Response of Barley Plants to Drought Might Be Associated with the Recruiting of Soil-Borne Endophytes
Abstract
1. Introduction
2. Material and Methods
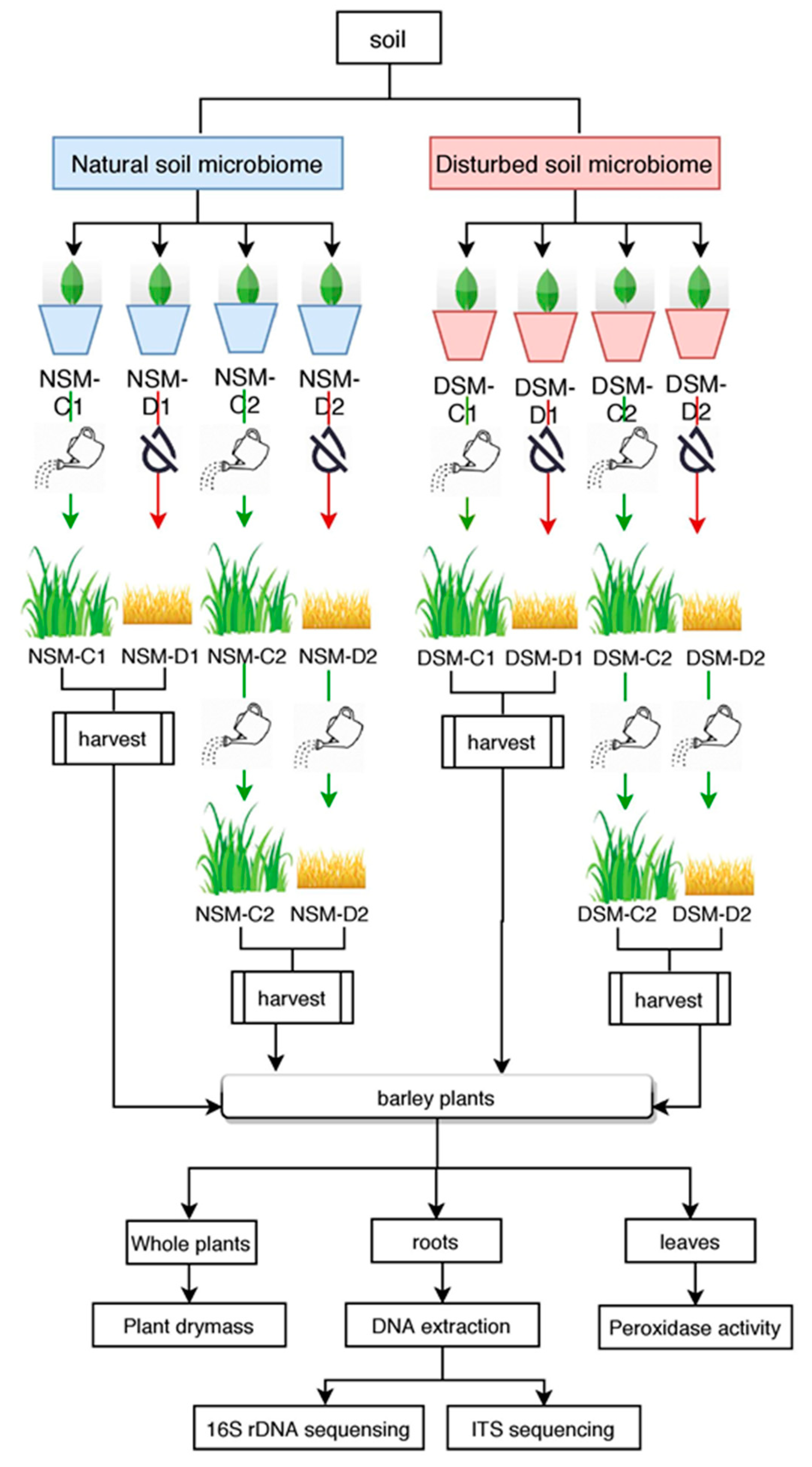
2.1. Experimental Design
2.2. Plants Harvesting
2.3. Peroxidase Activity in Barley Leaves
2.4. DNA Extraction and Sequencing Library Preparation
2.5. Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
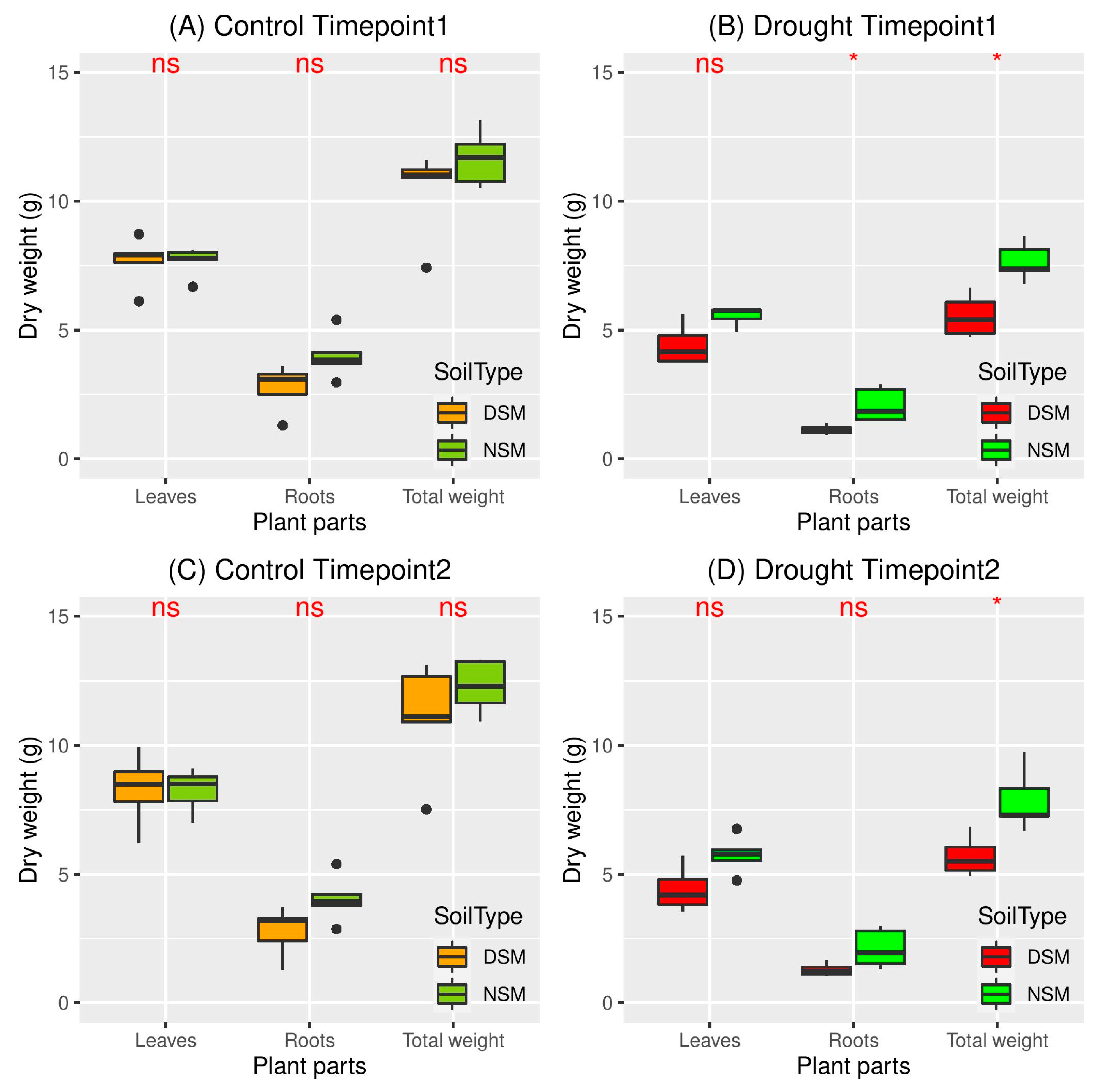
3.1. Effects of Drought on Plant Performance in Natural and Disturbed Soils
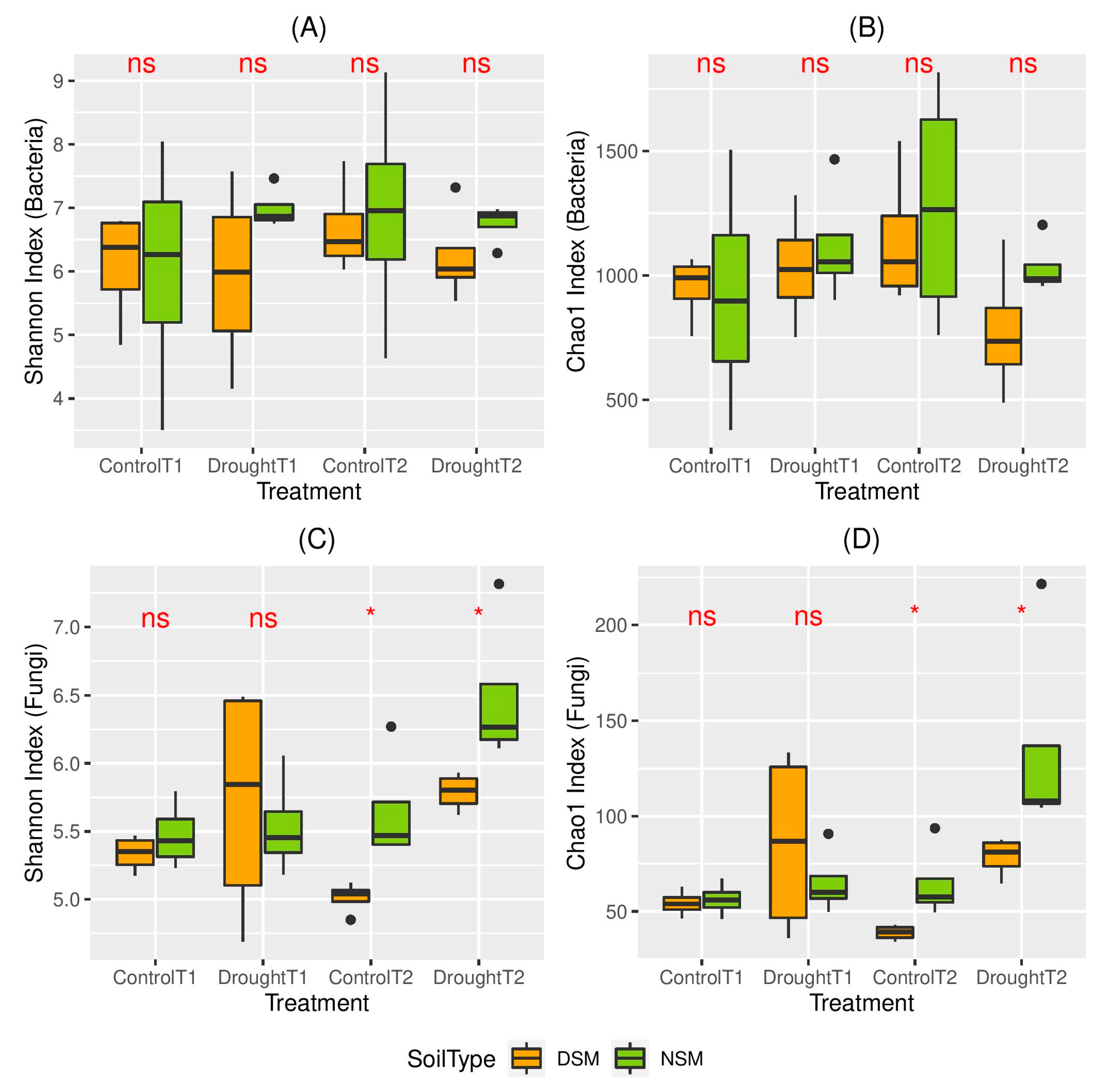
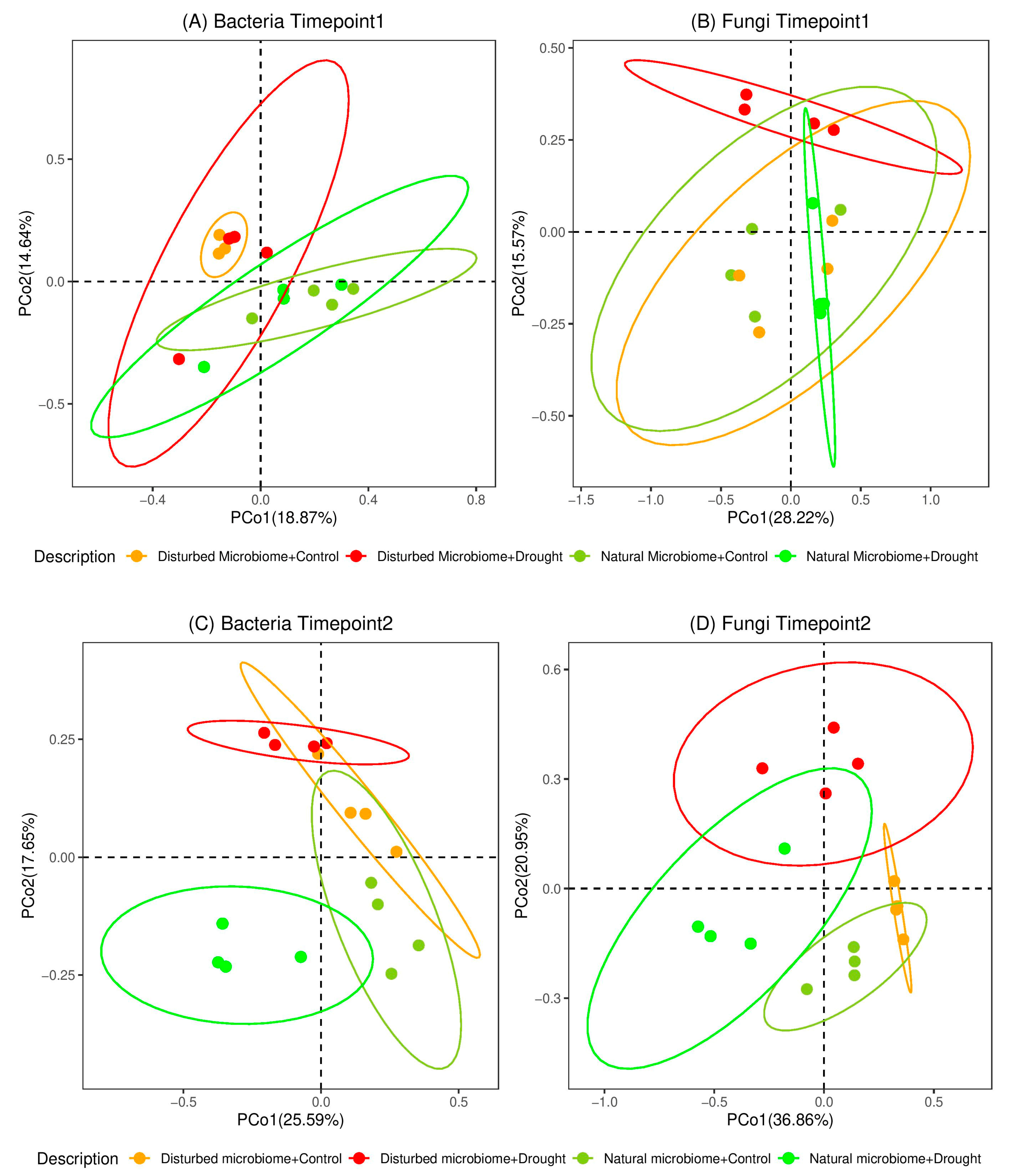
3.2. Effects of Drought on the Diversity of the Root-Associated Microbiome
3.3. Effects of Drought on Endophytic Bacteria
3.4. Effects of Drought on Endophytic Fungi
4. Discussion
4.1. Drought Stress Differently Affect Plant Growth in Soils with Natural and Disturbed Microbiome
4.2. The Diversity of the Root-Associated Microbiome Did Not Change Directly After Drought Stress
4.3. Enrichment of Soil Microbes in the Roots of Drought Stressed Plants
4.4. Seed-Borne Organisms Were the Major Drought Responders in Soils with Disturbed Microbiome
4.5. Changes in the Composition of Root-Associated Endophytes from Drought Alleviated Plants
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Hussain, M.; Wahid, A.; Siddique, K.H.M. Drought Stress in Plants: An Overview. In Plant Responses to Drought Stress from Morphological to Molecular Features; Aroca, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–33. [Google Scholar]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sehrish, S.; Wajid, N.; Steve, A.; Shah, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.R.; Pereira, A. Molecular and