Abstract
Streptococcus iniae is a major fish pathogen that contributes to large annual losses in the aquaculture industry, exceeding US$100 million. It is also reported to cause opportunistic infections in humans. We have recently identified two novel S. iniae virulence factors, an extracellular nuclease (SpnAi) and a secreted nucleotidase (S5nAi), and verified their predicted enzymatic activities using recombinant proteins. Here, we report the generation of green fluorescent S. iniae spnAi and s5nAi deletion mutants and their evaluation in a transgenic zebrafish infection model. Our results show nuclease and nucleotidase activities in S. iniae could be attributed to SpnAi and S5nAi, respectively. Consistent with this, larvae infected with the deletion mutants demonstrated enhanced survival and bacterial clearance, compared to those infected with wild-type (WT) S. iniae. Deletion of spnAi and s5nAi resulted in sustained recruitment of neutrophils and macrophages, respectively, to the site of infection. We also show that recombinant SpnAi is able to degrade neutrophil extracellular traps (NETs) isolated from zebrafish kidney tissue. Our results suggest that both enzymes play an important role in S. iniae immune evasion and might present potential targets for the development of therapeutic agents or vaccines.
1. Introduction
Streptococcus iniae is a major fish pathogen originally isolated from the Amazon fresh water dolphin, Inia geoffrensis [1]. Since then, a number of fish species have also been reported to be infected by S. iniae, including some economically important species, such as tilapia, trout, barramundi and salmon [2]. Globally, these infections contribute to annual losses exceeding US$100 million in the aquaculture industry [3]. S. iniae is also an opportunistic human pathogen with most cases reported among elderly Asian populations and among weak or immunocompromised individuals who have a history of handling fish [4,5]. Clinical symptoms of human disease including bacteremic cellulitis are similar to those caused by the important human pathogen, Streptococcus pyogenes, which can also subsequently develop into other conditions, such as meningitis, endocarditis, septic arthritis, osteomyelitis and toxic shock [4,6,7]. Following the complete sequencing of the genome of S. iniae strain 9117 [8], a number of virulence factors were discovered. Of those that have been documented, many are homologous to those expressed by S. pyogenes [5], such as S. iniae M-like protein [9,10], hyaluronic acid capsule [11,12,13], C5a peptidase [10] and streptolysin S [14,15].
Analyzing the complete genome of S. iniae 9117, we recently identified two S. iniae proteins that share some amino acid sequence homology with two important S. pyogenes immune evasion factors, Streptococcus pyogenes nuclease A (SpnA) [16] and streptococcal 5′-nucleotidase A (S5nA) [17]. These were termed SpnAi and S5nAi, respectively [18]. These S. iniae proteins are expressed as precursor proteins with an N-terminal signal peptide and a C-terminal cell wall anchor domain, including the canonical sortase A recognition motif LPXTG [16,17,18]. Our previous biochemical analysis using a recombinant form of SpnAi (rSpnAi) showed that the enzyme is a calcium and magnesium ion-dependent nuclease that cleaves double stranded linear DNA and chromosomal DNA [18]. Extracellular nucleases produced by S. pyogenes (SpnA) [16] and other Gram-positive bacterial species, such as Staphylococcus aureus (Nuc) [19], S. pyogenes (Sda1) [20] and S. pneumoniae (EndA) [21] have been shown to play an important role in immune evasion by degrading the DNA framework of neutrophil extracellular traps (NETs). NETs are released from activated neutrophils and comprise of DNA fibers with antimicrobial proteins such as neutrophil elastase, cathepsin G, and myeloperoxidase. Hence, extracellular nucleases play a crucial role in host immune evasion [22].
Similarly, we have shown that S5nAi is a 5′-nucleotidase that hydrolyzes adenosine monophosphate (AMP), adenosine diphosphate (ADP) and deoxyadenosine monophosphate (dAMP) to produce the immunomodulatory molecules, adenosine (Ado) and deoxyadenosine (dAdo) [18]. Ado is a purine nucleoside that plays important roles in cell signaling. Ado is typically produced under conditions of metabolic stress such as hypoxia and inflammatory tissue damage following an increase in the ATP production [23]. Ado binds specifically to Ado receptors (e.g., A1, A2A, A2B and A3) that are differentially expressed on various innate immune cells, such as neutrophils and macrophages, and have different thresholds for activation [24]. It has been shown that Ado decreases the phagocytic activity of macrophages by suppressing the production of various components required for the macrophage phagocytic action, including proinflammatory cytokines [25], superoxide [26] and nitric oxide [27]. In addition, the binding of Ado to A2 and A3 receptors on neutrophils inhibit neutrophil degranulation [28].
Several 5′-nucleotidases have also been identified in other Gram-positive bacteria, and have been shown to play an important role in evasion of the host immune response [29]. Staphylococcus aureus produces adenosine synthase (AdsA) that hydrolyzes AMP, ADP and ATP to produce Ado [30]. AdsA is also able to convert dAMP into dAdo, which activates caspase-3-mediated apoptosis of macrophages and monocytes [31]. S. pyogenes S5nA hydrolyzes AMP and ADP, but not ATP, to generate Ado, and has been shown to increase the survival of the non-virulent bacterium, Lactococcus lactis, in whole human blood killing assays [17]. The role of another 5′-nucleotidase, ecto-5′nucleotidase (Nt5e) from Streptococcus sanguinis was confirmed in a platelet aggregation assay and an experimental rabbit endocarditis model [32]. Another nucleotidase, Streptococcus suis adenosine synthase (Ssads) was reported to impair polymorphonuclear cell-mediated innate immunity [33] and was demonstrated to be an important virulence factor in piglet and mouse infection models [33,34].
Being a major fish pathogen, S. iniae has been the most extensively studied streptococcal species in the zebrafish infection model. A major advantage of using the larval zebrafish infection model is that the optical transparency of zebrafish larvae enables visualization of the host immune response to infection in vivo. In zebrafish, innate immunity develops during early stages of development, with fully functional macrophages and neutrophils present as early as 2 days post-fertilization (dpf) [35,36,37]. A morphologically and functionally mature adaptive immune system is not present until 4–6 weeks post-fertilization [38,39,40]. This temporal separation allows the study of the innate immune response independently from the adaptive immune response.
In this study, we show that the deletion of the spnAi and s5nAi genes in S. iniae significantly decreased the nuclease and nucleotidase activity, respectively. We also demonstrated the virulence role of SpnAi and S5nAi in vivo using a S. iniae-zebrafish infection model. Our findings suggest that SpnAi and S5nAi contribute to the virulence of S. iniae by suppressing innate immune cell function.
2. Materials and Methods
2.1. Bacterial Strains, Media, Growth Conditions and Electroporation
S. iniae strain 9117, a human clinical isolate from a patient with cellulitis (kindly provided by Dr. Sarah Highlander, JVCI, La Jolla, CA), was grown in Todd Hewitt broth medium supplemented with 0.2% yeast extract and 2% proteose peptone (THY+P, BD Biosciences, San Jose, CA, USA) at 37 °C without agitation. Escherichia coli DH5α (ATCC® 53868™) was grown in Luria Bertani (LB, BD Biosciences) broth at 37 °C with aeration. Solid THY+P or LB plates were made by adding 1.5% Bacto agar (BD Biosciences) to liquid medium. When appropriate, antibiotics were added to the THY+P or LB medium: spectinomycin at 50 μg/mL for S. iniae and 100 μg/mL for E. coli; or kanamycin at 200 μg/mL for S. iniae and 50 μg/mL for E. coli. For all experiments, overnight cultures of S. iniae were diluted 1:10 in fresh THY+P and grown to the mid-log phase at an optical density (OD600nm) of 0.6.
Electrocompetent S. iniae strains were made by growing the bacteria in THY+P media containing 0.6% sterile glycine overnight. The overnight culture was diluted in fresh THY+P supplemented with 0.6% sterile glycine and grown to OD600nm of 0.3–0.35. The bacteria were harvested by centrifugation (6000 × g, 10 min, 4 °C) in a chilled rotor, and resuspended in 40 mL ice-cold sterile electroporation medium (5% glucose, 1 mM MgCl2, pH 6.5). The bacterial cells were pelleted and washed twice by centrifugation before being resuspended in 150 μL of ice-cold sterile electroporation medium. The resulting bacterial competent cell suspension in 50 µL was mixed with 1–10 µg purified plasmid and transferred into a prechilled Micropulser™ electroporation cuvette (Bio-Rad, Hercules, CA, USA). A single pulse of 2.1 kV, capacitance at 25 µF and resistance at 200 Ω was applied to the bacterial cell suspension by using a Gene Pulser Xcell™ (Bio-Rad), and 1 mL THY+P was added immediately into the cuvette. The bacterial cell suspension was transferred into an Eppendorf tube, and allowed to recover at 37 °C for 3 h. The bacteria cells were then centrifuged and resuspended in 100 µL THY+P. The resuspended bacterial cells were plated on a THY+P agar plate containing an appropriate amount of antibiotic and incubated at 37 °C overnight.
2.2. Allelic Exchange Mutagenesis
The individual S. iniae spnAi and s5nAi gene-knockout mutants (∆spnAi and ∆s5nAi) were created by allelic replacement of the target gene on the wild-type (WT) S. iniae genome with a spectinomycin resistance gene, aad9 (Figure S1). Primers used to generate and confirm the allelic exchange are listed in Table 1. Approximately 1000-bp of the upstream flanking region (FR1) and downstream FR2 of the spnAi or s5nAi genes were amplified by 25 cycles of PCR using iProof™ high-fidelity DNA polymerase (Bio-Rad, Hercules, CA, USA) at an annealing temperature of 56 °C. The amplified fragments were cloned into the multiple cloning sites flanking the aad9 gene on the pFW11 plasmid [41]. The generated plasmid construct was then transformed into WT S. iniae by electroporation, and the successful transformants were selected using a THY+P agar plate containing spectinomycin. The targeted in-frame replacement of spnAi and s5nAi were confirmed by PCR using the aad9 forward primer (aad9.fw) and a reverse diagnostic primer (SpnAi_DP.rv for spnAi and S5nAi_DP.rv for s5nAi) that anneal downstream of the FR2 region.

Table 1.
Primers used in this study. The restriction site in each primer is underlined.
2.3. Green Fluorescence Labelling of Wild-Type S. iniae, Isogenic Mutants and Complementation Strains
Green fluorescent S. iniae, S. iniae ∆spnAi and S. iniae ∆s5nAi were generated by transformation with the toxin-antitoxin stabilized green fluorescent reporter plasmid pLZ12Km2-P23R:TA:gfpmut2 [42] by electroporation. The generation of green fluorescent complementation strains (S. iniae ∆spnAi:spnAi and S. iniae ∆s5nAi:s5nAi) were achieved by inserting the spnAi and s5nAi genes into the green fluorescent reporter plasmid and transforming the S. iniae ∆spnAi and S. iniae ∆s5nAi deletion strains, respectively. A list of primers used to generate and confirm the complementation strains is provided in Table 1. The full length spnAi gene including its ribosome binding site was amplified from the genome of the WT S. iniae, and cloned into pLZ12Km2-P23R:TA:gfpmut2 downstream of the original streptococcal promoter sequence (Figure S2). The resulting reporter plasmid carrying the spnAi or s5nAi gene was then introduced into ∆spnAi or ∆s5nAi by electroporation. These complementation strains simultaneously had their spnAi or s5nAi genes restored along with green fluorescence reporter. Successful S. iniae transformants were first selected on THY+P agar plates containing kanamycin, then screened for the ability to fluoresce.
2.4. DNA-Methyl Green Assay
The DNA-methyl green assay [43] was used to quantify the DNase activity of the generated S. iniae strains. Bacterial cells (106 colony-forming units, CFU) were resuspended in 50 µL of nuclease reaction buffer (25 mM Tris-HCl, pH 7.0, 1 mM CaCl2 and 3 mM MgCl2) and transferred into a clear-bottom Corning® 96-well plate (Sigma-Aldrich, St. Louis, MO, USA). An equal volume of DNA-methyl green substrate solution was then added and the plate was incubated at 37 °C for 20 h before measuring the absorbance at A492nm and A620nm using an EnSpire™ 2300 plate reader (Perkin Elmer, Waltham, MA, USA). The corrected absorbance was calculated by subtracting the A492nm reading from the A620nm reading.
