Genetic Characterization of Fungal Biodiversity in Storage Grains: Towards Enhancing Food Safety in Northern Uganda
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of Study Area
2.2. Sample Collection
2.3. Isolation of Fungi from Grains
2.4. DNA Extraction
2.5. PCR Reaction and Sequencing
2.6. Sequence Processing and Analysis
2.7. Phylogenetic Tree Construction
2.8. Genetic Differentiation of the Fungi Using TEF-α1 Gene
2.9. Haplotype Clustering
2.10. Detection of Fumonisin Gene Cluster among Fusarium Isolates
2.11. Mycotoxins Extraction and Quantifications
2.12. Data Analysis
3. Results
3.1. Fungal Genera and Species Associated with Cereal Grains in Northern Uganda
3.2. Distribution of the Fungi among the Different Grain Types
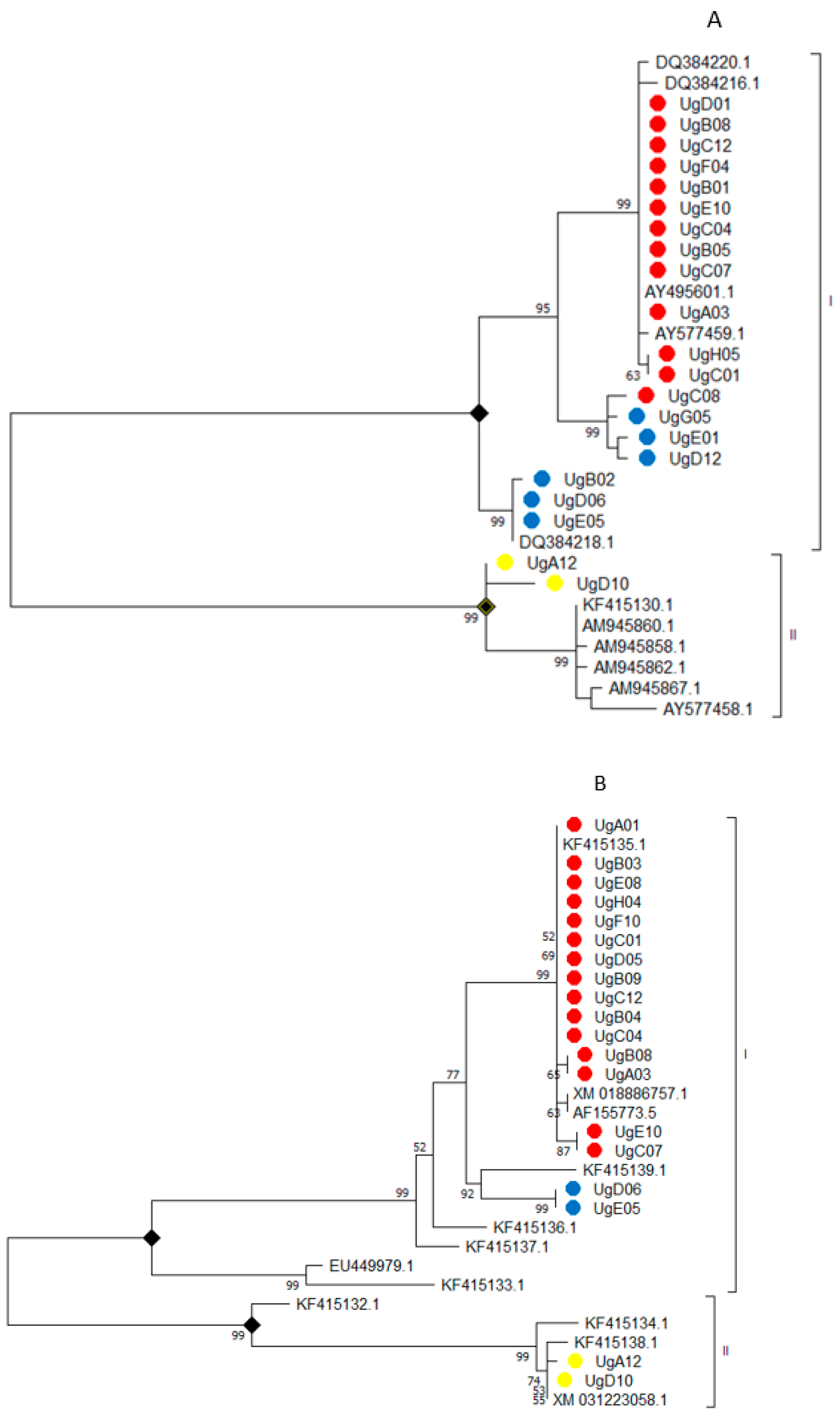
3.3. Genetic Differentiation between Isolates from Uganda and Isolates Reported in Other Countries
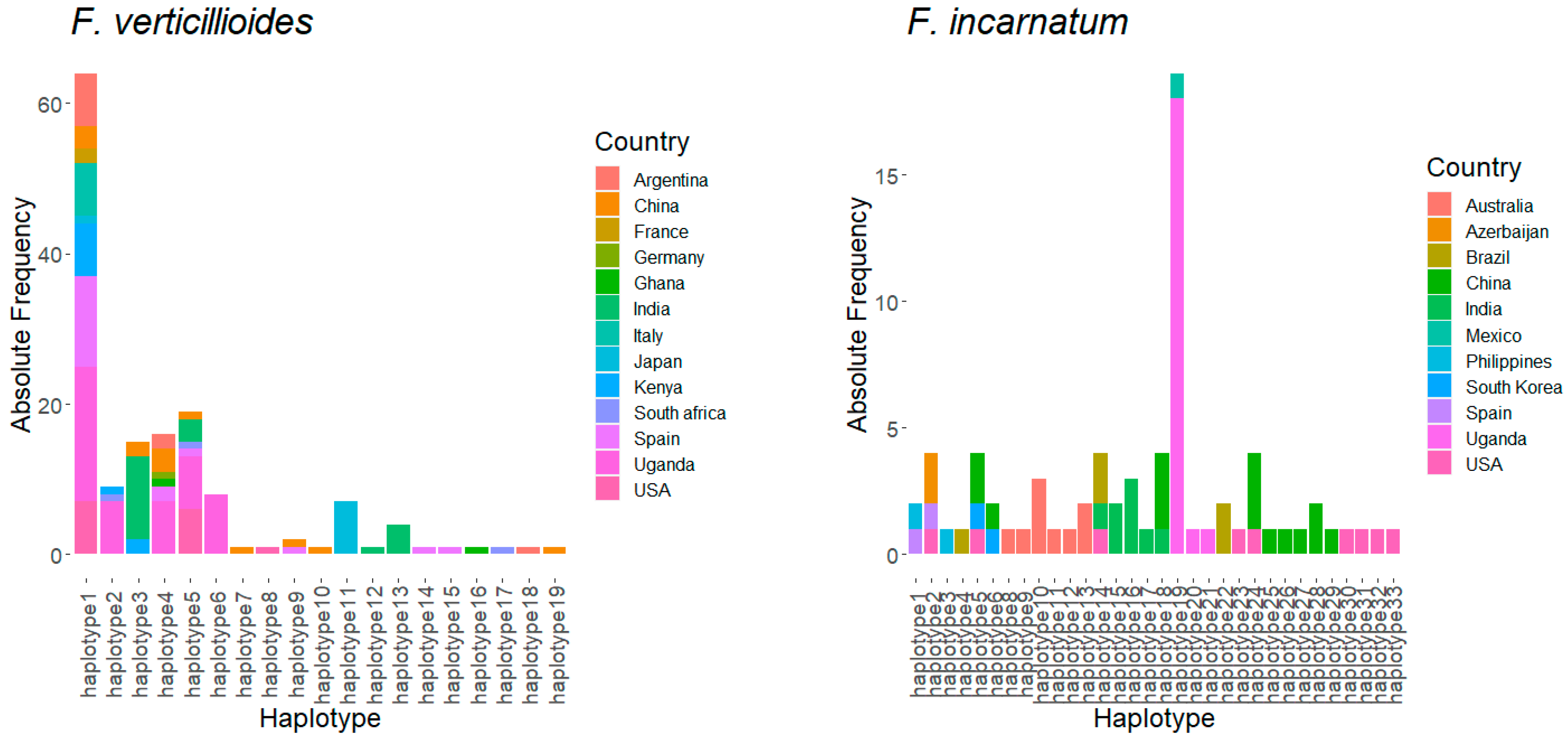
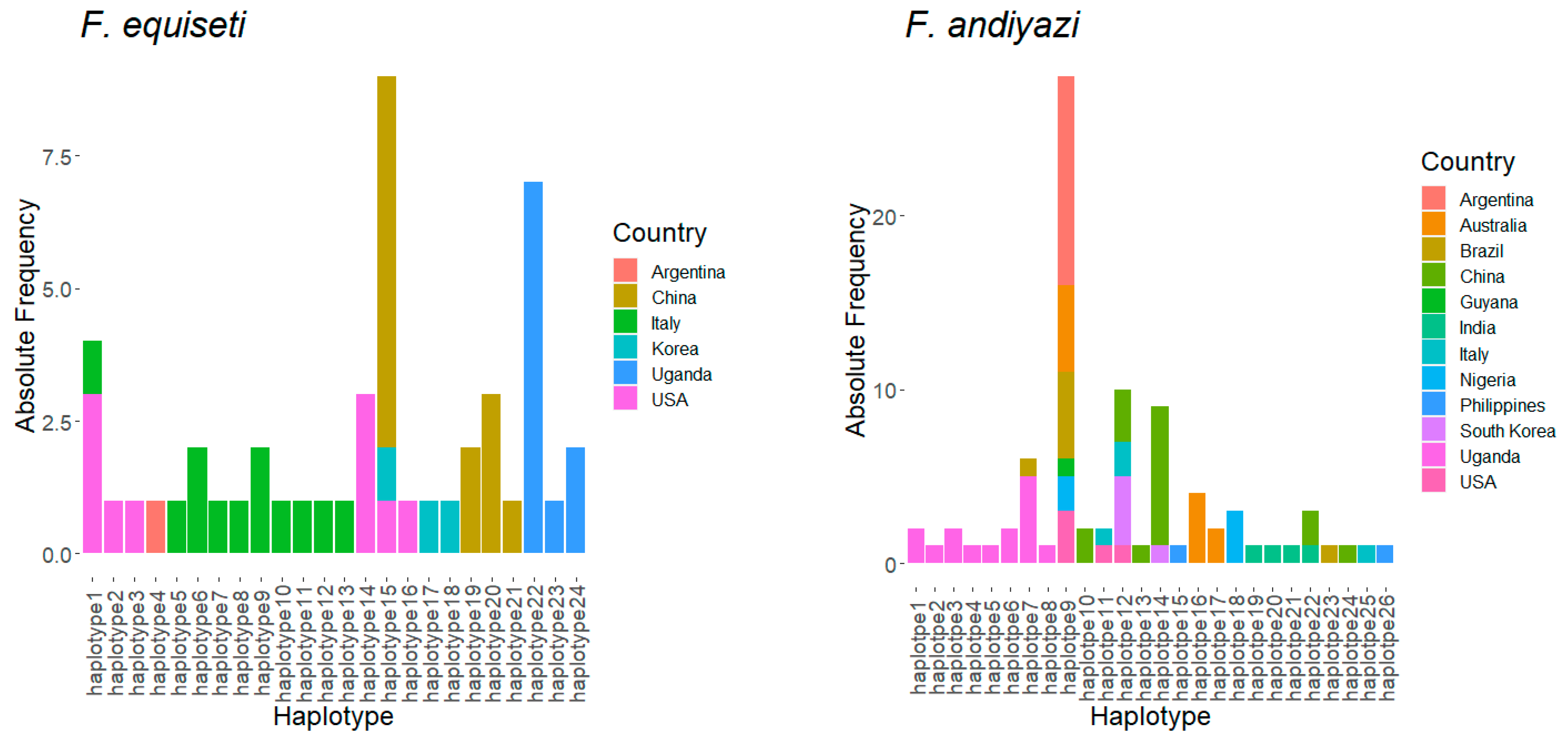
3.4. Haplotype Structure of Fusarium Species in Uganda Relative to Other Countries
3.5. Detection of Fumonisins Gene Clusters (FUM1 and FUM3) among Fusarium Isolates
3.6. Genetic Variability within Fumonisin Gene Clusters (FUM1 and FUM3)
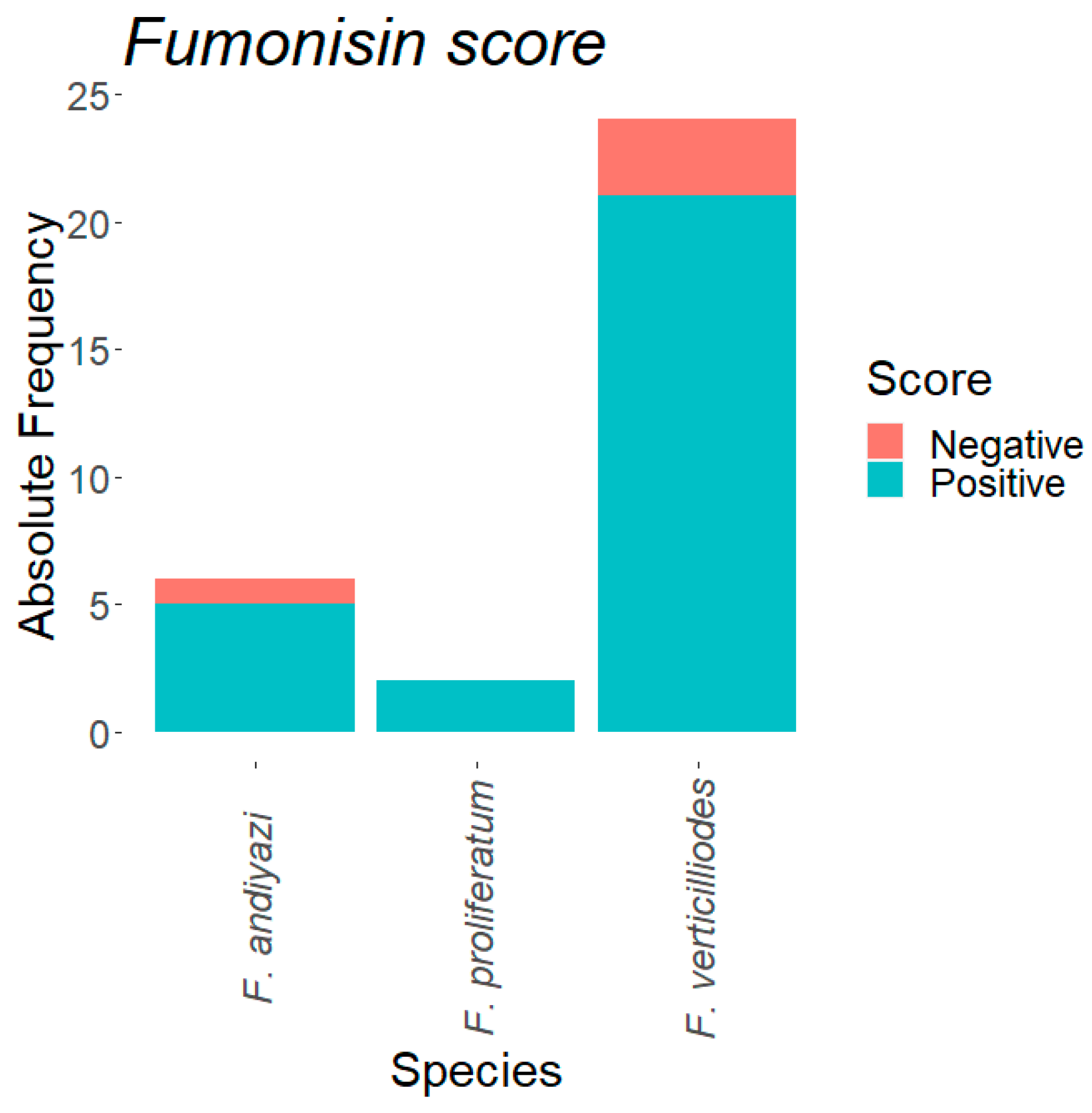
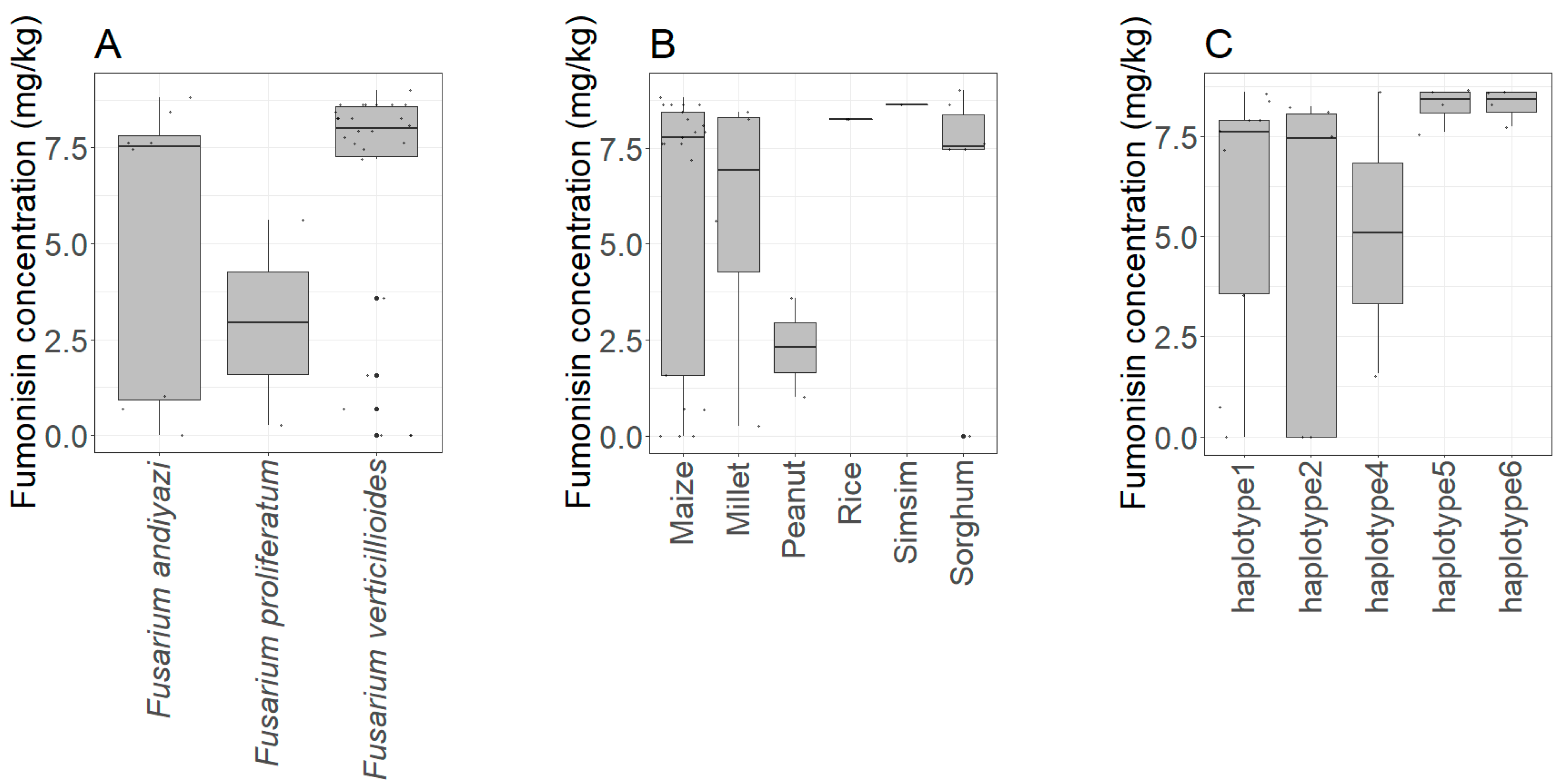
3.7. Mycotoxin Production Capability of the Fusarium Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, P.M.; Zannini, E.; Arendt, E.K. Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: From crop farming to cereal products. Food Microbiol. 2014, 37, 78–95. [Google Scholar] [CrossRef]
- Choi, J.-H.; Lee, S.; Nah, J.-Y.; Kim, H.-K.; Paek, J.-S.; Lee, S.; Ham, H.; Hong, S.K.; Yun, S.-H.; Lee, T. Species composition of and fumonisin production by the Fusarium fujikuroi species complex isolated from Korean cereals. Int. J. Food Microbiol. 2018, 267, 62–69. [Google Scholar] [CrossRef]
- Xu, X.; Nicholson, P.; Ritieni, A. Effects of fungal interactions among Fusarium head blight pathogens on disease development and mycotoxin accumulation. Int. J. Food Microbiol. 2007, 119, 67–71. [Google Scholar] [CrossRef]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 2002, 108, 611–624. [Google Scholar] [CrossRef]