Physiological Analysis of Drought Stress in Arabidopsis Reveals Early Responses Leading to Acclimation in Plant Growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Gemenet, D.C.; Villordon, A. Root System Architecture and Abiotic Stress Tolerance: Current Knowledge in Root and Tuber Crops. Front. Plant Sci. 2016, 7, 1584. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Rev. Environ. Sci. Bio/Technol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Cruz de Carvalho, M.H. Drought stress and reactive oxygen species. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef]
- Ngumbi, E.; Kloepper, J. Bacterial-mediated drought tolerance: Current and future prospects. Appl. Soil Ecol. 2016, 105, 109–125. [Google Scholar] [CrossRef]
- Naseem, H.; Bano, A. Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J. Plant Interact. 2014, 9, 689–701. [Google Scholar] [CrossRef]
- Naveed, M.; Hussain, M.B.; Zahir, Z.A.; Mitter, B.; Sessitsch, A. Drought stress amelioration in wheat through inoculation with Burkholderia phytofirmans strain PsJN. Plant Growth Regul. 2014, 73, 121–131. [Google Scholar] [CrossRef]
- Vardharajula, S.; Zulfikar Ali, S.; Grover, M.; Reddy, G.; Bandi, V. Drought-tolerant plant growth promoting Bacillus spp.: Effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 2011, 6, 1–14. [Google Scholar] [CrossRef]
- Castillo, P.; Escalante, M.; Gallardo, M.; Alemano, S.; Abdala, G. Effects of bacterial single inoculation and co-inoculation on growth and phytohormone production of sunflower seedlings under water stress. Acta Physiol. Plant 2013, 35, 2299–2309. [Google Scholar] [CrossRef]
