Conservation and Loss of a Putative Iron Utilization Gene Cluster among Genotypes of Aspergillus flavus
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Culture Conditions
2.2. Gene Identification and Characterization
2.3. Neighbor-Net Network
2.4. Phylogenetic Analysis
2.5. PCR Profiling of the IUC
3. Results
3.1. Identification, Characterization, and Validation of IUC
3.2. Polymorphisms within IUC among A. flavus Isolates
3.3. Production of Siderophores
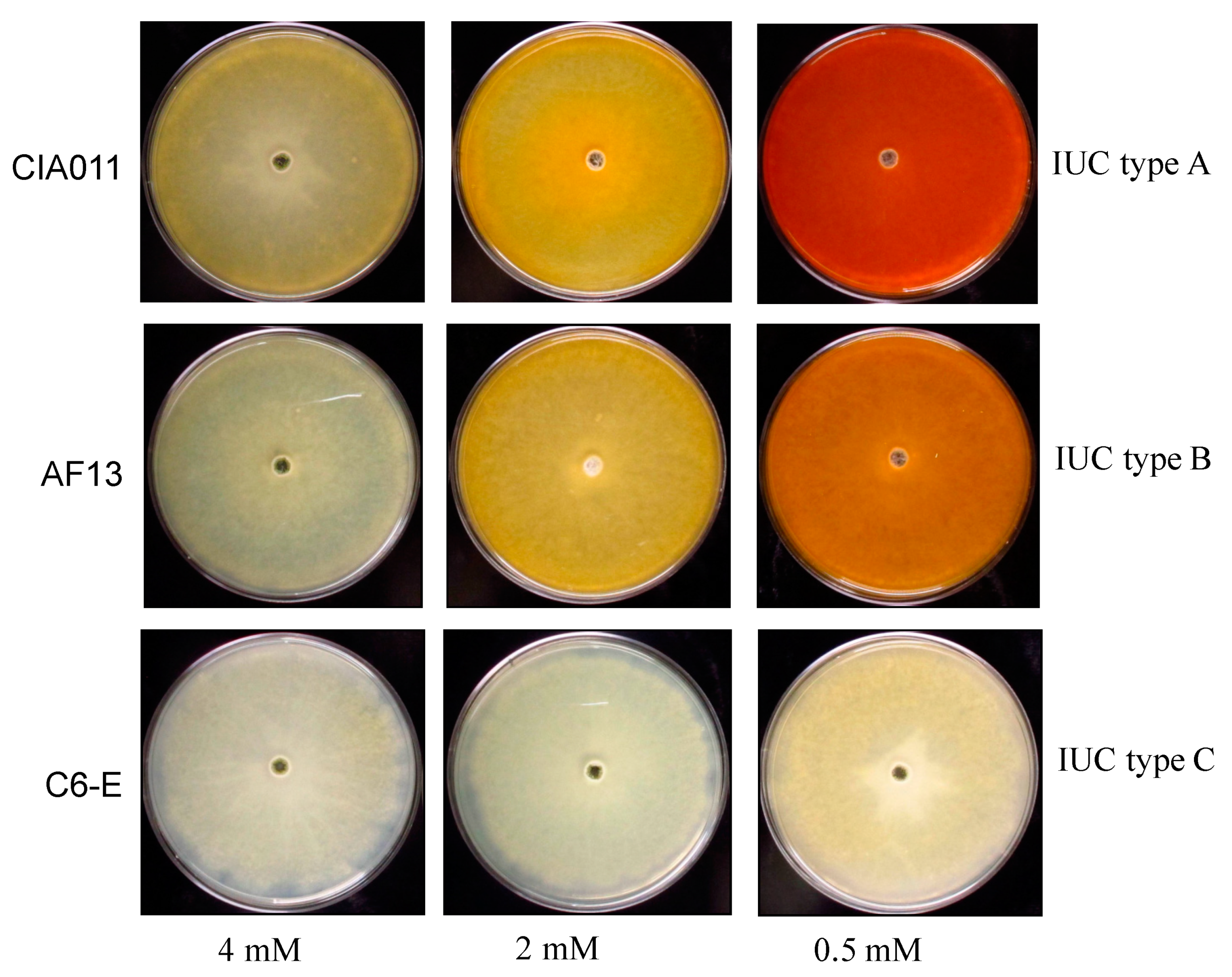
3.4. Response to Reduced Iron Media
3.5. Presence of Reductive Iron Assimilation (RIA) Pathway
3.6. Lineage-Specific Loss of IUC
3.7. Evolutionary Relationship of Iron Permease Gene
3.8. Presence of IUC in Other Fungi
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haber, F.; Weiss, J. The catalytic decomposition of hydrogen peroxide by iron salts. Proc. R. Soc. Lond. 1934, 147, 332–351. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. Role of iron in oxygen radical reactions. Methods Enzymol. 1984, 105, 47–56. [Google Scholar] [CrossRef]
- Kehrer, J.P. The Haber–Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Andrews, N.C. Forging a field: The golden age of iron biology. Blood 2008, 112, 219–230. [Google Scholar] [CrossRef]
- De Domenico, I.; Ward, D.M.V.; Kaplan, J. Regulation of iron acquisition and storage: Consequences for iron-linked disorders. Nat. Rev. Mol. Cell Biol. 2008, 9, 72–81. [Google Scholar] [CrossRef]
- Schrettl, M.; Beckmann, N.; Varga, J.; Heinekamp, T.; Jacobsen, I.D.; Jöchl, C.; Moussa, T.A.; Wang, S.; Gsaller, F.; Blatzer, M.; et al. HapX-Mediated Adaption to Iron Starvation Is Crucial for Virulence of Aspergillus fumigatus. PLoS Pathog. 2010, 6, e1001124. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.D. The role of iron in protozoan and fungal infectious diseases. J. Eukaryot. Microbiol. 1999, 46, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kosman, D.J. Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 2003, 47, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Guerinot, M.L.; Meidl, E.J.; Plessner, O. Citrate as a siderophore in Bradyrhizobium japonicum. J. Bacteriol. 1990, 172, 3298–3303. [Google Scholar] [CrossRef]
- Neilands, J.B. Siderophores: Structure and Function of Microbial Iron Transport Compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef]
- Haas, H.; Eisendle, M.; Turgeon, B.G. Siderophores in Fungal Physiology and Virulence. Annu. Rev. Phytopathol. 2008, 46, 149–187. [Google Scholar] [CrossRef] [PubMed]
- Haas, H. Molecular genetics of fungal siderophore biosynthesis and uptake: The role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 2003, 62, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Matzanke, B.F.; Bill, E.; Trautwein, A.X.; Winkelmann, G. Role of siderophores in iron storage in spores of Neurospora crassa and Aspergillus ochraceus. J. Bacteriol. 1987, 169, 5873–5876. [Google Scholar] [CrossRef] [PubMed]
- Schrettl, M.; Kim, H.S.; Eisendle, M.; Kragl, C.; Nierman, W.C.; Heinekamp, T.; Werner, E.R.; Jacobsen, I.; Illmer, P.; Yi, H.; et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 2008, 70, 27–43. [Google Scholar] [CrossRef]