- Yard, E.E.; Daniel, J.H.; Lewis, L.S.; Rybak, M.E.; Paliakov, E.M.; Kim, A.A.; Montgomery, J.M.; Bunnell, R.; Abudo, M.U.; Akhwale, W.; et al. Human aflatoxin exposure in Kenya, 2007: A cross-sectional study. Food Addit. Contam. Part A 2013, 30, 1322–1331. [Google Scholar] [CrossRef]
- Kumar, D.; Kalita, P. Reducing postharvest losses during storage of grain crops to strengthen food security in developing countries. Foods 2017, 6, 8. [Google Scholar] [CrossRef]
- Subramanyam, B.; Mahroof, R.; Brijwani, M. Heat treatment of grain-processing facilities for insect management: A historical overview and recent advances. Stewart Postharvest Rev. 2011, 7, 1–11. [Google Scholar] [CrossRef]
- Wambugu, P.; Mathenge, P.; Auma, E.O.; van Rheenen, H.A. Efficacy of traditional maize (Zea mays L.) seed storage methods in western Kenya. Afr. J. Food Agric. Nutr. Dev. 2009, 9, 1110–1128. [Google Scholar] [CrossRef]
- Abass, A.B.; Ndunguru, G.; Mamiro, P.; Alenkhe, B.; Mlingi, N.; Bekunda, M. Post-harvest food losses in a maize-based farming system of semi-arid savannah area of Tanzania. J. Stored Prod. Res. 2014, 57, 49–57. [Google Scholar] [CrossRef]
- Manjula, K.; Hell, K.; Fandohan, P.; Abass, A.; Bandyopadhyay, R. Aflatoxin and fumonisin contamination of cassava products and maize grain from markets in Tanzania and republic of the Congo. Toxin Rev. 2009, 28, 63–69. [Google Scholar] [CrossRef]
- Kamala, A.; Ortiz, J.; Kimanya, M.E.; Haesaert, G.; Donoso, S.; Tiisekwa, B.; De Meulenaer, B. Multiple mycotoxin co-occurrence in maize grown in three agro-ecological zones of Tanzania. Food Control. 2015, 54, 208–215. [Google Scholar] [CrossRef]
- Mutiga, S.K.; Hoffmann, V.; Harvey, J.W.; Milgroom, M.G.; Nelson, R.J. Assessment of aflatoxin and fumonisin contamination of maize in western Kenya. Phytopathology 2015, 105, 1250–1261. [Google Scholar] [CrossRef]