- Arshad, M.; Shaharoona, B.; Mahmood, T. Inoculation with Pseudomonas spp. Containing ACC-Deaminase Partially Eliminates the Effects of Drought Stress on Growth, Yield, and Ripening of Pea (Pisum sativum L.). Pedosphere 2008, 18, 611–620. [Google Scholar] [CrossRef]
- Pandey, V.; Ansari, M.W.; Tula, S.; Yadav, S.; Sahoo, R.K.; Shukla, N.; Bains, G.; Badal, S.; Chandra, S.; Gaur, A.K.; et al. Dose-dependent response of Trichoderma harzianum in improving drought tolerance in rice genotypes. Planta 2016, 243, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.A.; Lennon, J.T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. USA 2012, 109, 14058–14062. [Google Scholar] [CrossRef]
- Kaisermann, A.; de Vries, F.T.; Griffiths, R.I.; Bardgett, R.D. Legacy effects of drought on plant–soil feedbacks and plant–Plant interactions. New Phytol. 2017, 215, 1413–1424. [Google Scholar] [CrossRef]
- Rho, H.; Hsieh, M.; Kandel, S.L.; Cantillo, J.; Doty, S.L.; Kim, S. Do Endophytes Promote Growth of Host Plants Under Stress? A Meta-Analysis on Plant Stress Mitigation by Endophytes. Microb. Ecol. 2018, 75, 407–418. [Google Scholar] [CrossRef]
- Berns, A.E.; Philipp, H.; Narres, H.D.; Burauel, P.; Vereecken, H.; Tappe, W. Effect of gamma-sterilization and autoclaving on soil organic matter structure as studied by solid state NMR, UV and fluorescence spectroscopy. Eur. J. Soil Sci. 2008, 59, 540–550. [Google Scholar] [CrossRef]
- Mahmood, T.; Mehnaz, S.; Fleischmann, F.; Ali, R.; Hashmi, Z.H.; Iqbal, Z. Soil sterilization effects on root growth and formation of rhizosheaths in wheat seedlings. Pedobiologia 2014, 57, 123–130. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Strasser, F.; Woyke, T.; Schintlmeister, A.; Wagner, M.; Woebken, D. Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol. Ecol. 2015, 91, fiv106. [Google Scholar] [CrossRef]