2.5. Quantification and Visualization of NETs Destruction
Tg(lyz:EGFP)nz117 adult zebrafish [44] were anaesthetized in 0.168 mg/mL tricaine (Sigma-Aldrich) before dissection of the kidneys. The isolated kidney tissue was pooled and dissociated by gently teasing through a 40 µm Falcon™ cell strainer (BD Biosciences) in 2 mL ice-cold Hank’s balanced salt solution without phenol red (HBSS, Thermo Fisher Scientific, Waltham, MA, USA). The homogenized solution was again passed through the strainer twice and transferred to Falcon™ round-bottom polystyrene tubes (BD Biosciences). The enhanced green fluorescence protein (EGFP) labeled neutrophils were sorted using a FACSAria™ II cell-sorting system (BD Biosciences), and a volume containing 5 × 104 neutrophils in 100 µL HBSS was used in each reaction.
To quantify the NET destruction, the neutrophils were seeded in a solid-bottom Corning® 96-well plate and activated by the addition of phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) at a final concentration of 1 µg/mL at 37 °C for 2 h. Cytochalasin D (Sigma-Aldrich) in a final concentration of 2 µg/mL was added to the activated neutrophils before adding 2.5 × 106 bacterial cells. As a positive control, three wells of neutrophils were incubated with 2 U DNase I (Thermo Fisher Scientific). The reaction was incubated at 37 °C for a further 1 h. Sytox Orange (Thermo Fisher Scientific) was added to a final concentration of 0.1 µM and incubated at room temperature for 10 min before reading the fluorescence at excitation/emission at 530/590 nm using an EnSpire™ 2300 plate reader.
To visualize the degradation of NETs, a Nunc® Lab-Tek® Eight-well chamber slide (Sigma-Aldrich) was coated with 300 µL per well of 0.0001% poly-L-lysine (Sigma-Aldrich) at 37 °C for 1 h. Unbound poly-L-lysine was removed by washing once with UltraPure™ water (Invitrogen, Waltham, MA, USA). Each well of the slide was seeded with 5 × 104 neutrophils in 200 µL HBSS, and allowed to settle at 37 °C for 30 min. Neutrophils were then activated by adding PMA at a final concentration of 1 µg/mL and incubated at 37 °C for 2 h. The solution in the well was carefully removed before 2.5 × 106 bacterial cells and Cytochalasin D at a final concentration of 2 µg/mL were added. As a positive control, three wells of neutrophils were incubated with 2 U DNase I. The reaction mixture was incubated at 37 °C for a further 1 h, followed by washing once with HBSS. The slide was fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) at 4 °C for 1 h and washed 3 times with HBSS. The cells were permeabilized with 0.05% Triton X-100 and washed 3 times with HBSS. The slide was blocked with 300 µL of immunostaining blocking solution (1% bovine serum albumin in HBSS) at 4 °C overnight. The blocking solution was removed the next day and the slide was incubated with 200 µL of 1:1000 prediluted rabbit anti-human neutrophil elastase (Sigma-Aldrich) antibody in blocking solution at 37 °C for 1 h, followed by an incubation with 200 µL of 1:500 prediluted goat anti-rabbit IgG FITC (Abacus ALS, Queensland, Australia) in blocking solution at 37 °C for 1 h in the dark. The chamber frame was removed and ProLong® Gold Antifade Mountant containing DAPI (Molecular Probes, Eugene, OR, USA) was added to the slide and a coverslip was gently placed on top of the reagent. The slide was allowed to cure at room temperature for 24 h in the dark before being analyzed using a Nikon Eclipse E600 fluorescence microscope (Nikon, Tokyo, Japan).
2.6. Malachite Green Phosphate Colorimetric Assay
The malachite green phosphate colorimetric assay [43] was used to quantify the nucleotidase activity of the modified S. iniae strains. Bacterial cells (106 CFU) were resuspended in 98 µL of nucleotidase reaction buffer (50 mM Tris-HCl, pH 7.0 and 10 mM MgCl2) and transferred into a clear-bottom Corning® 96-well plate. A final concentration of 1 mM AMP (Sigma-Aldrich) substrate was added and incubated at 37 °C for 30 min. The reactions were stopped by adding EDTA to a final concentration of 50 mM and the release of inorganic phosphate (Pi) was then quantified using a malachite green phosphate colorimetric assay kit (Sigma-Aldrich) according to the manufacturer’s instruction. The release of Pi was measured at A650nm and the amount of Pi was calculated based on a standard curve of Pi.
2.7. Zebrafish Maintenance
Adult zebrafish were maintained in the commissioned zebrafish facility at The University of Auckland according to the standard operating procedures (SOP691) of the facility, with approval from the University of Auckland Animal Ethics Committee. The facility has a 14 h light and 10 h dark automated lighting cycle and the water conditions for the zebrafish maintenance are set to between pH 7.2 and 7.5, temperature 25.5 and 29.5 °C and conductivity 250 and 500 µS. Embryos were obtained by natural spawning after light exposure, and were then incubated in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgCl2) with 0.1% (v/v) methylene blue (PanReac AppliChem, Darmstadt, Germany) at 28 °C. At 1 day post-fertilization (dpf), embryos were manually dechorionated. The larvae were then transferred to a new Petri dish containing E3 medium supplemented with 0.003% phenylthiourea (PTU) to prevent pigmentation, and incubated at 28 °C. Wild-type (AB line) zebrafish larvae were used in survival analysis, bacterial load determination and time-lapse confocal imaging experiments. The previously published transgenic lines Tg(lyz:DsRED2)nz50 [44] and Tg(mpeg1:EGFP)gl22 [45] were used in the neutrophil and macrophage recruitment assays, respectively. Tg(lyz:DsRED2)nz50 is a transgenic zebrafish line in which the neutrophil-specific promoter lyz drives the neutrophil expression of DsRED2, rendering neutrophils red fluorescent [44]. Tg(mpeg1:EGFP)gl22 is a transgenic zebrafish line in which the macrophage-specific promoter of macrophage expressed gene 1 (mpeg1) drives macrophage expression of EGFP, rendering macrophages green fluorescent [45].
2.8. Preparation and Microinjection of S. iniae
One milliliter of mid-log phase bacterial cultures were centrifuged (2300 × g, 10 min, 4 °C) and resuspended in 1 mL sterile PBS. The OD600nm of resuspended bacterial cells was measured again and diluted to OD600nm of 0.1 in 1 mL sterile PBS. The bacterial cells were centrifuged again and resuspended with 0.25% phenol red in sterile PBS to a final concentration of 50 CFU/nL. Two dpf larvae were anesthetized in tricaine before being mounted in 3% methylcellulose (Sigma-Aldrich) in E3 medium. One nanoliter of bacterial suspension was microinjected into the hindbrain (Figure S3) of the larvae. The injection dose was validated by injecting one bolus into 10 µL sterile PBS, spot plated on the THY+P agar plate containing kanamycin and incubated at 37 °C overnight for enumeration. After injection, larvae were then transferred back to the Petri dish containing E3 medium supplemented with PTU and incubated at 28 °C. The larvae survival at indicated time-points was monitored. The determination of live or dead larvae was based on the presence of a heartbeat and response to gentle touching with a sterile transfer pipette.
2.9. S. iniae Enumeration From Infected Larvae
At the indicated times, infected larvae were anaesthetized and each larva was collected in a tube. Twenty microliters of sterile dissociation buffer (PBS with 1% Triton-X) was used to homogenize the larva by repeatedly pipetting up and down. For enumeration, serial dilution of the homogenized mixture was made in sterile PBS, spot plated on THY+P agar plate containing kanamycin and incubated at 37 °C overnight.
2.10. Mounting and Confocal Imaging of Infected Larvae
Infected larvae were anaesthetized before mounting in 0.8% UltraPure™ low melting point agarose (Invitrogen) in E3 medium supplemented with PTU. Z-Series time-lapse fluorescence images were acquired using a Fluoview FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan) with a 20× water immersion objective lens. For live cell imaging, the temperature of the incubation chamber was preadjusted to 28 °C. Z-stacks of the hindbrain ventricle in dorsal view (Figure S3, bottom panel) were taken with a total of 36 sections at 5 µm step size intervals every 5 min. Confocal imaging was carried out with the similar acquisition setting in each set of experiments. The resulting images were analyzed using the Volocity 3D image analysis software V6.1.1 (Quorum Technologies, Lewes, UK) and the images were made into movies using ImageJ V1.52k [46].
2.11. Immunofluorescence Detection of Innate Immune Cells in Infected Larvae
At the indicated time-points, infected larvae were anaesthetized and collected in a tube. One milliliter of ice-cold fixing solution (4% PFA and 4% sucrose in PBS) was added and incubated at 4 °C overnight. Larvae were dehydrated the next day with increasing successive concentrations of methanol in PBS with 0.1% Tween 20 (PBS-T) for 5 min each incubation and stored at −20 °C for up to a week. Dehydrated larvae were rehydrated with decreasing successive concentrations of methanol in PBS-T for 5 min each incubation and washed once with PBS-T for 5 min before being incubated in 1 mL of ice-cold pure acetone on a shaker for 7 min to allow tissue permeabilization. The larvae were washed with PBS-T for 5 min three times, and incubated in 1 mL of immunostaining blocking solution (10% goat serum, 2% blocking reagent and Roche in maleic acid buffer) on a shaker for 2 h. The immunostaining blocking solution was replaced with 200 µL of 1:500 prediluted primary antibody anti-DsRED2 (Clonetech, Mountain View, CA, USA) for detecting neutrophils, or with anti-GFP (Abcam, Cambridge, UK) for detecting macrophages in immunostaining blocking solution, and incubated on a shaker at 4 °C overnight. The larvae were washed with PBS-T for 5 min four times to remove unbound antibodies before incubated with 100 µL of 1:500 prediluted secondary antibody anti-rabbit IgG-Alexa Fluor 568 (Thermo Fisher Scientific, Waltham, MA, USA) or anti-chicken IgG-Alexa Fluor 488 (Thermo Fisher Scientific, Waltham, MA, USA) in immunostaining blocking solution in the dark on a shaker for 2 h. From this point onwards, the larvae were protected from light exposure to avoid photobleaching of the fluorophores. The larvae were washed with PBS-T again for 5 min four times and fixed with 1 mL ice-cold fixing solution and stored at 4 °C overnight. The larvae were mounted and imaged as described in the previous section.
2.12. Statistical Analyses
The statistical analyses were performed using GraphPad Prism V7.03 (GraphPad Software, San Diego, CA, USA). Statistical significance was calculated using either the unpaired two-tailed t-test or 2-way ANOVA with a Tukey’s multiple comparisons test. The statistical significance analysis for Kaplan–Meier curves was calculated using the Gehan–Breslow–Wilcoxon test. A p value of <0.05 was considered to be statistically significant. p value <0.05 was labeled with *, p value <0.01 was labeled with **, p value <0.001 was labeled with *** and p value <0.0001 was labeled with ****. A p value of >0.05 was considered not statistically significant and was labeled with ns.