- Jung, W.H.; Kronstad, J.W. Iron and fungal pathogenesis: A case study with Cryptococcus neoformans. Cell. Microbiol. 2008, 10, 277–284. [Google Scholar] [CrossRef]
- Nevitt, T.; Thiele, D.J. Host Iron Withholding Demands Siderophore Utilization for Candida glabrata to Survive Macrophage Killing. PLoS Pathog. 2011, 7, e1001322. [Google Scholar] [CrossRef]
- Philpott, C.C.; Protchenko, O. Response to Iron Deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 2008, 7, 20–27. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore Biosynthesis But Not Reductive Iron Assimilation Is Essential for Aspergillus fumigatus Virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef]
- Caza, M.; Kronstad, J.W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 3, 80. [Google Scholar] [CrossRef]
- Howard, D.H. Acquisition, Transport, and Storage of Iron by Pathogenic Fungi. Clin. Microbiol. Rev. 1999, 12, 394–404. [Google Scholar] [CrossRef]
- Morrissey, J.; Guerinot, M.L. Iron Uptake and Transport in Plants: The Good, the Bad, and the Ionome. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef] [PubMed]
- Philpott, C.C. Iron uptake in fungi: A system for every source. Biochim. Biophys. Acta 2006, 1763, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Ardon, O.; Nudelman, R.; Caris, C.; Libman, J.; Shanzer, A.; Chen, Y.; Hadar, Y. Iron Uptake in Ustilago maydis: Tracking the Iron Path. J. Bacteriol. 1998, 180, 2021–2026. [Google Scholar] [CrossRef] [PubMed]
- Dancis, A.; Yuan, D.S.; Haile, D.; Askwith, C.; Eide, D.; Moehle, C.; Kaplan, J.; Klausner, R.D. Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 1994, 76, 393–402. [Google Scholar] [CrossRef]
- Stearman, R.; Yuan, D.S.; Yamaguchi-Iwai, Y.; Klausner, R.D.; Dancis, A. A Permease-Oxidase Complex Involved in High-Affinity Iron Uptake in Yeast. Science 1996, 271, 1552–1557. [Google Scholar] [CrossRef] [PubMed]
- Heymann, P.; Ernst, J.F.; Winkelmann, G.; Winkelmann, G. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2000, 186, 221–227. [Google Scholar] [CrossRef][Green Version]
- Askwith, C.; Kaplan, J. An Oxidase-Permease-based Iron Transport System inSchizosaccharomyces pombeand Its Expression inSaccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 401–405. [Google Scholar] [CrossRef]
- Ecker, D.J.; Emery, T. Iron uptake from ferrichrome A and iron citrate in Ustilago sphaerogena. J. Bacteriol. 1983, 155, 616–622. [Google Scholar] [CrossRef]
- Fedorovich, D.; Protchenko, O.; Lesuisse, E. Iron uptake by the yeast Pichia guilliermondii. Flavinogenesis and reductive iron assimilation are co-regulated processes. BioMetals 1999, 12, 295–300. [Google Scholar] [CrossRef]
- Lesuisse, E.; Casterassimon, M.; Labbé, P. Ferrireductase Activity in Saccharomyces cerevisiae and Other Fungi: Colorimetric Assays on Agar Plates. Anal. Biochem. 1995, 226, 375–377. [Google Scholar] [CrossRef]
- Nyhus, K.J.; Jacobson, E.S. Genetic and Physiologic Characterization of Ferric/Cupric Reductase Constitutive Mutants of Cryptococcus neoformans. Infect. Immun. 1999, 67, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, M.M.; Woods, J.P. Ferric Reduction Is a Potential Iron Acquisition Mechanism for Histoplasma capsulatum. Infect. Immun. 1999, 67, 6403–6408. [Google Scholar] [CrossRef] [PubMed]