- Yang, L.; Danzberger, J.; Schöler, A.; Schröder, P.; Schloter, M.; Radl, V. Dominant groups of potentially active bacteria shared by barley seeds become less abundant in root associated microbiome. Front. Plant Sci. 2017, 8, 1005. [Google Scholar] [CrossRef]
- Schröder, P.; Maier, H.; Debus, R. Detoxification of Herbicides in Phragmites australis. Z. Nat. C 2005, 60, 317. [Google Scholar]
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [Google Scholar] [CrossRef]
- Dorn-In, S.; Bassitta, R.; Schwaiger, K.; Bauer, J.; Hölzel, C.S. Specific amplification of bacterial DNA by optimized so-called universal bacterial primers in samples rich of plant DNA. J. Microbiol. Methods 2015, 113, 50–56. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, H.; Hildebrand, F.; et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Sapkota, R.; Knorr, K.; Jørgensen, L.N.; O’Hanlon, K.A.; Nicolaisen, M. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol. 2015, 207, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Env. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Nowak, A. On the efficiency of soil sterilization in autoclave, combined with addition of antibiotics. Zent. Mikrobiol. 1991, 146, 267–270. [Google Scholar] [CrossRef]
- Kim, M.; Heo, E.; Kang, H.; Adams, J. Changes in Soil Bacterial Community Structure with Increasing Disturbance Frequency. Microb. Ecol. 2013, 66, 171–181. [Google Scholar] [CrossRef]
- Wertz, S.; Czarnes, S.; Bartoli, F.; Renault, P.; Commeaux, C.; Guillaumaud, N.; Clays-Josserand, A. Early-stage bacterial colonization between a sterilized remoulded soil clod and natural soil aggregates of the same soil. Soil Biol. Biochem. 2007, 39, 3127–3137. [Google Scholar] [CrossRef]