3. Results
3.1. Deletion of SpnAi Decreases DNase Activity in S. iniae
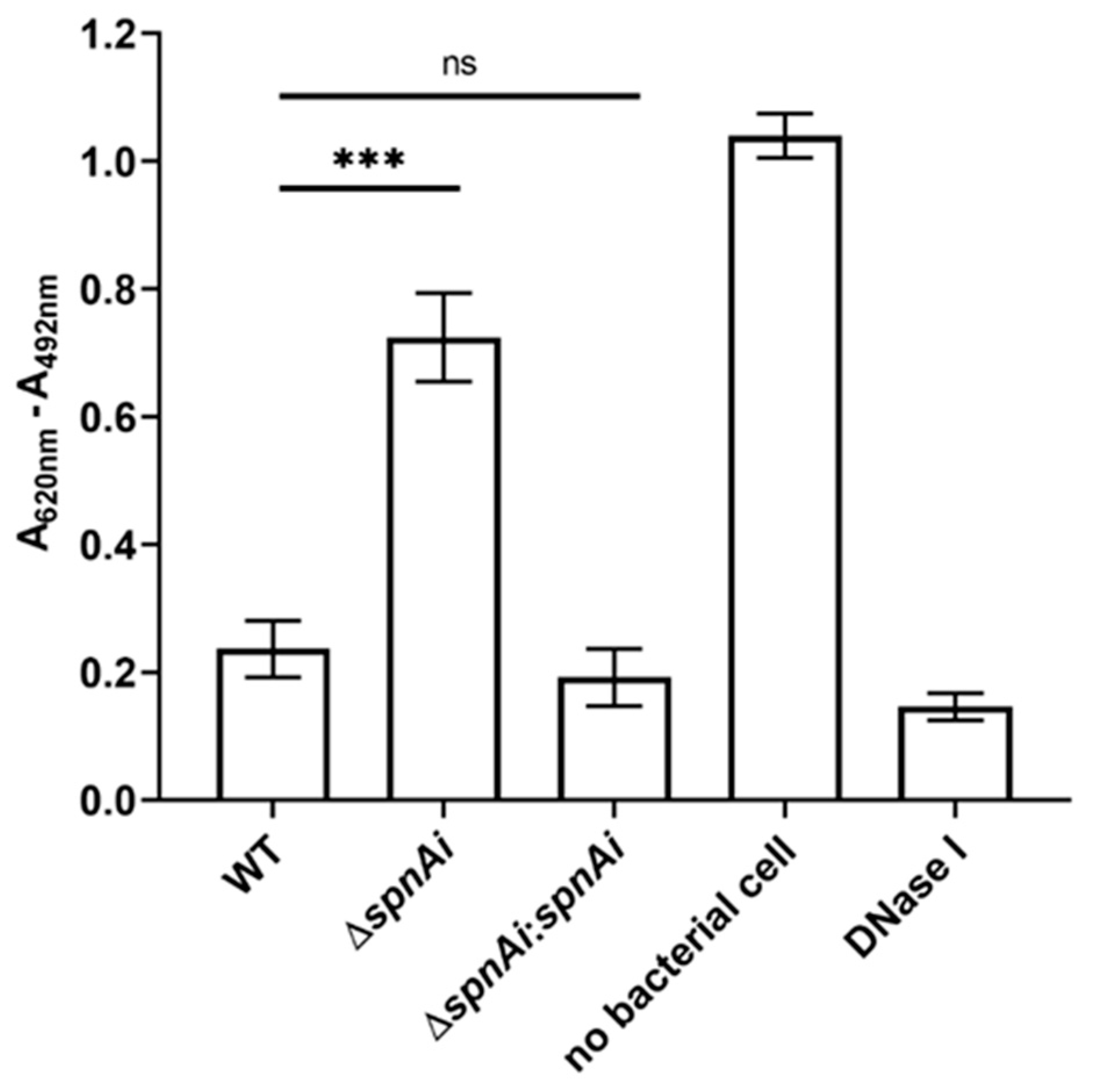
Our previous study showed that recombinant SpnAi is able to digest DNA in vitro [18]. To confirm that SpnAi also shows activity in vivo, a spnAi gene deletion mutant (S. iniae ∆spnAi) and the corresponding complementation strain (S. iniae ∆spnAi:spnAi) were analyzed for DNase activity using a DNA-methyl green assay. As shown in Figure 1, deletion of the spnAi gene resulted in significant loss of DNase activity (approximately 3-fold, p < 0.001). Complementation with the spnAi gene resulted in a DNase activity level similar to that of the WT S. iniae strain. This activity was also similar to 1 U of commercial DNase I.
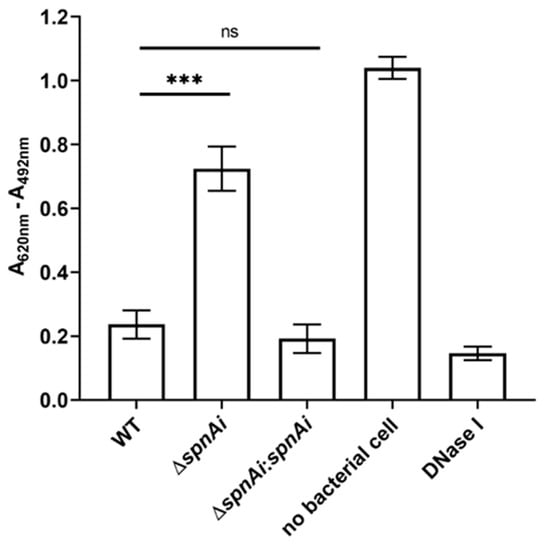
Figure 1.
DNase activity of Streptococcus iniae strains. The DNase activity against salmon sperm DNA was determined for wild-type (WT) S. inia, S. iniae ∆spnAi and S. iniae ∆spnAi:spnAi using a methyl green assay [43]. A decrease in the reading of A620nm - A492nm indicates DNA digestion. The error bars show the standard deviation of three experiments performed in triplicate. ***, p < 0.001; ns, not significant (as determined by an unpaired two-tailed t-test).
3.2. SpnAi Promotes Degradation of NETs
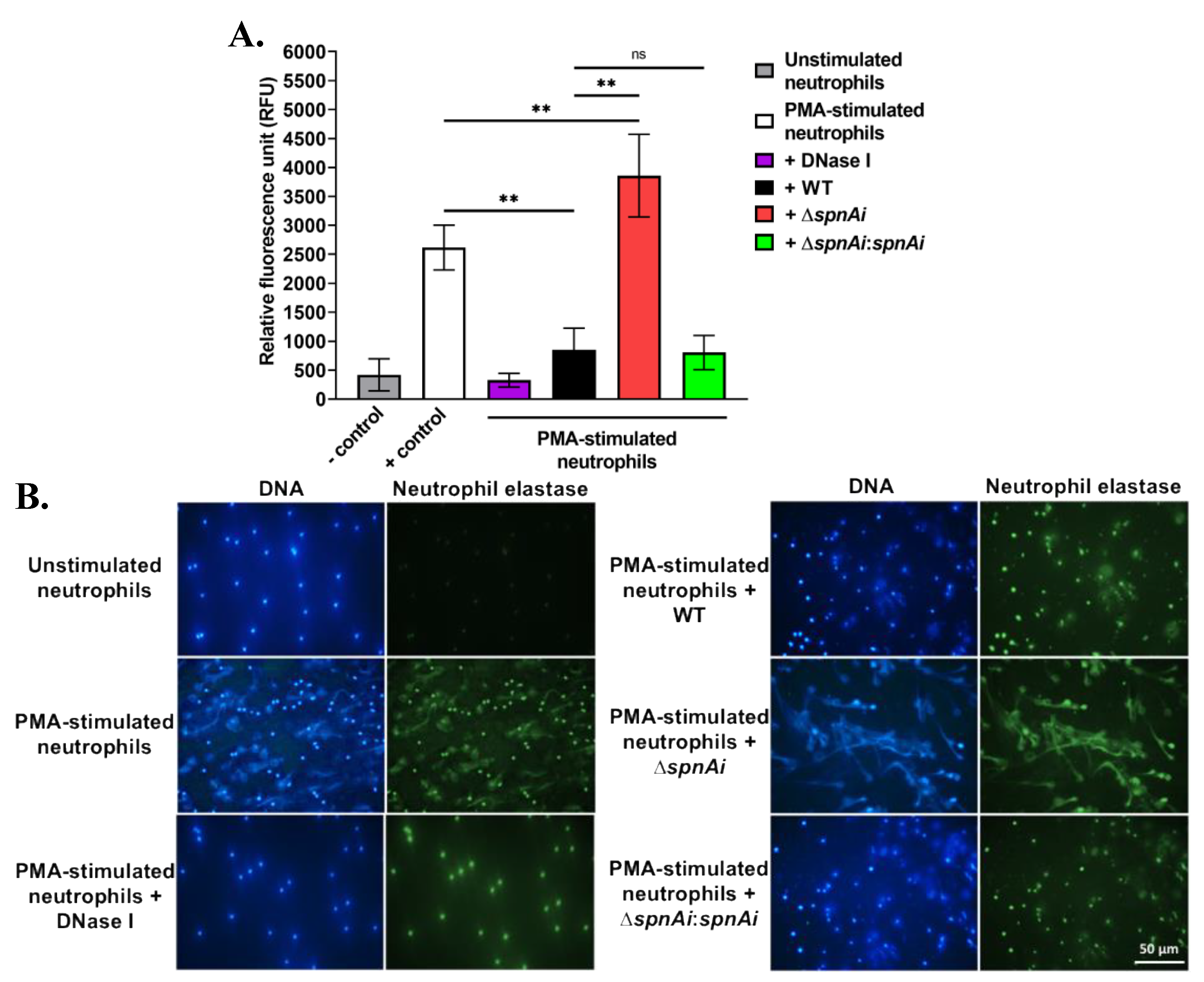
The destruction of NETs by extracellular nucleases has been demonstrated for other streptococci, including S. pyogenes SpnA [16], S. suis SsnA [47] and S. pneumoniae TatD [48]. The ability of SpnAi to digest the backbone DNA of NETs from fish neutrophils was therefore hypothesized and investigated. Neutrophils were isolated from the kidneys of adult zebrafish and stimulated with PMA for 2.5 h to generate NETs, before being incubated with S. iniae WT and mutant strains. DNA released from NETs was quantified using the extracellular DNA stain Sytox Orange (Figure 2A). Incubation of WT S. iniae with stimulated neutrophils substantially decreased the fluorescence signal (approximately 3-fold) indicating DNA degradation (p < 0.01). DNase I was used as a control and reduced fluorescence approximately 8-fold. Notably, the incubation with the spnAi deletion strain not only showed no reduction in NETs degradation, but also increased the amount of extracellular DNA (p < 0.01). As expected, incubation with the complementation strain with stimulated neutrophils degraded NETs at a similar level as the WT strain.
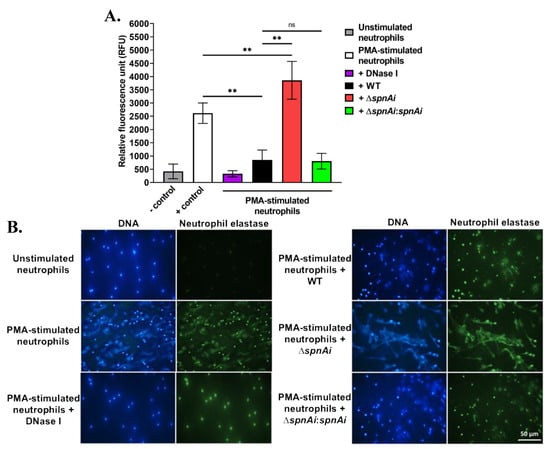
Figure 2.
Release and destruction of neutrophil extracellular traps (NETs) by various S. iniae strains. (A) The extracellular DNA stain Sytox Orange was used to quantify the NETs and is presented as relative fluorescence units (RFU). NETs were degraded in the presence of WT S. iniae and DNase I (positive control), but not in the presence of the spnAi deletion mutant. Complementation of the mutant strain with a functional spnAi gene restored the NET-degrading ability of the mutant strain. Error bars show the standard deviation of two experiments performed in triplicate. **, p < 0.01; ns, not significant (as determined by an unpaired two-tailed t-test). (B) Immunofluorescence microscopy of zebrafish neutrophils. DAPI stain (blue) indicates both the neutrophil nuclei and released NETs. A cross-reactive human antibody against zebrafish neutrophil elastase, a bactericidal enzyme associated with NETs, was used to label the released NETs (green). Activation of unstimulated neutrophils (top left) results in NET release (middle left), which could be degraded by adding DNase I (bottom left). Addition of WT S. iniae also led to NETs destruction (top right), which was abrogated in the spnAi deletion mutant (middle right) and restored by complementing the deletion mutant with the spnAi gene (bottom right).
The destruction of NETs by various S. iniae strains was also visualized using immunofluorescence microscopy (Figure 2B). The extracellular filamentous structures stained for DNA (blue) and neutrophil elastase (green) confirmed the presence of NETs. No NET-like structure was visible in the absence of PMA stimulant, but after PMA stimulation, extracellular structures were apparent. When the activated neutrophils were exposed to S. iniae WT or DNase I was added as a positive control, the web-like structures disappeared indicating degradation of NETs. In contrast, when the S. iniae ∆spnAi mutant was added to the activated neutrophils, the net-like structure remained visible, showing that spnAi was involved in the degradation of NETs. Complementation of the mutant strain with a functional spnAi gene restored the NET-degrading ability of the mutant strain.