- Franken, A.C.W.; Lechner, B.E.; Werner, E.R.; Haas, H.; Lokman, B.C.; Ram, A.F.J.; Van den Hondel, C.A.; De Weert, S.; Punt, P.J. Genome mining and functional genomics for siderophore production in Aspergillus niger. Brief. Funct. Genom. 2014, 13, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Haas, H. Iron—A Key Nexus in the Virulence of Aspergillus fumigatus. Front. Microbiol. 2012, 3, 28. [Google Scholar] [CrossRef]
- Schrettl, M.; Haas, H. Iron homeostasis—Achilles’ heel of Aspergillus fumigatus? Curr. Opin. Microbiol. 2011, 14, 400–405. [Google Scholar] [CrossRef]
- Eisendle, M.; Oberegger, H.; Zadra, I.; Haas, H. The siderophore system is essential for viability of Aspergillus nidulans: Functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 2003, 49, 359–375. [Google Scholar] [CrossRef]
- Haas, H.; Zadra, I.; Stöffler, G.; Angermayr, K. TheAspergillus nidulansGATA Factor SREA Is Involved in Regulation of Siderophore Biosynthesis and Control of Iron Uptake. J. Biol. Chem. 1999, 274, 4613–4619. [Google Scholar] [CrossRef]
- Oberegger, H.; Zadra, I.; Schoeser, M.; Abt, B.; Parson, W.; Haas, H. Identification of members of the Aspergillus nidulans SREA regulon: Genes involved in siderophore biosynthesis and utilization. Biochem. Soc. Trans. 2002, 30, 781–783. [Google Scholar] [CrossRef]
- Almeida, R.S.; Wilson, D.; Hube, B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009, 9, 1000–1012. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron Metabolism in Pathogenic Bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef]
- Khlangwiset, P.; Wu, F. Costs and efficacy of public health interventions to reduce aflatoxin-induced human disease. Food Addit. Contam. Part A 2010, 27, 998–1014. [Google Scholar] [CrossRef] [PubMed]
- Shephard, G. Impact of mycotoxins on human health in developing countries. Food Addit. Contam. Part A 2008, 25, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Probst, C.; Schulthess, F.; Cotty, P.J. Impact ofAspergillussectionFlavicommunity structure on the development of lethal levels of aflatoxins in Kenyan maize (Zea mays). J. Appl. Microbiol. 2010, 108, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Mehl, H.L.; Cotty, P.J. Variability in competitive ability among Aspergillus flavus vegetative compatibility groups during maize infection. Phytopathology 2008, 98, S103. [Google Scholar]
- Mehl, H.L.; Cotty, P.J. Variation in Competitive Ability among Isolates of Aspergillus flavus from Different Vegetative Compatibility Groups During Maize Infection. Phytopathology 2010, 100, 150–159. [Google Scholar] [CrossRef]
- Mehl, H.; Cotty, P.J. Influence of the Host Contact Sequence on the Outcome of Competition amongAspergillus flavusIsolates during Host Tissue Invasion. Appl. Environ. Microbiol. 2011, 77, 1691–1697. [Google Scholar] [CrossRef]
- Aziz, N.H.; Shahin, A.A.M.; Abou-Zeid, A.A.M.; El-Zeany, S.A. Correlation of growth and aflatoxin production by Asper-gillus flavus with some essential metals in gamma irradiated crushed corn. Food/Nahrung 2000, 44, 354–359. [Google Scholar] [CrossRef]