- Wokorach, G.; Landschoot, S.; Anena, J.; Audenaert, K.; Echodu, R.; Haesaert, G. Mycotoxin profile of staple grains in northern Uganda: Understanding the level of human exposure and potential risks. Food Control. 2021, 122, 107813. [Google Scholar] [CrossRef]
- Azziz-Baumgartner, E.; Lindblade, K.; Gieseker, K.; Rogers, H.S.; Kieszak, S.; Njapau, H.; Schleicher, R.; McCoy, L.F.; Misore, A.; DeCock, K.; et al. Case-control study of an acute aflatoxicosis outbreak, Kenya. Environ. Health Perspect. 2005, 113, 1779–1783. [Google Scholar] [CrossRef]
- Kamala, A.; Shirima, C.; Jani, B.; Bakari, M.; Sillo, H.; Rusibamayila, N.; De Saeger, S.; Kimanya, M.; Gong, Y.; Simba, A.; et al. Outbreak of an acute aflatoxicosis in Tanzania during 2016. World Mycotoxin J. 2018, 11, 311–320. [Google Scholar] [CrossRef]
- Serck-Hanssen, A. Aflatoxin-induced fatal hepatitis? Arch. Environ. Health Int. J. 1970, 20, 729–731. [Google Scholar] [CrossRef]
- Bernhoft, A.; Torp, M.; Clasen, P.E.; Løesk, A.K.; Kristoffersenk, A.B. Influence of agronomic and climatic factors on Fusarium infestation and mycotoxin contamination of cereals in Norway. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2012, 29, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, R.; Ortega-Beltran, A.; Akande, A.; Mutegi, C.; Atehnkeng, J.; Kaptoge, L.; Senghor, A.; Adhikari, B.; Cotty, P. Biological control of aflatoxins in Africa: Current status and potential challenges in the face of climate change. World Mycotoxin J. 2016, 9, 771–789. [Google Scholar] [CrossRef]
- Echodu, R.; Malinga, G.M.; Kaducu, J.M.; Ovuga, E.; Haesaert, G. Prevalence of aflatoxin, ochratoxin and deoxynivalenol in cereal grains in northern Uganda: Implication for food safety and health. Toxicol. Rep. 2019, 6, 1012–1017. [Google Scholar] [CrossRef]
- Kaaya, A.N.; Harris, C.; Eigel, W. Peanut aflatoxin levels on farms and in markets of Uganda. Peanut Sci. 2006, 33, 68–75. [Google Scholar] [CrossRef]
- Olewnik, M. FSMA: Regulating prevention, detection and response, and imported foods. Cereal Foods World 2012, 57, 111–114. [Google Scholar] [CrossRef]