3.3. Deletion of S5nAi Decreases Nucleotidase Activity in S. iniae
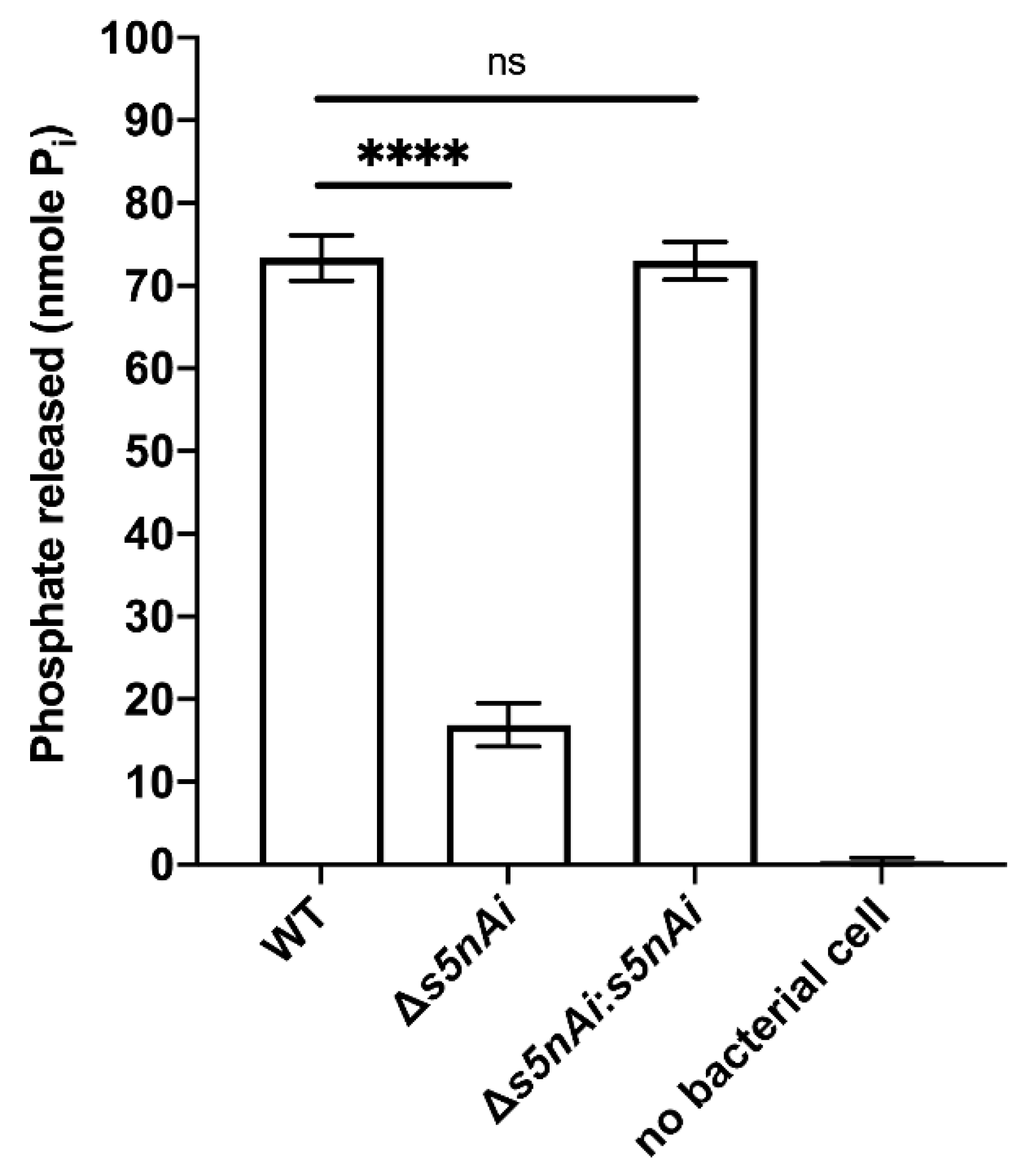
We have previously shown that recombinant S5nAi is able to generate adenosine from ADP and AMP [18]. To show that the bacteria produce the active enzyme, a s5nAi deletion mutant (∆s5nAi) was generated along with a complementation strain (∆s5nAi:s5nAi). The nucleotidase activity of S. iniae WT and the mutant strains was tested by quantifying the amount of Pi released from 1 mM AMP (Figure 3). Incubation of WT S. iniae resulted in the release of about 70 nmole Pi indicating nucleotidase activity. In contrast, AMP hydrolysis with the ∆s5nAi deletion mutant resulted in significantly lower amounts of Pi (approximately 4.3-fold, p < 0.0001). No measurable levels of Pi were found when no bacteria were added excluding spontaneous hydrolysis of AMP. Full nucleotidase activity could be restored in the ∆s5nAi:s5nAi complementation mutant.
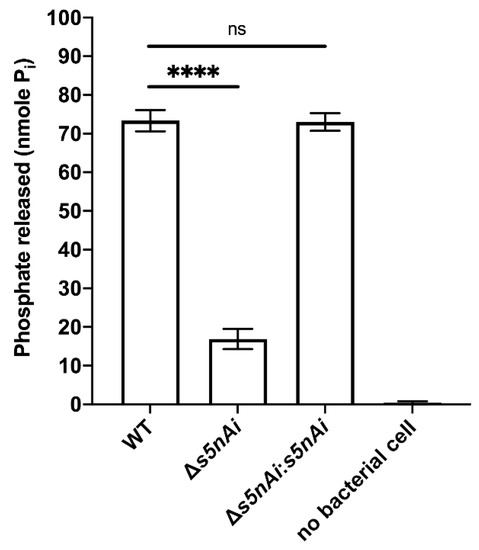
Figure 3.
Nucleotidase activity of S. iniae strains. WT, ∆s5nAi and ∆s5nAi:s5nAi were incubated with 1 mM AMP as a substrate and the Pi released was measured using a malachite green phosphate colorimetric assay. Error bars show the standard deviation of three experiments performed in triplicate. ****, p < 0.0001; ns, not significant (as determined by an unpaired two-tailed t-test).
3.4. SpnAi and S5nAi Contribute to S. iniae Virulence in Zebrafish Larvae
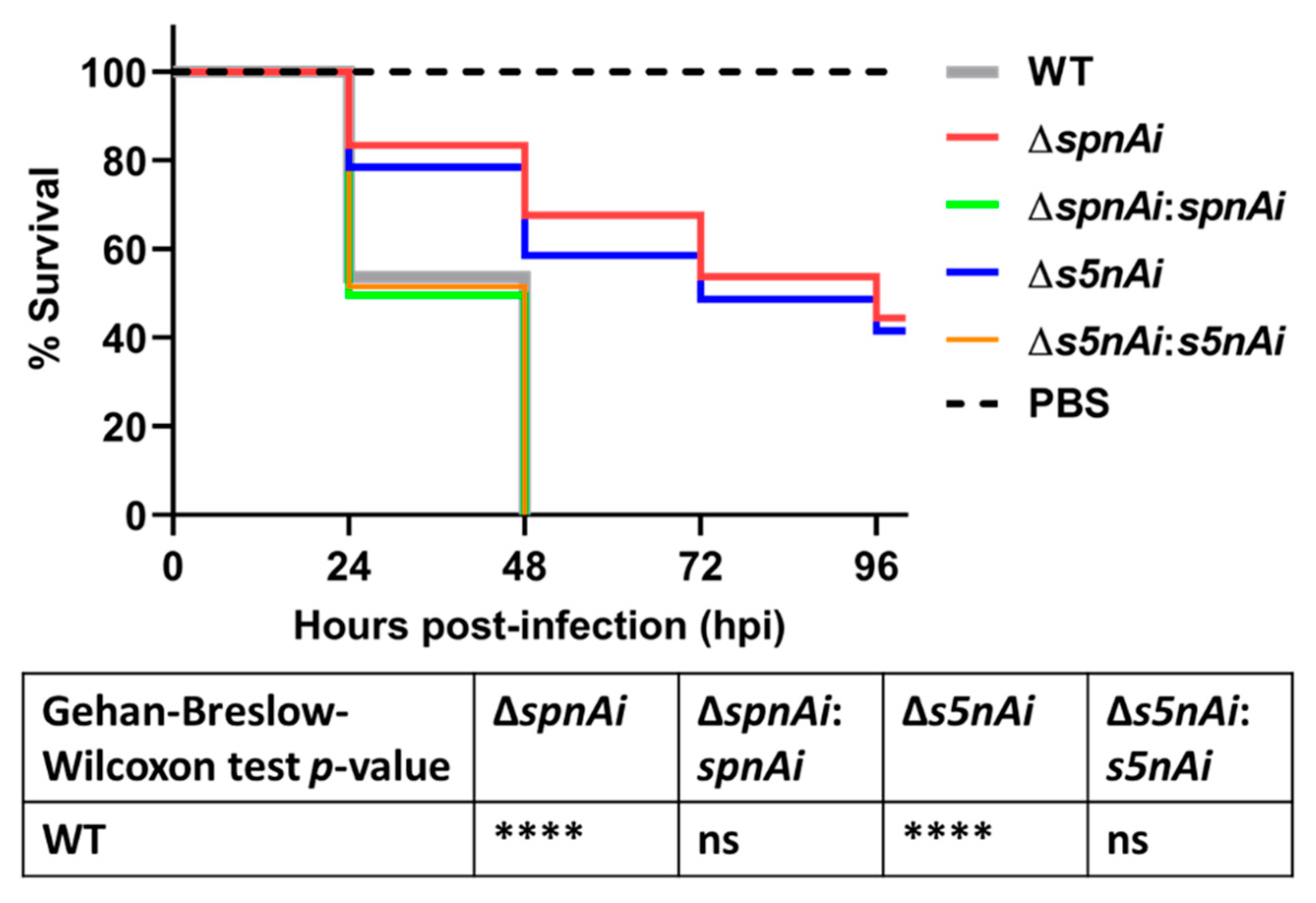
Next, we investigated the virulence attributes of SpnAi and S5nAi using a zebrafish infection model. The contribution of the two enzymes to S. iniae survival in the zebrafish host was analyzed by injecting 2 days post-fertilization (dpf) larvae (wild-type AB line) with 50 CFU of WT and mutant S. iniae strains. The survival of larvae was recorded every 24 h over a 96 h time-course (Figure 4). Infection with WT S. iniae killed all larvae within 48 h, whereas infection with the deletion mutants, ∆spnAi and ∆s5nAi, showed increased survival rates 48 h post-infection (hpi) of approximately 63% and 60%, respectively, and about 40% survival at 96 hpi. Zebrafish infected with the complementation strains with spnAi and s5nAi genes restored, displayed a similar mortality rate as the WT.
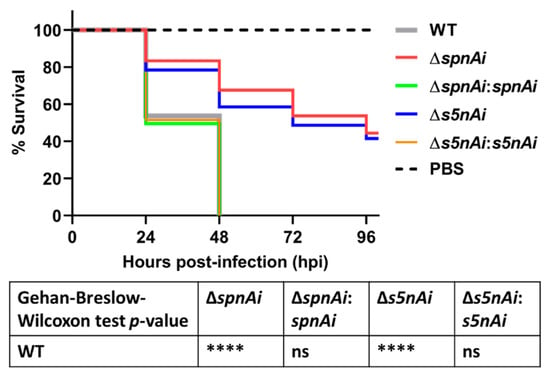
Figure 4.
Survival analysis of zebrafish larvae infected with WT and mutant S. iniae strains. WT, ∆spnAi, ∆s5nAi and their complementation strains were each injected into 2 days post-fertilization (dpf) larvae (wild-type AB line). The survival of infected larvae was recorded over a 96 h-period. This data is the combined result from 3 independent experiments (n = 35 per group in each experiment). ****, p < 0.0001; ns, not significant (as determined by the a Gehan–Breslow–Wilcoxon test).
3.5. SpnAi and S5nAi Support the Growth of S. iniae in Zebrafish Larvae
As a measurement of disease progression, the bacterial load in whole zebrafish larvae (wild-type AB line) were monitored using the number of bacteria recovered from homogenized larvae (Figure 5). The growth rate of the S. iniae strains were first tested in vitro in order to investigate whether the deletion of spnAi or s5nAi on the S. iniae WT genome and the complementation of ∆spnAi or ∆s5nAi would affect their growth compared to their parental strain (Figure S4). Overall, no significant difference was seen between the growth rate of WT, ∆spnAi or ∆s5nAi and the complementation strains (Figure S4), confirming that the gene deletions and complementations had no effect on bacterial growth.
