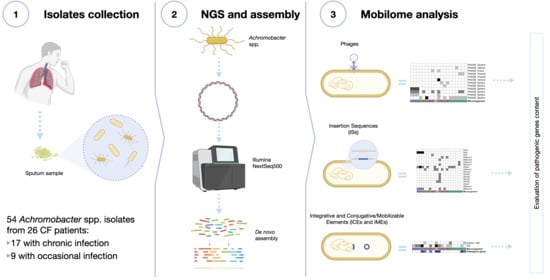
Mobilome Analysis of Achromobacter spp. Isolates from Chronic and Occasional Lung Infection in Cystic Fibrosis Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection and Identification
2.2. Genome Sequencing
2.3. Mobilome Analysis
3. Results
3.1. Phage Analysis
3.2. Insertion Sequences (ISs) Analysis
3.3. Integrative and Conjugative Elements (ICEs) and Integrative and Mobilizable Elements (IMEs) Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lederberg, J.; Tatum, E.L. Gene recombination in Escherichia coli. Nature 1946, 158, 558. [Google Scholar] [CrossRef] [PubMed]
- Zinder, N.D.; Lederberg, J. Genetic exchange in Salmonella. J. Bacteriol. 1952, 64, 679–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Kurland, C.G.; Canback, B.; Berg, O.G. Horizontal gene transfer: A critical view. Proc. Natl. Acad. Sci. USA 2003, 100, 9658–9662. [Google Scholar] [CrossRef] [Green Version]
- Bordenstein, S.R.; Reznikoff, W.S. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 2005, 3, 688–699. [Google Scholar] [CrossRef]
- Sorensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Goldenfeld, N.; Woese, C. Biology’s next revolution. Nature 2007, 445, 369. [Google Scholar] [CrossRef] [Green Version]
- Soucy, S.M.; Huang, J.L.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004, 2, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Rankin, D.J.; Rocha, E.P.C.; Brown, S.P. What traits are carried on mobile genetic elements, and why? Heredity 2011, 106, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniak, R.A.F.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Grossman, A.D. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botelho, J.; Schulenburg, H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2020. [Google Scholar] [CrossRef]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guedon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar]
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 2002, 46, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yates, C.M.; Shaw, D.J.; Roe, A.J.; Woolhouse, M.E.J.; Amyes, S.G.B. Enhancement of bacterial competitive fitness by apramycin resistance plasmids from non-pathogenic Escherichia coli. Biol. Lett. UK 2006, 2, 463–465. [Google Scholar] [CrossRef] [Green Version]
- Martinez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef]
- Martinez, J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Biol. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [Green Version]
- Senadheera, D.B.; Cordova, M.; Ayala, E.A.; de Paz, L.E.C.; Singh, K.; Downey, J.S.; Svensater, G.; Goodman, S.D.; Cvitkovitch, D.G. Regulation of Bacteriocin Production and Cell Death by the VicRK Signaling System in Streptococcus mutans. J. Bacteriol. 2012, 194, 1307–1316. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.P.; Inglis, R.F.; Taddei, F. Evolutionary ecology of microbial wars: Within-host competition and (incidental) virulence. Evol. Appl. 2009, 2, 32–39. [Google Scholar] [CrossRef]
- Botelho, J.; Mourao, J.; Roberts, A.P.; Peixe, L. Comprehensive genome data analysis establishes a triple whammy of carbapenemases, ICEs and multiple clinically relevant bacteria. Microb. Genom. 2020, 6. [Google Scholar] [CrossRef]
- Hansen, C.R.; Pressler, T.; Nielsen, K.G.; Jensen, P.O.; Bjarnsholt, T.; Hoiby, N. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. 2010, 9, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Firmida, M.C.; Pereira, R.H.V.; Silva, E.A.S.R.; Marques, E.A.; Lopes, A.J. Clinical impact of Achromobacter xylosoxidans colonization/infection in patients with cystic fibrosis. Braz. J. Med. Biol. Res. 2016, 49, e5097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridderberg, W.; Nielsen, S.M.; Norskov-Lauritsen, N. Genetic Adaptation of Achromobacter sp. during Persistence in the Lungs of Cystic Fibrosis Patients. PLoS ONE 2015, 10, e0136790. [Google Scholar] [CrossRef] [Green Version]
- Oliver, A.; Canton, R.; Campo, P.; Baquero, F.; Blazquez, J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2000, 288, 1251–1254. [Google Scholar] [CrossRef] [PubMed]
- Lambiase, A.; Catania, M.R.; del Pezzo, M.; Rossano, F.; Terlizzi, V.; Sepe, A.; Raia, V. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur. J. Clin. Microbiol. 2011, 30, 973–980. [Google Scholar] [CrossRef] [Green Version]
- Pereira, R.H.V.; Carvalho-Assef, A.P.; Albano, R.M.; Folescu, T.W.; Jones, M.C.M.F.; Leao, R.S.; Marques, E.A. Achromobacter xylosoxidans: Characterization of Strains in Brazilian Cystic Fibrosis Patients. J. Clin. Microbiol. 2011, 49, 3649–3651. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhu, Y.; Ma, Y.; Liu, F.; Lu, N.; Yang, X.; Luan, C.; Yi, Y.; Zhu, B. Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 2015, 59, 1152–1161. [Google Scholar] [CrossRef] [Green Version]
