fish Products. Microorganisms, 8(9), 1346. https://doi.org/10.3390/microorganisms8091346