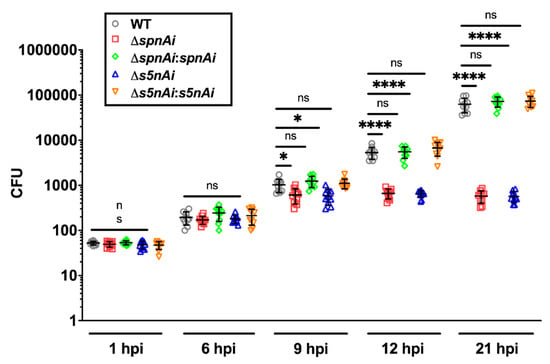
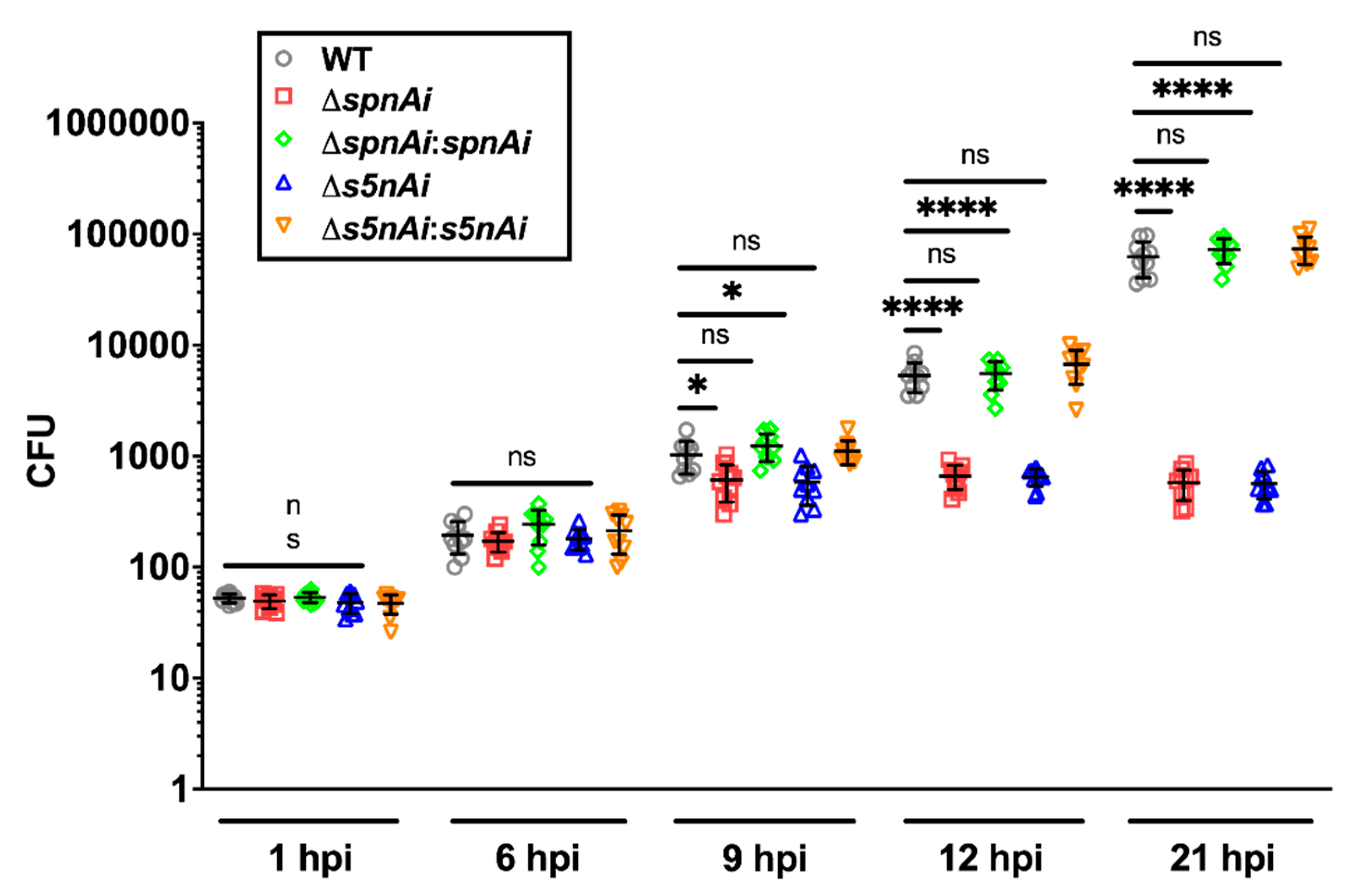
Figure 5.
Bacterial load of zebrafish larvae infected with WT and mutant S. iniae strains. WT, ∆spnAi, ∆s5nAi and their complementation strains were each injected into 2 dpf larvae (wild-type AB line). The bacterial load of infected larvae (n = 5 per group in each experiment) at indicated time-points were quantified by plating on kanamycin-containing THY+P agar plates. This data is the combined result from 2 independent experiments (n = 5 per group in each experiment). The error bars show the standard deviation of two experiments. *, p < 0.05; ****, p < 0.0001; ns, not significant (as determined by a 2-way ANOVA with a Tukey’s multiple comparisons test).
The disease progression experiment showed that the amount of WT bacteria recovered from infected larvae increased 2000-fold over a 21 h time-course, from an initial inoculum of 50 CFU to 1 × 105 CFU at 21 hpi, which was similar to the bacterial load in larvae infected with spnAi and s5nAi complementation strains. Single gene-knockout S. iniae showed lower bacterial counts, as the number of bacteria recovered from larvae infected with ∆spnAi and ∆s5nAi mutants increased 20-fold from an initial inoculum of 50 CFU to a maximum of 1000 CFU at 9 hpi that was maintained until the end of the experiment. An additional group of larvae was injected with sterile PBS as a negative control and showed no colony formation on the agar plates (data not shown). These results explained the high mortality rate of larvae infected with WT and complementation strains, and higher survival rate of larvae infected with ∆spnAi and ∆s5nAi mutants at 24 h.
3.6. Wild-type S. iniae, but Not SpnAi and S5nAi Deletion Mutants Were Able to Proliferate and Disseminate in Zebrafish Larvae
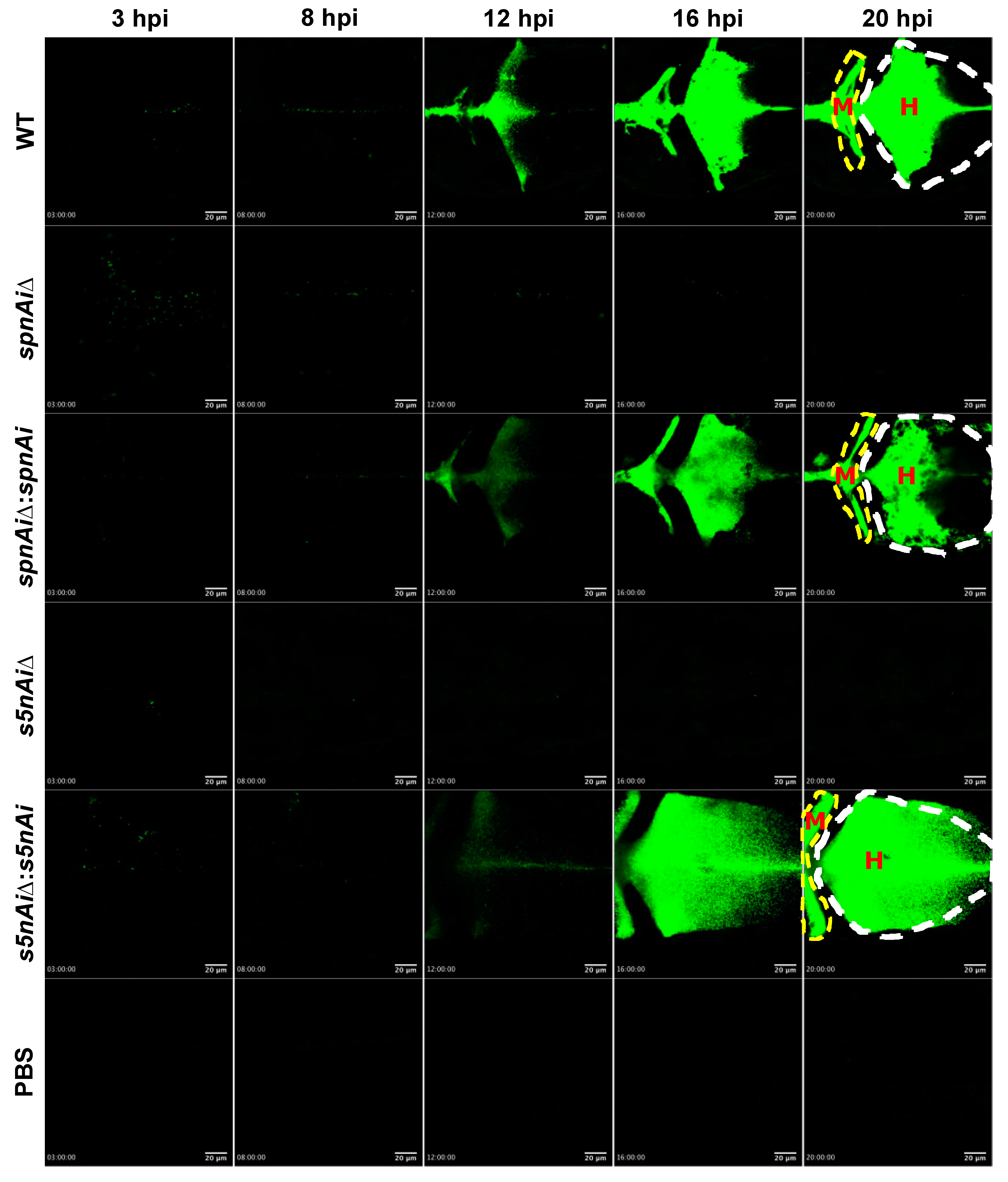
To visualize the survival of S. iniae in the presence or absence of SpnAi or S5nAi at the site of infection, approximately 50 CFU of green fluorescent-labeled S. iniae or mutant strains were individually injected into the hindbrain ventricle of 2 dpf larvae (wild-type AB line). An additional group of larvae were injected with sterile PBS as a control for this experiment. The hindbrain ventricle of each infected or control larvae in dorsal view (Figure S3, bottom panel) was imaged using a confocal microscope from 3 to 21 hpi (Figure 6; see Movies S1 in the supplemental materials). Overall, the area of green fluorescence increased in WT S. iniae-infected larvae over the 21 h time-course, particularly starting from 12 hpi (Figure 6). Similar observations were made in larvae infected with ∆spnAi:spnAi and ∆s5nAi:s5nAi but not ∆spnAi and ∆s5nAi. Furthermore, it was also observed that the WT and ∆spnAi:spnAi and ∆s5nAi:s5nAi strains disseminated from the hindbrain ventricle (Figure 6, labeled as H) to the midbrain ventricle (Figure 6, labeled as M) of the larvae.
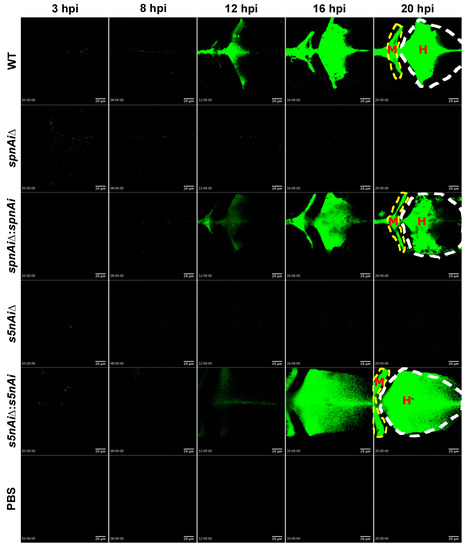
Figure 6.
Time-lapse confocal imaging of 2 dpf zebrafish larvae infected with green fluorescent-labeled S. iniae or mutant strains. Confocal microscopy was used to observe the proliferation and dissemination of green fluorescent-labeled S. iniae at the infection site. Larvae (wild-type AB line) were injected with 50 colony-forming units (CFU) of gfpmut2-labelled WT, ∆spnAi, ∆spnAi:spnAi, ∆s5nAi, ∆s5nAi:s5nAi and sterile PBS at the hindbrain ventricle. Images of the hindbrain ventricle in dorsal view were taken from 3 to 21 hpi using a confocal microscope with a 20× objective lens. Five frames at 3, 8, 12, 16 and 20 hpi were extracted from the individual movies. H, hindbrain ventricle; M, midbrain ventricle. Scale bar, 20 µm. In the supplemental materials, see Movies S1. Results are representative of 2 independent experiments.