- Geiser, D.M.; Jiménez-Gasco, M.D.M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’donnell, K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Funnell-Harris, D.L.; Sattler, S.E.; O’Neill, P.M.; Eskridge, K.M.; Pedersen, J.F. Effect of waxy (low amylose) on fungal infection of sorghum grain. Phytopathology 2015, 105, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Bolchacova, E.; Voigt, K.; Crous, P.W.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Amatulli, M.T.; Spadaro, D.; Gullino, M.L.; Garibaldi, A. Conventional and real-time PCR for the identification of Fusarium fujikuroi and Fusarium proliferatum from diseased rice tissues and seeds. Eur. J. Plant Pathol. 2012, 134, 401–408. [Google Scholar] [CrossRef]
- Uganda Bureau of Statistics. National Population and Housing Census 2014—Main Report; Bureau of Statistics: Kampala, Uganda, 2014; pp. 1–209. [Google Scholar]
- Levine, S. Poverty and Human Development in Northern Uganda: Notes for the Development Partners Group for Northern Uganda Recovery and Development; UNDP: Kampala, Uganda, 2009. [Google Scholar]
- Ssewanyana, S.; Kasirye, I. Food security in Uganda: A dilemma to achieving the millennium development goal. No. 677-2016-46693. Res. Ser. 2010. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.O.; Cigelnik, E.L.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Proctor, R.H.; Plattner, R.D.; Brown, D.W.; Seo, J.-A.; Lee, Y.-W. Discontinuous distribution of fumonisin biosynthetic genes in the Gibberella fujikuroi species complex. Mycol. Res. 2004, 108, 815–822. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2017, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Hum. Genet. 1954, 1, 1–8. [Google Scholar]