- Cuero, R.; Ouellet, T.; Yu, J.; Mogongwa, N. Metal ion enhancement of fungal growth, gene expression and aflatoxin synthesis in Aspergillus flavus: RT-PCR characterization. J. Appl. Microbiol. 2003, 94, 953–961. [Google Scholar] [CrossRef]
- LaRochelle, O.; Gagné, V.; Charron, J.; Soh, J.-W.; Séguin, C. Phosphorylation Is Involved in the Activation of Metal-regulatory Transcription Factor 1 in Response to Metal Ions. J. Biol. Chem. 2001, 276, 41879–41888. [Google Scholar] [CrossRef]
- Cuero, R.; Ouellet, T. Metal ions modulate gene expression and accumulation of the mycotoxins aflatoxin and zearalenone. J. Appl. Microbiol. 2005, 98, 598–605. [Google Scholar] [CrossRef]
- Liu, J.; Sun, L.; Zhang, N.; Zhang, J.; Guo, J.; Li, C.; Rajput, S.A.; Qi, D. Effects of Nutrients in Substrates of Different Grains on Aflatoxin B1 Production by Aspergillus flavus. Biomed. Res. Int. 2016, 2016, 7232858. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.N.; Bandyopadhyay, R.; Cotty, P.J. Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America. AMB Express 2016, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Cotty, P.J. Virulence and Cultural Characteristics of TwoAspergillus flavusStrains Pathogenic on Cotton. Phytopathology 1989, 79, 808–814. [Google Scholar] [CrossRef]
- Pitt, J.; Hocking, A.D.; Glenn, D.R. An improved medium for the detection ofAspergillus flavusandA. parasiticus. J. Appl. Bacteriol. 1983, 54, 109–114. [Google Scholar] [CrossRef]
- Abramoff, M.; Magalhaes, P.; Ram, S. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.-I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- Yu, J.; Fedorova, N.D.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W.; Nierman, W.C. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 2011, 322, 145–149. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinform. 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007, 35, D237–D240. [Google Scholar] [CrossRef]
- Li, H.; Ruan, J.; Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008, 18, 1851–1858. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Grubisha, L.C.; Cotty, P.J. Twenty-four microsatellite markers for the aflatoxin-producing fungusAspergillus flavus. Mol. Ecol. Resour. 2009, 9, 264–267. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker® HID: A Reliable Software Tool for the Analysis of Forensic STR Data. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef]
- Grubisha, L.C.; Cotty, P.J. Genetic isolation among sympatric vegetative compatibility groups of the aflatoxin-producing fungus Aspergillus flavus. Mol. Ecol. 2010, 19, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.; Feil, E.J.; Chan, M.-S.; Maiden, M.C.J. Sequence type analysis and recombinational tests (START). Bioinformatics 2001, 17, 1230–1231. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Assante, G.; Camarda, L.; Locci, R.; Merlini, L.; Nasini, G.; Papadopoulos, E. Isolation and structure of red pigments from Aspergillus flavus and related species, grown on a differential medium. J. Agric. Food Chem. 1981, 29, 785–787. [Google Scholar] [CrossRef]