3.7. Zebrafish Larvae Infected with S. iniae ∆SpnAi Showed an Increasing Number of Neutrophils at the Site of Infection over Time
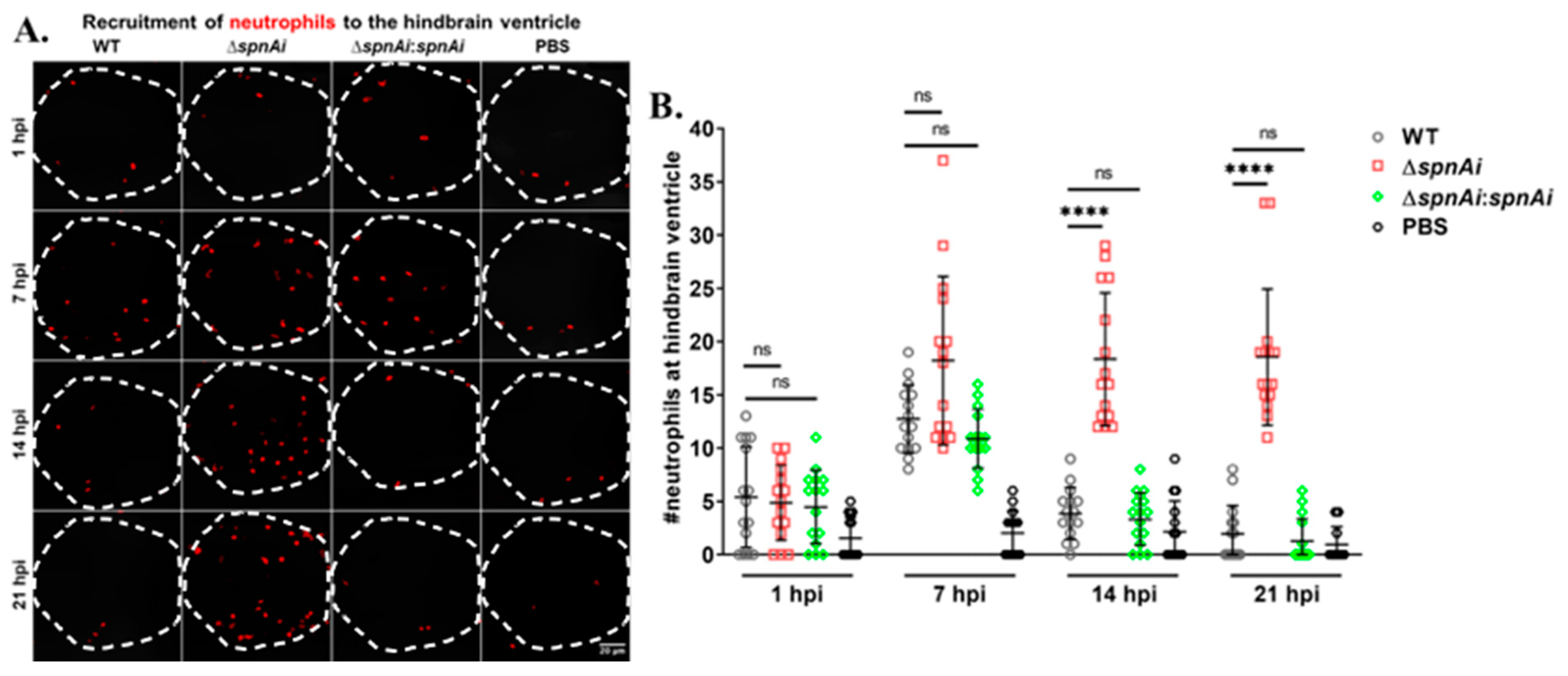
SpnAi was shown to digest NETs released from activated neutrophils (Figure 2). Therefore, we were interested to investigate the effect of SpnAi on neutrophil recruitment to the site of infection. In order to quantify the number of neutrophils recruited to the hindbrain ventricle at various time-points after infection, Tg(lyz:DsRED2)nz50 larvae infected with WT or mutant S. iniae strains were fixed at 1, 7, 14 and 21 hpi. An additional group of larvae were injected with sterile PBS as a control for this experiment. Neutrophils were rendered red by immunostaining, and neutrophils recruited to the hindbrain ventricle were quantified by confocal microscopy (Figure 7). The results showed a slight increase in the number of neutrophils recruited to the S. iniae infection site as compared to control larvae at 1 hpi, indicating that neutrophils responded to S. iniae infection immediately at the hindbrain ventricle. At 7 hpi, the number of neutrophils recruited to the hindbrain ventricle of infected larvae continued to increase with no significant difference in the neutrophils number between the groups of S. iniae infected larvae. A significant difference in the number of neutrophils at the hindbrain ventricles was seen between larvae infected with WT and ∆spnAi at 14 hpi and 21 hpi (p < 0.0001). The number of neutrophils at the hindbrain ventricle of larvae infected with WT and ∆spnAi:spnAi was significantly lower as compared to ∆spnAi at 14 hpi and 21 hpi.
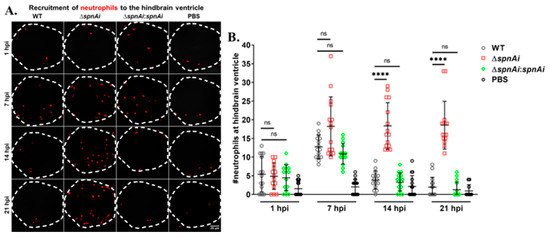
Figure 7.
Quantification of neutrophils recruited at the site of S. iniae infection. (A) Immunofluorescence detection of neutrophils recruited to the hindbrain ventricle of Tg(lyz:DsRED2)nz50 larvae infected with 50 CFU of WT (first column), ∆spnAi (second column), ∆spnAi:spnAi (third column) or sterile PBS (last column) at 1 hpi (first row), 7 hpi (second row), 14 hpi (third row) and 21 hpi (last row). The larvae (n = 15 per group per time-point) were infected at 2 dpf and fixed at the indicated time-points. A confocal microscope was used to image immunostained neutrophils at the hindbrain ventricle of each larvae in dorsal view (white dotted line). Scale bar, 20 µm. (B) Quantification of neutrophils in the hindbrain ventricle of individual larvae. ****, p < 0.0001; ns, not significant (as determined by a 2-way ANOVA with a Tukey’s multiple comparisons test).
3.8. Zebrafish Larvae Infected with S. iniae ∆S5nAi Showed an Increasing Number of Macrophages at the Site of Infection over Time
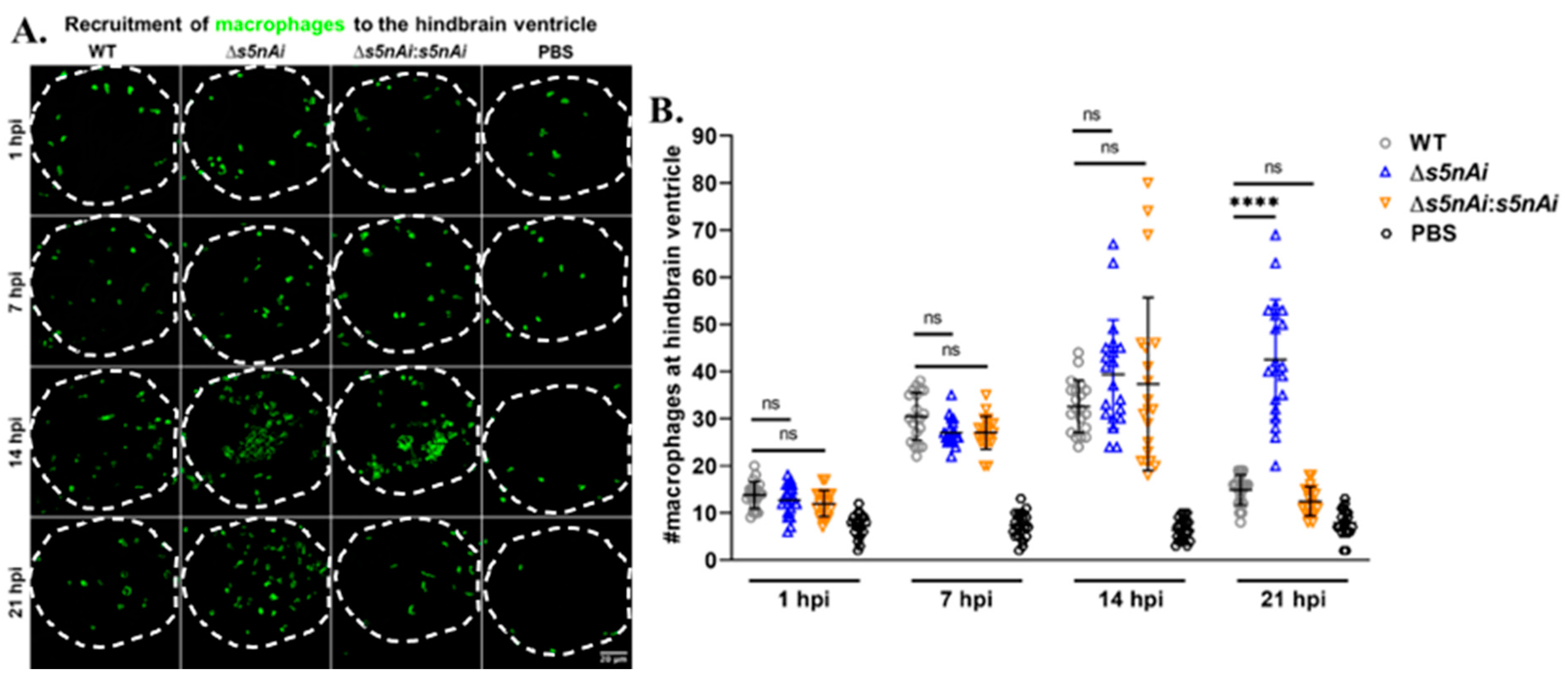
We have previously shown that S5nAi is able to convert dAMP into dAdo [18], which is known to be cytotoxic for macrophages [49]. Therefore, we analyzed the in vivo function of S5nAi in relation to the recruitment of macrophages to the infection site. The number of macrophages recruited to the hindbrain ventricle of Tg(mpeg1:EGFP)gl22 larvae infected with WT or mutant strains at various time-points post-infection was quantified by immunostaining and confocal microscopy (Figure 8). During the first hour post-infection, an increase in the number of macrophages recruited to the infection site was observed in comparison to control larvae, but no significant difference was seen between all infected larvae. As the time progressed, the number of macrophages at the infection site increased as seen at 7 hpi and 14 hpi for all infected larvae. No significant difference in the number of macrophages recruited to the hindbrain ventricle of all infected larvae was observed up to 14 hpi, though at 21 hpi the number of macrophages decreased in larvae infected with WT and ∆s5nAi:s5nAi, but not with ∆s5nAi.
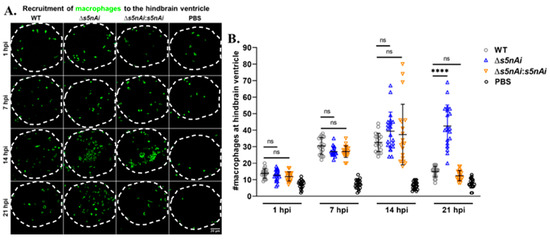
Figure 8.
Quantification of macrophages recruited to the site of S. iniae infection. (A) Immunofluorescence detection of macrophage recruited to the hindbrain ventricle of Tg(mpeg1:EGFP)gl22 larvae infected with 50 CFU of WT (first column), Δs5nAi (second column), ∆s5nAi:s5nAi (third column) or sterile PBS (last column) at 1 hpi (first row), 7 hpi (second row), 14 hpi (third row) and 21 hpi (last row). The larvae (n = 20 per group per time-point) were infected at 2 dpf and fixed at indicated time-points. Confocal microscopy was used to image immunostained macrophages at the hindbrain ventricle of each larvae in the dorsal view (white dotted line). Scale bar, 20 μm. (B) Quantification of macrophages in the hindbrain ventricle of individual larvae. An increase in the number of macrophages recruited to the hindbrain ventricle of infected larvae was seen compared to larvae injected with sterile PBS. The number of macrophages at the infection site for all infected larvae peaked at 14 hpi. At 21 hpi, the number of macrophages recruited to the infection site of larvae infected with WT and ∆s5nAi:s5nAi was significantly lower as compared to Δs5nAi. ****, p < 0.0001; ns, not significant (as determined by a 2-way ANOVA with a Tukey’s multiple comparisons test).