- Saleh, A.A.; Esele, J.P.; Logrieco, A.; Ritieni, A.; Leslie, J.F. Fusarium verticillioides from finger millet in Uganda. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2012, 29, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, M.; Zani, A.; Rickerts, V.; McCormick, I.; Desnos-Ollivier, M.; Velegraki, A.; Escandon, P.; Ichikawa, T.; Ikeda, R.; Bienvenue, A.-L.; et al. Multilocus sequence typing analysis reveals that Cryptococcus neoformans var. neoformans is a recombinant population. Fungal Genet. Biol. 2016, 87, 22–29. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bengyella, L.; Yekwa, E.L.; Nawaz, K.; Iftikhar, S.; Tambo, E.; Alisoltani, A.; Feto, N.A.; Roy, P. Global invasive Cochliobolus species: Cohort of destroyers with implications in food losses and insecurity in the twenty-first century. Arch. Microbiol. 2017, 200, 119–135. [Google Scholar] [CrossRef]
- Kiss, N.; Homa, M.; Manikandan, P.; Mythili, A.; Krizsán, K.; Revathi, R.; Varga, M.; Papp, T.; Vágvölgyi, C.; Kredics, L.; et al. New species of the genus Curvularia: C. tamilnaduensis and C. coimbatorensis from fungal keratitis cases in South India. Pathogens 2019, 9, 9. [Google Scholar] [CrossRef]
- Gao, J.-X.; Chen, J. Transcriptome analysis identifies candidate genes associated with melanin and toxin biosynthesis and pathogenicity of the maize pathogen, Curvularia lunata. J. Phytopathol. 2018, 166, 233–241. [Google Scholar] [CrossRef]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.N.; Lysøe, E.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef]
- Pena, G.A.; Cavaglieri, L.R.; Chulze, S.N. Fusarium species and moniliformin occurrence in sorghum grains used as ingredient for animal feed in Argentina. J. Sci. Food Agric. 2018, 99, 47–54. [Google Scholar] [CrossRef]
- Wakelin, S.A.; Warren, R.A.; Kong, L.; Harvey, P.R. Management factors affecting size and structure of soil Fusarium communities under irrigated maize in Australia. Appl. Soil Ecol. 2008, 39, 201–209. [Google Scholar] [CrossRef]
- Nayaka, S.C.; Shankar, A.C.U.; Niranjana, S.R.; Wulff, E.G.; Mortensen, C.N.; Prakash, H.S. Detection and quantification of fumonisins from Fusarium verticillioides in maize grown in southern India. World J. Microbiol. Biotechnol. 2009, 26, 71–78. [Google Scholar] [CrossRef]