- Murphy, B.R.; Doohan, F.M.; Hodkinson, T.R. Fungal endophytes of barley roots. J. Agric. Sci. 2014, 152, 602–615. [Google Scholar] [CrossRef]
- Rahman, M.M.; Flory, E.; Koyro, H.-W.; Abideen, Z.; Schikora, A.; Suarez, C.; Schnell, S.; Cardinale, M. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst. Appl. Microbiol. 2018, 41, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Ali, S.; Qi, L.K.; Zahoor, R.; Tian, Z.; Jiang, D.; Snider, J.L.; Dai, T. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4615. [Google Scholar]
- Torun, H. Time-course analysis of salicylic acid effects on ROS regulation and antioxidant defense in roots of hulled and hulless barley under combined stress of drought, heat and salinity. Physiol. Plant 2019, 165, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.R.; Mirzaei, M.; Ghabooli, M.; Khatabi, B.; Wu, Y.; Zabet-Moghaddam, M.; Mohammadi-Nejad, G.; Haynes, P.A.; Hajirezaei, M.R.; Sepehri, M.; et al. Root endophytic fungus Piriformospora indica improves drought stress adaptation in barley by metabolic and proteomic reprogramming. Environ. Exp. Bot. 2019, 157, 197–210. [Google Scholar] [CrossRef]
- De Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; Degraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellín, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought Stress Results in a Compartment-Specific Restructuring of the rice root-associated microbiomes. MBio 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Bor, B.; McLean, J.S.; Foster, K.R.; Cen, L.; To, T.T.; Serrato-Guillen, A.; Dewhirst, F.E.; Shi, W.; He, X. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl. Acad. Sci. USA 2018, 115, 12277–12282. [Google Scholar] [CrossRef]