- Doring, G.; Conway, S.P.; Heijerman, H.G.M.; Hodson, M.E.; Hoiby, N.; Smyth, A.; Touw, D.J.; Comm, C. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: A European consensus. Eur. Respir. J. 2000, 16, 749–767. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [Green Version]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [Green Version]
- Veschetti, L.; Sandri, A.; Johansen, H.K.; Lleo, M.M.; Malerba, G. Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection. Pathogens 2020, 9, 72. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.J.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007, 1, 127. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Li, X.B.; Xie, Y.Z.; Bi, D.X.; Sun, J.Y.; Li, J.; Tai, C.; Deng, Z.X.; Ou, H.Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32-6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traglia, G.M.; Almuzara, M.; Merkier, A.K.; Adams, C.; Galanternik, L.; Vay, C.; Centron, D.; Ramirez, M.S. Achromobacter xylosoxidans: An emerging pathogen carrying different elements involved in horizontal genetic transfer. Curr. Microbiol. 2012, 65, 673–678. [Google Scholar] [CrossRef] [Green Version]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [Green Version]
- Jeukens, J.; Freschi, L.; Vincent, A.T.; Emond-Rheault, J.G.; Kukavica-Ibrulj, I.; Charette, S.J.; Levesque, R.C. A Pan-Genomic Approach to Understand the Basis of Host Adaptation in Achromobacter. Genome Biol. Evol. 2017, 9, 1030–1046. [Google Scholar] [CrossRef]
- Palazzo, A.; Lorusso, P.; Miskey, C.; Walisko, O.; Gerbino, A.; Marobbio, C.M.T.; Ivics, Z.; Marsano, R.M. Transcriptionally promiscuous “blurry” promoters in Tc1/mariner transposons allow transcription in distantly related genomes. Mob. DNA 2019, 10, 13. [Google Scholar] [CrossRef] [Green Version]
- Ramage, H.R.; Connolly, L.E.; Cox, J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009, 5, e1000767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrethsen, J.; Agner, J.; Piersma, S.R.; Hojrup, P.; Pham, T.V.; Weldingh, K.; Jimenez, C.R.; Andersen, P.; Rosenkrands, I. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol. Cell Proteom. 2013, 12, 1180–1191. [Google Scholar] [CrossRef] [Green Version]
- Miallau, L.; Jain, P.; Arbing, M.A.; Cascio, D.; Phan, T.; Ahn, C.J.; Chan, S.; Chernishof, I.; Maxson, M.; Chiang, J.; et al. Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes. Structure 2013, 21, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Korch, S.B.; Contreras, H.; Clark-Curtiss, J.E. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 2009, 191, 1618–1630. [Google Scholar] [CrossRef] [Green Version]
- Korch, S.B.; Malhotra, V.; Contreras, H.; Clark-Curtiss, J.E. The Mycobacterium tuberculosis relBE toxin:antitoxin genes are stress-responsive modules that regulate growth through translation inhibition. J. Microbiol. 2015, 53, 783–795. [Google Scholar] [CrossRef]
- De la Cruz, M.A.; Zhao, W.D.; Farenc, C.; Gimenez, G.; Raoult, D.; Cambillau, C.; Gorvel, J.P.; Meresse, S. A Toxin-Antitoxin Module of Salmonella Promotes Virulence in Mice. PLoS Pathog. 2013, 9, e1003827. [Google Scholar] [CrossRef]
- Kedzierska, B.; Hayes, F. Emerging Roles of Toxin-Antitoxin Modules in Bacterial Pathogenesis. Molecules 2016, 21, 790. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.J.; Hergenrother, P.J. Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol. 2012, 20, 291–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 2013, 3, e26219. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, M.G.; Pandey, D.P.; Jaskolska, M.; Gerdes, K. HicA of Escherichia coli Defines a Novel Family of Translation-Independent mRNA Interferases in Bacteria and Archaea. J. Bacteriol. 2009, 191, 1191–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Pogliano, J.; Helinski, D.R.; Konieczny, I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002, 44, 971–979. [Google Scholar] [CrossRef]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veschetti, L.; Sandri, A.; Patuzzo, C.; Melotti, P.; Malerba, G.; Lleò, M.M. Mobilome Analysis of Achromobacter spp. Isolates from Chronic and Occasional Lung Infection in Cystic Fibrosis Patients. Microorganisms 2021, 9, 130. https://doi.org/10.3390/microorganisms9010130
Veschetti L, Sandri A, Patuzzo C, Melotti P, Malerba G, Lleò MM. Mobilome Analysis of Achromobacter spp. Isolates from Chronic and Occasional Lung Infection in Cystic Fibrosis Patients. Microorganisms. 2021; 9(1):130. https://doi.org/10.3390/microorganisms9010130
Chicago/Turabian StyleVeschetti, Laura, Angela Sandri, Cristina Patuzzo, Paola Melotti, Giovanni Malerba, and Maria M. Lleò. 2021. "Mobilome Analysis of Achromobacter spp. Isolates from Chronic and Occasional Lung Infection in Cystic Fibrosis Patients" Microorganisms 9, no. 1: 130. https://doi.org/10.3390/microorganisms9010130
APA StyleVeschetti, L., Sandri, A., Patuzzo, C., Melotti, P., Malerba, G., & Lleò, M. M. (2021). Mobilome Analysis of Achromobacter spp. Isolates from Chronic and Occasional Lung Infection in Cystic Fibrosis Patients. Microorganisms, 9(1), 130. https://doi.org/10.3390/microorganisms9010130