- Brown, D.W.; Cheung, F.; Proctor, R.H.; Butchko, R.A.; Zheng, L.; Lee, Y.; Utterback, T.; Smith, S.; Feldblyum, T.; Glenn, A.E.; et al. Comparative analysis of 87,000 expressed sequence tags from the fumonisin-producing fungus Fusarium verticillioides. Fungal Genet. Biol. 2005, 42, 848–861. [Google Scholar] [CrossRef]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Wilke, A.L.; Bronson, C.R.; Tomas, A.; Munkvold, G.P. Seed transmission of Fusarium verticillioides in maize plants grown under three different temperature regimes. Plant Dis. 2007, 91, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Maiorano, A.; Blandino, M.; Reyneri, A.; Vanara, F. Effects of maize residues on the Fusarium spp. infection and deoxynivalenol (DON) contamination of wheat grain. Crop. Prot. 2008, 27, 182–188. [Google Scholar] [CrossRef]
- Trenholm, H.L.; Prelusky, D.B.; Young, J.C.; Miller, J.D. A practical guide to the prevention of Fusarium mycotoxins in grain and animal feedstuffs I. Arch. Environ. Contam. Toxicol. 1989, 18, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, G.P. Cultural and genetic approaches to managing mycotoxins in maize. Annu. Rev. Phytopathol. 2003, 41, 99–116. [Google Scholar] [CrossRef]
- Stępień, Ł.; Gromadzka, K.; Chełkowski, J. Polymorphism of mycotoxin biosynthetic genes among Fusarium equiseti isolates from Italy and Poland. J. Appl. Genet. 2012, 53, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.A.; Mohd, M.H.; Nor, N.M.I.M.; Zakaria, L. Fumonisin B1-producing Fusarium species from agricultural crops in Malaysia. Crop. Prot. 2017, 98, 70–75. [Google Scholar] [CrossRef]
- Susca, A.; Proctor, R.H.; Butchko, R.A.; Haidukowski, M.; Stea, G.; Logrieco, A.; Moretti, A. Variation in the fumonisin biosynthetic gene cluster in fumonisin-producing and nonproducing black aspergilli. Fungal Genet. Biol. 2014, 73, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Thakur, R.P.; Senthilvel, S.; Nayak, S.; Reddy, S.V.; Rao, V.P.; Varshney, R.K.; Senapathy, S. Identification and characterization of toxigenic Fusaria associated with sorghum grain mold complex in