- Dutcher, J.D. Aspergillic acid; an antibiotic substance produced by Aspergillus flavus. J. Biol. Chem. 1958, 232, 785–795. [Google Scholar] [CrossRef]
- Lesuisse, E.; Labbe, P. Reductive and Non-reductive Mechanisms of Iron Assimilation by the Yeast Saccharomyces cerevisiae. J. Gen. Microbiol. 1989, 135, 257–263. [Google Scholar] [CrossRef]
- Nierman, W.C.; Yu, J.; Fedorova-Abrams, N.D.; Losada, L.; Cleveland, T.E.; Bhatnagar, D.; Bennett, J.W.; Dean, R.; Payne, G.A. Genome Sequence of Aspergillus flavus NRRL 3357, a Strain That Causes Aflatoxin Contamination of Food and Feed. Genome Announc. 2015, 3, e00168-15. [Google Scholar] [CrossRef]
- Eichhorn, H.; Lessing, F.; Winterberg, B.; Schirawski, J.; Kämper, J.; Müller, P.; Kahmann, R. A Ferroxidation/Permeation Iron Uptake System Is Required for Virulence in Ustilago maydis. Plant Cell 2006, 18, 3332–3345. [Google Scholar] [CrossRef]
- Kombrink, A.; Thomma, B.P.H.J. LysM Effectors: Secreted Proteins Supporting Fungal Life. PLoS Pathog. 2013, 9, e1003769. [Google Scholar] [CrossRef]
- Linz, J.E.; Wee, J.; Roze, L.V. Aspergillus parasiticus SU-1 Genome Sequence, Predicted Chromosome Structure, and Comparative Gene Expression under Aflatoxin-Inducing Conditions: Evidence that Differential Expression Contributes to Species Phenotype. Eukaryot. Cell 2014, 13, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef]
- Woloshuk, C.P.; Foutz, K.R.; Brewer, J.F.; Bhatnagar, D.; E Cleveland, T.; A Payne, G. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 1994, 60, 2408–2414. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.; Oliveira, D.; Hu, G.; Kronstad, J.W. Role of Ferric Reductases in Iron Acquisition and Virulence in the Fungal Pathogen Cryptococcus neoformans. Infect. Immun. 2014, 82, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Tamayo-Ramos, J.A.; Barends, S.; Verhaert, R.M.D.; De Graaff, L.H. The Aspergillus niger multicopper oxidase family: Analysis and overexpression of laccase-like encoding genes. Microb. Cell Fact. 2011, 10, 78. [Google Scholar] [CrossRef]
- Kues, U.; Ruhl, M. Multiple Multi-Copper Oxidase Gene Families in Basidiomycetes—What for? Curr. Genom. 2011, 12, 72–94. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Gebremariam, T.; Lin, L.; Luo, G.; Husseiny, M.I.; Skory, C.D.; Fu, Y.; French, S.W.; Jr, J.E.E.; Spellberg, B. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol. Microbiol. 2010, 77, 587–604. [Google Scholar] [CrossRef]
- Lian, T.; Simmer, M.I.; D’Souza, C.A.; Steen, B.R.; Zuyderduyn, S.D.; Jones, S.J.M.; Marra, M.A.; Kronstad, J.W. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 2005, 55, 1452–1472. [Google Scholar] [CrossRef]
- Kosman, D.J. Redox Cycling in Iron Uptake, Efflux, and Trafficking. J. Biol. Chem. 2010, 285, 26729–26735. [Google Scholar] [CrossRef]
- Singh, A.; Severance, S.; Kaur, N.; Wiltsie, W.; Kosman, D.J. Assembly, Activation, and Trafficking of the Fet3p·Ftr1p High Affinity Iron Permease Complex inSaccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 13355–13364. [Google Scholar] [CrossRef]