4. Discussion
S. iniae is a pathogenic and zoonotic bacterium that threatens both human health and aquaculture industries. Numerous S. iniae virulence factors have been identified with the advances in molecular biology techniques. Elucidating the function of these virulence factors in S. iniae pathogenesis is essential to control infection and for identifying effective vaccine targets. We have recently reported the functional characterization of recombinant forms of two novel S. iniae virulence factors; an extracellular nuclease (SpnAi) and a secreted 5′-nucleotidase (S5Ai) [18]. Here, we have shown that both enzymatic activities can be found in live S. iniae. Furthermore, both activities could be reduced by deletion of the corresponding genes in the S. iniae genome by allelic replacement, confirming the function previously evaluated using recombinant proteins [18].
Deletion of spnAi in the S. iniae strain 9117 did not completely inactivate DNase activity suggesting that S. iniae is able to produce other DNase(s). This has also been reported in other Gram-positive cocci such as the production of Nuc1 [50] and Nuc2 [51] by S. aureus, and SpnA [16,52] and Sda1 [53] by S. pyogenes. However, the major decrease of S. iniae DNase activity seen with the spnAi deletion mutant suggests that SpnAi is the main enzyme responsible for the nuclease activity. Like other streptococcal DNases, such as EndA of S. pneumoniae [21], SpnA of S. pyogenes [16] and SsnA of S. suis [47], SpnAi also cleaves NETs released from neutrophils, confirming a role as an immune evasion virulence factor. Notably, the deletion of spnAi not only showed the absence of NET degradation activity, but also an increase in the production of NETs when the deletion strain was added to neutrophils. This is expected as known triggers of NET release (NETosis), include PMA, lipopolysaccharide and living/dead bacterial cells [54]. Individual stimulus triggers diverse NETosis pathways, and it has recently been shown that Streptococcus agalactiae uses a similar NETosis pathway to PMA [55].
The role in virulence and immune evasion of these novel virulence factors was further highlighted in a zebrafish infection model, which has been widely used to investigate S. iniae [56] and other pathogenic bacteria [57,58,59]. Advantages of this infection model include a fully sequenced genome showing significant homology with the human genome [60,61], rapid breeding and transparency of larvae during early life stages. The transparency of the larvae allows time-lapse imaging of biological and disease processes to be carried out. Zebrafish infection studies have shown that both neutrophils and macrophages play important roles in the innate immune response against S. iniae [56,62]. However, most infection studies with S. iniae uses wild-type bacteria, which are not visible using standard imaging methods. A previous study establishing a S. iniae-zebrafish infection model used the CellTracker Red CMPTX dye (Thermo Fisher Scientific, Waltham, MA, USA) for staining, but reported the inability to reliably track S. iniae in vivo because the dye became diluted or disappeared as bacteria divided [56]. Furthermore, the same study was unable to accurately measure the S. iniae burden in the host since non-selective agar plates were used [56], which allowed the growth of other Gram-positive bacteria that were present as part of the normal zebrafish gut microbiota [63]. Here, we reported for the first time the stable green fluorescence labeling of S. iniae, by introducing a novel toxin-antitoxin (TA) stabilized green fluorescent reporter plasmid [42,64,65]. The fluorescent labeling of S. iniae enables the reliable and non-invasive tracking of S. iniae in vivo, due to the post-segregational killing of those bacteria that have lost the plasmid. In addition, the reporter plasmid also contained a kanamycin resistance gene that allowed for the selective growth of S. iniae.
In vivo survival analysis using the zebrafish larvae infection model demonstrated a significant reduction in the virulence of the S. iniae ΔspnAi and ∆s5nAi mutants compared to the parental strain. The reintroduction of the wild-type genes into the S. iniae gene deletion strains effectively restored the enzymatic activity, suggesting that gene deletion did not cause any down-stream effects. The attenuated virulence of S. iniae ΔspnAi and ∆s5nAi in the larval zebrafish model was also evident in the relatively constant bacterial load, compared to the WT S. iniae and the complementation strains where an increase in abundance was seen in the first 21 hpi. These results corroborate the confocal time-lapse imaging, where increased bacterial load was seen throughout the 21 hpi in larvae infected with S. iniae WT and complementation strains, but not the ∆spnAi and ∆s5nAi mutants. These results suggest that WT S. iniae was able to escape from killing by innate immune cells, allowing the bacterium to multiply at the infection site and disseminate to the midbrain of the larvae or even to other parts of the body. These results again indicate that SpnAi and S5nAi play an important role in contributing to S. iniae virulence.
Notably, the number of recoverable S. iniae ∆spnAi and ∆s5nAi bacteria from the whole larvae was maintained at 1000 CFU at 9–21 hpi, which was higher in contrast to the number of ∆spnAi and ∆s5nAi mutants that were rapidly cleared from the hindbrain ventricle. This indicates that some S. iniae ∆spnAi and ∆s5nAi mutants were able to somehow survive and disseminate from the infection site to other parts of the larvae, but not proliferate. It has previously been shown that S. iniae is able to survive intracellularly in macrophages of mice [66] and salmon [67], thereby providing an opportunity for bacterial survival and dissemination throughout the host. However, any details of this proposed mechanism in vivo have yet to be elucidated.
To take further advantage of the optically transparent properties of larval zebrafish, the transgenic zebrafish strain Tg(lyz:DsRED2)nz50 possessing red fluorescent neutrophils were used to investigate the in vivo function of SpnAi. There was no detectable difference between WT and mutant strains in stimulating infiltration of neutrophils to the site of infection at 1 hpi and 7 hpi. Based on our in vitro results, it is possible that destruction of NETs plays an important role in the low neutrophil phagocytosis rate of WT S. iniae. Notably, the number of neutrophils at the site of infection decreased at 14 hpi with WT, but not the spnAi deletion mutant suggesting an additional function of SpnAi apart from nuclease activity. We have recently shown that S. pyogenes SpnA catalytic site mutants lacking nuclease activity were still virulent in a Galleria mellonella (wax worm) infection model indicating a nuclease-independent virulence mechanism [68]. Like SpnA [16], SpnAi consists of two large domains, a C-terminal nuclease domain and a N-terminal domain of unknown function. Further evaluations are required to decipher a potential role of the N-terminal domain in prevention of neutrophil recruitment or induction of neutrophil death.
It has previously been reported that adenosine synthase A (AdsA), a secreted 5′-nucleotidase expressed by S. aureus, generates dAdo from dAMP, which causes apoptotic killing of macrophages [31]. We have therefore used a transgenic zebrafish strain Tg(mpeg1:EGFP)gl22 that possesses green fluorescent macrophages to investigate the in vivo function of S5nAi. Similar to the effect of SpnAi on neutrophils, no difference in macrophage recruitment was detected at 7 hpi between WT and s5nAi mutant strains. However, WT S. iniae, but not the ∆s5nAi mutant, reduced macrophage infiltration at 21 hpi. This is in line with previous observations that AdsA prevented macrophage infiltration into staphylococcal abscesses in a mouse infection model due to apoptotic killing mediated by dAdo [69]. S5nAi might have a similar role and the delayed effect may be due to a slow decay of the fluorescent signal after macrophage killing.
A number of virulence factors were discovered following the complete sequencing of the S. iniae 9117 genome [8]. In addition to SpnAi and S5nAi, many other documented S. iniae virulence factors are also homologous to those expressed by the important solely human pathogen, S. pyogenes [5], such as α-enolase [70], streptolysin S [14] and the hyaluronic acid capsule [11] that showed 97%, 73% and 71% in amino acid sequence identity to S. iniae, respectively. Therefore, the establishment of a S. iniae-zebrafish infection model with the use of fluorescently labeled S. iniae in this study also allows for modeling homologues of S. pyogenes virulence factors in a natural host system. This would therefore also aid the discovery and exploration of functional relationships of streptococcal virulence factors.
5. Conclusions
We have shown that the previously reported in vitro enzymatic activities of recombinant S. iniae SpnAi and S5nAi are responsible for virulence traits in S. iniae using zebrafish infection models. Our work further establishes SpnAi and S5nAi as important immune evasion factors that might be potential targets for the development of therapeutic agents or vaccines against S. iniae infections, both in the aquaculture industry and in human health.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/9/1361/s1, Figure S1: Allelic exchange mutagenesis of spnAi and s5nAi, Figure S2: Plasmid construction for the generation of green fluorescent S. iniae spnAi and s5nAi complementation strains, Figure S3: Lateral and dorsal view of a 2 dpf zebrafish larva, Figure S4: Growth rates of S. iniae strains. Movie S1: Time-lapse confocal imaging of 2 dpf zebrafish larvae infected with stably green fluorescent labeled S. iniae or mutant strains.
Author Contributions
All authors have contributed substantially to the work reported. Conceptualization, T.P. and C.H.; methodology, K.Y.S.; formal analysis, K.Y.S., C.H., J.M.S.L. and T.P.; investigation, K.Y.S.; resources, T.P. and C.H.; data curation, K.Y.S., C.H., J.M.S.L. and T.P.; writing—original draft preparation, K.Y.S.; writing—review and editing, K.Y.S., T.P., C.H., and J.M.S.L.; supervision, T.P., C.H., and J.M.S.L.; project administration, T.P.; funding acquisition, T.P. and K.Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by a Graduate Student fund from The University of Auckland. K.Y.S. was supported by The University of Auckland Doctoral Scholarship. J.M.S.L. is the recipient of a New Zealand National Heart Foundation Senior Research Fellowship.