- Starr, E.P.; Shi, S.; Blazewicz, S.J.; Probst, A.J.; Herman, D.J.; Firestone, M.K.; Banfield, J.F. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Wang, P.; Marsh, E.L.; Kruger, G.A.; Schachtman, D.P. Belowground microbial communities respond to water deficit and are shaped by decades of maize hybrid breeding. Env. Microbiol. 2020, 22, 889–904. [Google Scholar] [CrossRef]
- Pandey, S.; Chakraborty, D. Salicylic Acid and Drought Stress Response: Biochemical to Molecular Crosstalk. In Stress Responses in Plants; Tripathi, B., Müller, M., Eds.; Springer: Cham, Switzerland, 2015; pp. 247–265. [Google Scholar]
- Lebeis, S.L.; Herrera Paredes, S.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef]
- Kia, S.H.; Glynou, K.; Nau, T.; Thines, M.; Piepenbring, M.; Maciá-Vicente, J.G. Influence of phylogenetic conservatism and trait convergence on the interactions between fungal root endophytes and plants. ISME J. 2017, 11, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum wheat stress tolerance induced by endophyte pantoea agglomerans with genes contributing to plant functions and secondary metabolite Arsenal. Int. J. Mol. Sci. 2019, 20, 3989. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef] [PubMed]
- Redman Regina, S.; Dunigan David, D.; Rodriguez Rusty, J. Fungal symbiosis from mutualism to parasitism: Who controls the outcome, host or invader? New Phytol. 2001, 151, 705–716. [Google Scholar] [CrossRef]
- Fuchslueger, L.; Bahn, M.; Fritz, K.; Hasibeder, R.; Richter, A. Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow. New Phytol. 2014, 201, 916–927. [Google Scholar] [CrossRef]