- Urbanowski, J.L.; Piper, R.C. The Iron Transporter Fth1p Forms a Complex with the Fet5 Iron Oxidase and Resides on the Vacuolar Membrane. J. Biol. Chem. 1999, 274, 38061–38070. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kaur, N.; Kosman, D.J. The Metalloreductase Fre6p in Fe-Efflux from the Yeast Vacuole. J. Biol. Chem. 2007, 282, 28619–28626. [Google Scholar] [CrossRef] [PubMed]
- Rees, E.M.; Thiele, D.J. Identification of a Vacuole-associated Metalloreductase and Its Role in Ctr2-mediated Intracellular Copper Mobilization. J. Biol. Chem. 2007, 282, 21629–21638. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.A.; Williams, P.H.; Cashmore, A.M. Candida Albicans has a Cell-Associated Ferric-Reductase Activity which is Regulated in Response to Levels of Iron and Copper. Microbiology 1996, 142, 485–492. [Google Scholar] [CrossRef][Green Version]
- Osbourn, A. Gene Clusters for Secondary Metabolic Pathways: An Emerging Theme in Plant Biology. Plant Physiol. 2010, 154, 531–535. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Inglis, D.; Binkley, J.; Skrzypek, M.S.; Arnaud, M.B.; Cerqueira, G.C.; Shah, P.; Wymore, F.; Wortman, J.R.; Sherlock, G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013, 13, 91. [Google Scholar] [CrossRef]
- Andersen, M.R.; Nielsen, J.B.; Klitgaard, A.; Petersen, L.M.; Zachariasen, M.; Hansen, T.J.; Blicher, L.H.; Gotfredsen, C.H.; Larsen, T.O.; Nielsen, K.F.; et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. USA 2012, 110, E99–E107. [Google Scholar] [CrossRef]
- Chang, P.-K.; Abbas, H.K.; Weaver, M.A.; Ehrlich, K.C.; Scharfenstein, L.L.; Cotty, P.J. Identification of genetic defects in the atoxigenic biocontrol strain Aspergillus flavus K49 reveals the presence of a competitive recombinant group in field populations. Int. J. Food Microbiol. 2012, 154, 192–196. [Google Scholar] [CrossRef]
- Chang, P.-K.; Horn, B.W.; Dorner, J.W. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005, 42, 914–923. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Tominaga, M.; Hayashi, R.; Sakamoto, K.; Yamada, O.; Akita, O. Aspergillus oryzae strains with a large deletion of the aflatoxin biosynthetic homologous gene cluster differentiated by chromosomal breakage. Appl. Microbiol. Biotechnol. 2006, 72, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Hsiang, T.; Baillie, D.L. Comparison of the Yeast Proteome to Other Fungal Genomes to Find Core Fungal Genes. J. Mol. Evol. 2005, 60, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Cherayil, B.J. The role of iron in the immune response to bacterial infection. Immunol. Res. 2010, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Beach, R.H.; Sulser, T.B.; Crimmins, A.; Cenacchi, N.; Cole, J.; Fukagawa, N.K.; Mason-D’Croz, D.; Myers, S.; Sarofim, M.C.; Smith, M.; et al. Combining the effects of increased atmospheric carbon dioxide on protein, iron, and zinc availability and projected climate change on global diets: A modelling study. Lancet Planet. Health 2019, 3, e307–e317. [Google Scholar] [CrossRef]
- Greenshields, D.L.; Liu, G.; Wei, Y. Roles of Iron in Plant Defence and Fungal Virulence. Plant Signal. Behav. 2007, 2, 300–302. [Google Scholar] [CrossRef]
- Ramanan, N.; Wang, Y.A. High-Affinity Iron Permease Essential for Candida albicans Virulence. Science 2000, 288, 1062–1064. [Google Scholar] [CrossRef]