Acknowledgments
We thank Sarah Highlander (JCVI, La Jolla, CA, USA) for sending us the S. iniae 9117 strain, Andreas Podbielski (University of Rostock, Rostock, Germany) for sending us the pFW11 vector and Jason McArthur (University of Wollongong, Wollongong, NSW, Australia) for the pLZ12Km2-P23R:TA:gfpmut2 plasmid. We also thank Hannah Darroch and Pramuk Keerthisinghe for providing the zebrafish larvae in this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pier, G.B.; Madin, S.H. Streptococcus iniae sp. nov., a beta-hemolytic streptococcus isolated from an Amazon. freshwater dolphin, Inia geoffrensis. Int. J. Syst. Evol. Microbiol. 1976, 26, 545–553. [Google Scholar] [CrossRef]
- Agnew, W.; Barnes, A.C. Streptococcus iniae: An aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet. Microbiol. 2007, 122, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.A.; Klesius, P.H.; Evans, J.J. Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States. Am. J. Vet. Res. 2001, 62, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Luk, W.-k.; Fung, A.M.; Hui, W.-t.; Fong, A.H.; Chow, C.-w.; Wong, S.S.; Yuen, K.-y. Clinical isolates of Streptococcus iniae from Asia are more mucoid and β-hemolytic than those from North. America. Diagn. Microbiol. Infect. Dis. 2006, 54, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Baiano, J.C.; Barnes, A.C. Towards control of Streptococcus iniae. Emerg. Infect. Dis. 2009, 15, 1891–1896. [Google Scholar] [CrossRef]
- McMillan, D.J.; Sanderson-Smith, M.L.; Smeesters, P.R.; Sriprakash, K.S. Molecular Markers for the Study of Streptococcal Epidemiology. In Host-Pathogen Interactions in Streptococcal Diseases; Springer: Berlin/Heidelberg, Germany, 2012; pp. 29–48. [Google Scholar]
- Sun, J.-R.; Yan, J.-C.; Yeh, C.-Y.; Lee, S.-Y.; Lu, J.-J. Invasive infection with Streptococcus iniae in Taiwan. J. Med. Microbiol. 2007, 56, 1246–1249. [Google Scholar] [CrossRef]
- Richards, V.P.; Palmer, S.R.; Bitar, P.D.P.; Qin, X.; Weinstock, G.M.; Highlander, S.K.; Town, C.D.; Burne, R.A.; Stanhope, M.J. Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol. Evol. 2014, 6, 741–753. [Google Scholar] [CrossRef]
- Baiano, J.C.; Tumbol, R.A.; Umapathy, A.; Barnes, A.C. Identification and molecular characterisation of a fibrinogen binding protein from Streptococcus iniae. BMC Microbiol. 2008, 8, 1–16. [Google Scholar] [CrossRef]
- Locke, J.B.; Aziz, R.K.; Vicknair, M.R.; Nizet, V.; Buchanan, J.T. Streptococcus iniae M-like protein contributes to virulence in fish and is a target for live attenuated vaccine development. PLoS ONE 2008, 3, e2824. [Google Scholar] [CrossRef]
- Locke, J.B.; Colvin, K.M.; Datta, A.; Patel, S.K.; Naidu, N.N.; Neely, M.N.; Nizet, V.; Buchanan, J.T. Streptococcus iniae Capsule Impairs Phagocytic Clearance and Contributes to Virulence in Fish. J. Bacteriol. 2006, 189, 1279–1287. [Google Scholar] [CrossRef]
- Lowe, B.A.; Miller, J.D.; Neely, M.N. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect. Immun. 2007, 75, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Neely, M.N. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect. Immun. 2005, 73, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, S.; Fuller, J.D.; Bast, D.J.; de Azavedo, J.C. The two-component system sivS/R regulates virulence in Streptococcus iniae. FEMS Immunol. Med. Microbiol. 2007, 51, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Fuller, J.D.; Camus, A.C.; Duncan, C.L.; Nizet, V.; Bast, D.J.; Thune, R.L.; Low, D.E.; de Azavedo, J.C. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect. Immun. 2002, 70, 5730–5739. [Google Scholar] [CrossRef]
- Chang, A.; Khemlani, A.; Kang, H.; Proft, T. Functional analysis of Streptococcus pyogenes nuclease A (SpnA), a novel group A streptococcal virulence factor. Molec. Microbiol. 2011, 79, 1629–1642. [Google Scholar] [CrossRef]
- Zheng, L.; Khemlani, A.; Lorenz, N.; Loh, J.M.; Langley, R.J.; Proft, T. Streptococcal 5′-Nucleotidase A (S5nA), a novel Streptococcus pyogenes virulence factor that facilitates immune evasion. J. Biol. Chem. 2015, 290, 31126–31137. [Google Scholar] [CrossRef]
- Soh, K.Y.; Loh, J.M.S.; Proft, T. Orthologues of Streptococcus pyogenes nuclease A (SpnA) and Streptococcal 5′-nucleotidase A (S5nA) found in Streptococcus iniae. J. Biochem. 2018, 164, 165–171. [Google Scholar] [CrossRef]
- Berends, E.T.; Horswill, A.R.; Haste, N.M.; Monestier, M.; Nizet, V.; von Köckritz-Blickwede, M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2010, 2, 576–586. [Google Scholar] [CrossRef]
- Buchanan, J.T.; Simpson, A.J.; Aziz, R.K.; Liu, G.Y.; Kristian, S.A.; Kotb, M.; Feramisco, J.; Nizet, V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 2006, 16, 396–400. [Google Scholar] [CrossRef]
- Beiter, K.; Wartha, F.; Albiger, B.; Normark, S.; Zychlinsky, A.; Henriques-Normark, B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 2006, 16, 401–407. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Cekic, C.; Linden, J. Purinergic regulation of the immune system. Nat. Rev. Immun. 2016, 16, 177. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, A. Adenosine: An endogenous modulator of innate immune system with therapeutic potential. Eur. J. Pharm. 2009, 616, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Hasko, G.; Szabo, C.; Nemeth, Z.H.; Kvetan, V.; Pastores, S.M.; Vizi, E.S. Adenosine receptor agonists differentially regulate IL-10, TNF-alpha, and nitric oxide production in RAW 264.7 macrophages and in endotoxemic mice. J. Immun. 1996, 157, 4634–4640. [Google Scholar] [PubMed]
- Cronstein, B.N.; Kramer, S.B.; Weissmann, G.; Hirschhorn, R. Adenosine: A physiological modulator of superoxide anion generation by human neutrophils. J. Exp. Med. 1983, 158, 1160–1177. [Google Scholar] [CrossRef]
- Xaus, J.; Mirabet, M.; Lloberas, J.; Soler, C.; Lluis, C.; Franco, R.; Celada, A. IFN-γ up-regulates the A2B adenosine receptor expression in macrophages: A mechanism of macrophage deactivation. J. Immun. 1999, 162, 3607–3614. [Google Scholar] [PubMed]
- Bouma, M.G.; Jeunhomme, T.; Boyle, D.L.; Dentener, M.A.; Voitenok, N.N.; Van den Wildenberg, F.; Buurman, W.A. Adenosine inhibits neutrophil degranulation in activated human whole blood: Involvement of adenosine A2 and A3 receptors. J. Immun. 1997, 158, 5400–5408. [Google Scholar]
- Soh, K.Y.; Loh, J.M.; Proft, T. Cell wall-anchored 5′-nucleotidases in Gram-positive cocci. Mol. Microbiol. 2019, 113, 691–698. [Google Scholar] [CrossRef]
- Thammavongsa, V.; Schneewind, O.; Missiakas, D.M. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA). BMC Biochem. 2011, 12, 56. [Google Scholar] [CrossRef]
- Thammavongsa, V.; Missiakas, D.M.; Schneewind, O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 2013, 342, 863–866. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, Y.; Chuang-Smith, O.N.; Frank, K.L.; Guenther, B.D.; Kern, M.; Schlievert, P.M.; Herzberg, M.C. Ecto-5′-nucleotidase: A candidate virulence factor in Streptococcus sanguinis experimental endocarditis. PLoS ONE 2012, 7, e38059. [Google Scholar] [CrossRef]
- Liu, P.; Pian, Y.; Li, X.; Liu, R.; Xie, W.; Zhang, C.; Zheng, Y.; Jiang, Y.; Yuan, Y. Streptococcus suis adenosine synthase functions as an effector in evasion of PMN-mediated innate immunity. J. Infect. Dis. 2014, 210, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Lai, L.; Tang, H.; Wang, W.; Wang, S.; Lu, C.; Yao, H.; Fan, H.; Wu, Z. Streptococcus suis synthesizes deoxyadenosine and adenosine by 5′-nucleotidase to dampen host immune responses. Virulence 2018, 9, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Herbomel, P.; Thisse, B.; Thisse, C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Blood 1999, 126, 3735–3745. [Google Scholar]
- Hermann, A.C.; Millard, P.J.; Blake, S.L.; Kim, C.H. Development of a respiratory burst assay using zebrafish kidneys and embryos. J. Immunol. Methods 2004, 292, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Le Guyader, D.; Redd, M.J.; Colucci-Guyon, E.; Murayama, E.; Kissa, K.; Briolat, V.; Mordelet, E.; Zapata, A.; Shinomiya, H.; Herbomel, P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 2008, 111, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Zon, L.I. The ‘definitive’(and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 2004, 23, 7233–7246. [Google Scholar] [CrossRef]
- Lam, S.; Chua, H.; Gong, Z.; Lam, T.; Sin, Y. Development and maturation of the immune system in zebrafish, Danio rerio: A gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 2004, 28, 9–28. [Google Scholar] [CrossRef]
- Trede, N.S.; Langenau, D.M.; Traver, D.; Look, A.T.; Zon, L.I. The use of zebrafish to understand immunity. Immunity 2004, 20, 367–379. [Google Scholar] [CrossRef]
- Podbielski, A.; Spellerberg, B.; Woischnik, M.; Pohl, B.; Lütticken, R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 1996, 177, 137–147. [Google Scholar] [CrossRef]
- Loh, J.M.; Proft, T. Toxin–antitoxin-stabilized reporter plasmids for biophotonic imaging of Group A streptococcus. Appl. Microbiol. Biotechnol. 2013, 97, 9737–9745. [Google Scholar] [CrossRef] [PubMed]
- Sinicropi, D.; Baker, D.L.; Prince, W.S.; Shiffer, K.; Shak, S. Colorimetric determination of DNase I activity with a DNA-methyl green substrate. Anal. Biochem. 1994, 222, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Flores, M.V.; Storm, T.; Crosier, K.; Crosier, P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, 49–56. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- de Buhr, N.; Neumann, A.; Jerjomiceva, N.; von Köckritz-Blickwede, M.; Baums, C.G. Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 2014, 160, 385–395. [Google Scholar] [CrossRef]
- Jhelum, H.; Sori, H.; Sehgal, D. A novel extracellular vesicle-associated endodeoxyribonuclease helps Streptococcus pneumoniae evade neutrophil extracellular traps and is required for full virulence. Sci. Rep. 2018, 8, 7985. [Google Scholar] [CrossRef]
- Fox, I.H.; Kelley, W.N. The role of adenosine and 2’-deoxyadenosine in mammalian cells. Annu. Rev. Biochem. 1978, 47, 655–686. [Google Scholar] [CrossRef]
- Tucker, P.W.; Hazen, E.E.; Cotton, F.A. Staphylococcal nuclease reviewed: A prototypic study in contemporary enzymology. I isolation; physical and enzymatic properties. Mol. Cell. Biochem. 1978, 22, 67–78. [Google Scholar] [CrossRef]
- Hu, Y.; Meng, J.; Shi, C.; Hervin, K.; Fratamico, P.M.; Shi, X. Characterization and comparative analysis of a second thermonuclease from Staphylococcus aureus. Microbiol. Res. 2013, 168, 174–182. [Google Scholar] [CrossRef]
- Hasegawa, T.; Minami, M.; Okamoto, A.; Tatsuno, I.; Isaka, M.; Ohta, M. Characterization of a virulence-associated and cell-wall-located DNase of Streptococcus pyogenes. Microbiology 2010, 156, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Ismail, S.A.; Park, H.W.; Kotb, M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 2004, 54, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Hoppenbrouwers, T.; Autar, A.S.; Sultan, A.R.; Abraham, T.E.; van Cappellen, W.A.; Houtsmuller, A.B.; van Wamel, W.J.; van Beusekom, H.M.; van Neck, J.W.; de Maat, M.P. In vitro induction of NETosis: Comprehensive live imaging comparison and systematic review. PLoS ONE 2017, 12, e0176472. [Google Scholar] [CrossRef] [PubMed]
- Kenny, E.F.; Herzig, A.; Krüger, R.; Muth, S.; Mondal, A.; Thompson, P.R.; Brinkmann, V.; Von Bernuth, H.; Zychlinsky, A. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife 2017, 6, e24437. [Google Scholar] [CrossRef]
- Harvie, E.A.; Green, J.M.; Neely, M.N.; Huttenlocher, A. Innate immune response to Streptococcus iniae infection in zebrafish larvae. Infect. Immun. 2013, 81, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.N.; Pfeifer, J.D.; Caparon, M. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 2002, 70, 3904–3914. [Google Scholar] [CrossRef]
- Astin, J.; Keerthisinghe, P.; Du, L.; Sanderson, L.; Crosier, K.; Crosier, P.; Hall, C. Innate immune cells and bacterial infection in zebrafish. Methods Cell Biol. 2017, 138, 31–60. [Google Scholar]
- Miller, J.D.; Neely, M.N. Zebrafish as a model host for streptococcal pathogenesis. Acta Trop. 2004, 91, 53–68. [Google Scholar] [CrossRef]
- Shin, J.T.; Priest, J.R.; Ovcharenko, I.; Ronco, A.; Moore, R.K.; Burns, C.G.; MacRae, C.A. Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res. 2005, 33, 5437–5445. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Vincent, W.J.; Harvie, E.A.; Sauer, J.-D.; Huttenlocher, A. Neutrophil derived LTB4 induces macrophage aggregation in response to encapsulated Streptococcus iniae infection. PLoS ONE 2017, 12, e0179574. [Google Scholar] [CrossRef] [PubMed]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.M.; Proft, T. Stabilized plasmid-based system for bioluminescent labeling of multiple streptococcal species. Biotechnol. Lett. 2016, 38, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S.J.G. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar] [CrossRef]
- Allen, J.P.; Neely, M.N. The Streptococcus iniae transcriptional regulator CpsY is required for protection from neutrophil-mediated killing and proper growth in vitro. Infect. Immun. 2011, 79, 4638–4648. [Google Scholar] [CrossRef]
- Zlotkin, A.; Chilmonczyk, S.; Eyngor, M.; Hurvitz, A.; Ghittino, C.; Eldar, A. Trojan horse effect: Phagocyte-mediated Streptococcus iniae infection of fish. Infect. Immun. 2003, 71, 2318–2325. [Google Scholar] [CrossRef]
- Chalmers, C.; Khemlani, A.; Sohn, C.R.; Loh, J.M.; Proft, T. Streptococcus pyogenes nuclease A (SpnA) mediated virulence does not exclusively depend on nuclease activity. J. Microbiol. Immunol. Infect. 2020, 53, 42–48. [Google Scholar] [CrossRef]
- Thammavongsa, V.; Kern, J.W.; Missiakas, D.M.; Schneewind, O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J. Exp. Med. 2009, 206, 2417–2427. [Google Scholar] [CrossRef]
- Kim, M.S.; Choi, S.H.; Lee, E.H.; Nam, Y.K.; Kim, S.K.; Kim, K.H. α-enolase, a plasmin (ogen) binding and cell wall associating protein from a fish pathogenic Streptococcus iniae strain. Aquaculture 2007, 265, 55–60. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).