- Redman, R.S.; Sheehan, K.B.; Stout, R.G.; Rodriguez, R.J.; Henson, J.M. Thermotolerance generated by plant/fungal symbiosis. Science 2002, 298, 1581. [Google Scholar] [CrossRef]
- De Oliveira, T.B.; de Lucas, R.C.; de Almeida Scarcella, A.S.; Contato, A.G.; Pasin, T.M.; Martinez, C.A.; de Moraes Polizeli, M.d.L.T. Fungal communities differentially respond to warming and drought in tropical grassland soil. Mol. Ecol. 2020, 29, 1550–1559. [Google Scholar] [CrossRef]
- Mahoney, D.P.; LaFavre, J.S. Coniochaeta extramundana, with a Synopsis of Other Coniochaeta Species. Mycologia 1981, 73, 931–952. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Schröder, P.; Vestergaard, G.; Schloter, M.; Radl, V. Response of Barley Plants to Drought Might Be Associated with the Recruiting of Soil-Borne Endophytes. Microorganisms 2020, 8, 1414. https://doi.org/10.3390/microorganisms8091414
Yang L, Schröder P, Vestergaard G, Schloter M, Radl V. Response of Barley Plants to Drought Might Be Associated with the Recruiting of Soil-Borne Endophytes. Microorganisms. 2020; 8(9):1414. https://doi.org/10.3390/microorganisms8091414
Chicago/Turabian StyleYang, Luhua, Peter Schröder, Gisle Vestergaard, Michael Schloter, and Viviane Radl. 2020. "Response of Barley Plants to Drought Might Be Associated with the Recruiting of Soil-Borne Endophytes" Microorganisms 8, no. 9: 1414. https://doi.org/10.3390/microorganisms8091414
APA StyleYang, L., Schröder, P., Vestergaard, G., Schloter, M., & Radl, V. (2020). Response of Barley Plants to Drought Might Be Associated with the Recruiting of Soil-Borne Endophytes. Microorganisms, 8(9), 1414. https://doi.org/10.3390/microorganisms8091414