- Schrettl, M.; Winkelmann, G.; Haas, H. Ferrichrome in Schizosaccharomyces pombe—An iron transport and iron storage compound. BioMetals 2004, 17, 647–654. [Google Scholar] [CrossRef]




| Isolate | Substrate | Aflatoxin * | Culture Accession/Source | Reference |
|---|---|---|---|---|
| AF13 | Cotton | Toxigenic | ATCC 96044 | [53] |
| BY18-A | Maize | Atoxigenic | USDA-ARS, Tucson | [52] |
| CIA011 | Maize | Atoxigenic | USDA-ARS, Tucson | [52] |
| DO114-A | Maize | Atoxigenic | USDA-ARS, Tucson | [52] |
| EC69-E | Maize | Atoxigenic | USDA-ARS, Tucson | [52] |
| Protein ID | Length ¶ | Homologue $ | Nucleotide Identity (%), Coverage (%) | Functional annotation £ | Protein Domain § |
|---|---|---|---|---|---|
| BAE62583.1 | 459 | Hypothetical protein, Aspergillus oryzae RIB40 | 96, 99 | C6 transcription factor | DUF1421 |
| BAE62584.1 | 229 | Hypothetical protein, Aspergillus oryzae RIB40 | 99, 86 | Predicted protein | LysM |
| BAE62585.1 | 225 | Hypothetical protein, Aspergillus oryzae RIB40 | 99, 98 | Unnamed protein | GAL4 |
| BAE62586.1 | 825 | Ferric reductase, Aspergillus oryzae RIB40 | 94, 93 | Ferric reductase | NOX_Duox_like NAD_NADP, NAD_binding_1 |
| BAE62587.1 | 621 | Ferrooxidoreductase, Aspergillus oryzae RIB40 | 89, 69 | Multicopper oxidase | CuRO_1_Fet3p, Las1 and Cupredoxin superfamily |
| BAE62588.1 | 370 | Iron permease, Aspergillus oryzae RIB40 | 91, 82 | Iron permease | Iron permease Ftr1 family |
| Cluster Type § | Genes Deleted ¶ | Sequence End (5′-3′) ⌘ | Sequence End (3′-5′) | Group ∞ | Proportion of Isolates $ |
|---|---|---|---|---|---|
| Type C | None | CGGGCAACTCTCCCCTTGCG | TATGTTCCAGTCAAAGACGT | I | 79% |
| Type B | FeRed, MCO | CTGGCAGTTCATCGTTTGCG | TATGTTCCAGTCAAAGACGT | II | 1% |
| Type A | IrPm (partial), MCO, FeRed, GAL4 | TGTCCTATCTATCTCTGGTG | CTGGTGTTGCAGTAGGCTAT | III | 11% |
| Type B | IrPm (partial), MCO, FeRed, GAL4, LysM | TGTCCTATCTATCTCTGGTG | CTGGTGTTGCAGTAGGCTAT | II | 1% |
| Type B | FeRed, MCO (partial) | CTGGCAGTTCTTCGTTTGCG | T-TGATCATGCTGCGGAAGG | IV | 2% |
| Type B | FeRed, MCO (partial) | CTGGCAGTTCATCTCTTGCG | T-TGATCATGCTGCGGGAGG | V | 6% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adhikari, B.N.; Callicott, K.A.; Cotty, P.J. Conservation and Loss of a Putative Iron Utilization Gene Cluster among Genotypes of Aspergillus flavus. Microorganisms 2021, 9, 137. https://doi.org/10.3390/microorganisms9010137
Adhikari BN, Callicott KA, Cotty PJ. Conservation and Loss of a Putative Iron Utilization Gene Cluster among Genotypes of Aspergillus flavus. Microorganisms. 2021; 9(1):137. https://doi.org/10.3390/microorganisms9010137
Chicago/Turabian StyleAdhikari, Bishwo N., Kenneth A. Callicott, and Peter J. Cotty. 2021. "Conservation and Loss of a Putative Iron Utilization Gene Cluster among Genotypes of Aspergillus flavus" Microorganisms 9, no. 1: 137. https://doi.org/10.3390/microorganisms9010137
APA StyleAdhikari, B. N., Callicott, K. A., & Cotty, P. J. (2021). Conservation and Loss of a Putative Iron Utilization Gene Cluster among Genotypes of Aspergillus flavus. Microorganisms, 9(1), 137. https://doi.org/10.3390/microorganisms9010137