India. Mycopathologia 2010, 171, 223–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tancic, S.; Stankovi, S.; Levic, J.; Krnjaja, V.; Vukojevi, J. Diversity of the Fusarium verticillioides and F. proliferatum isolates according to their fumonisin b1 production potential and origin. Genetika 2012, 44, 163–176. [Google Scholar] [CrossRef]
- Terciolo, C.; Bracarense, A.P.; Souto, P.C.; Cossalter, A.-M.; Dopavogui, L.; Loiseau, N.; Oliveira, C.A.F.; Pinton, P.; Oswald, I.P. Fumonisins at doses below EU regulatory limits induce histological alterations in piglets. Toxins 2019, 11, 548. [Google Scholar] [CrossRef]
- Kabak, B.; Dobson, A.D.W.; Var, I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006, 46, 593–619. [Google Scholar] [CrossRef]
| Genus Classification Based on ITS Region | N | Percentage (%) |
|---|---|---|
| Aspergillus | 1 | 0.63 |
| Bipolaris | 18 | 11.25 |
| Cladosporium | 2 | 1.25 |
| Curvularia | 13 | 8.13 |
| Diaporthe | 1 | 0.63 |
| Epicoccum | 12 | 7.50 |
| Exserohilum | 1 | 0.63 |
| Fusarium | 85 | 51.25 |
| Lasiodiplodia | 8 | 5.00 |
| Macrophomina | 3 | 1.88 |
| Nigrospora | 1 | 0.63 |
| Periconia | 1 | 0.63 |
| Sarocladium | 2 | 1.25 |
| Trametes | 2 | 1.25 |
| Trichoderma | 1 | 0.63 |
| Species classification based on TEF1-α | ||
| Aspergillus spp. | 1 | 0.63 |
| Fusarium andiyazi | 18 | 11.25 |
| Fusarium equiseti | 11 | 6.88 |
| Fusarium incarnatum | 21 | 13.13 |
| Fusarium proliferatum | 2 | 1.25 |
| Fusarium solani | 4 | 2.50 |
| Fusarium thapsinum | 2 | 1.25 |
| Fusarium verticillioides | 32 | 20.00 |
| Trichoderma gamsii | 1 | 0.63 |
| No sequence | 4 | 2.50 |
| Fungus | Maize (n = 44) | Millet (n = 12) | Peanut (n = 11) | Rice (n = 5) | Simsim (n = 2) | Sorghum (n = 22) |
|---|---|---|---|---|---|---|
| Fusarium andiyazi | 7.50 | 1.87 | 0.63 | 0.00 | 0.00 | 1.25 |
| Fusarium equiseti | 1.25 | 0.00 | 3.75 | 0.00 | 0.00 | 1.87 |
| Fusarium incarnatum | 3.13 | 3.13 | 0.00 | 1.87 | 0.00 | 5.00 |
| Fusarium proliferatum | 0.00 | 1.24 | 0.00 | 0.00 | 0.00 | 0.00 |
| Fusarium solani | 0.63 | 0.00 | 1.87 | 0.00 | 0.00 | 0.00 |
| Fusarium thapsinum | 0.00 | 0.00 | 0.00 | 0.00 | 0.63 | 0.63 |
| Fusarium verticillioides | 13.13 | 1.25 | 0.63 | 1.25 | 0.63 | 3.13 |
| Trichoderma gamsii | 0.63 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Fungal Species | Country of Origin | Fst | Chi-Square | p-Value |
|---|---|---|---|---|
| Fusarium verticillioides | Kenya (n = 10) | 0.13 | 19.87 | 0.00 |
| Eastern Uganda (n = 11) | 0.11 | 10.30 | 0.07 | |
| china (n = 13) | 0.03 | 20.75 | 0.02 | |
| USA (n = 14) | 0.12 | 23.75 | 0.00 | |
| Italy (n = 7) | 0.30 | 6.96 | 0.14 | |
| India (n = 19) | 0.21 | 38.93 | 0.00 | |
| Argentina (n = 10) | 0.04 | 9.51 | 0.09 | |
| Japan (n = 11) | 0.43 | 29.67 | 0.00 | |
| Spain (n = 17) | 0.01 | 12.20 | 0.09 | |
| Fusarium andiyazi | Argentina (n = 21) | 0.33 | 34.00 | 0.00 |
| USA (n = 6) | 0.24 | 19.00 | 0.04 | |
| China (n = 19) | 0.29 | 32.00 | 0.00 | |
| Australia | 0.32 | 24.00 | 0.01 | |
| Nigeria (n = 5) | 0.16 | 18.00 | 0.02 | |
| India (n = 4) | 0.05 | 17.00 | 0.07 | |
| Brazil (n = 9) | 0.10 | 18.55 | 0.07 | |
| Korea (n = 5) | 0.35 | 18.00 | 0.04 | |
| Italy (n = 5) | 0.36 | 18.00 | 0.06 | |
| Fusarium equiseti | USA (n = 10) | 0.36 | 20.00 | 0.01 |
| Italy (n = 12) | 0.26 | 22.00 | 0.04 | |
| Korea (n = 3) | 0.18 | 13.00 | 0.02 | |
| China (n = 13) | 0.38 | 23.00 | 0.00 | |
| Fusarium incarnatum | Spain (n = 5) | 0.19 | 24.00 | 0.00 |
| Australia (n = 10) | 0.47 | 29.00 | 0.00 | |
| India (n = 8) | 0.77 | 27.00 | 0.00 | |
| USA (n = 4) | 0.51 | 28.00 | 0.00 | |
| Brazil (n = 5) | 0.33 | 24.00 | 0.00 | |
| China (n = 15) | 0.64 | 34.00 | 0.00 |
| Fusarium Species | N | Absent (%) | Present (%) | |
|---|---|---|---|---|
| FUM1 gene | Fusarium andiyazi | 18 | 38.89 | 61.11 |
| Fusarium proliferatum | 2 | 0.00 | 100.00 | |
| Fusarium verticillioides | 32 | 3.13 | 96.88 | |
| FUM3 gene | Fusarium andiyazi | 18 | 50.00 | 50.00 |
| Fusarium proliferatum | 2 | 0.00 | 100.00 | |
| Fusarium verticillioides | 32 | 3.13 | 96.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wokorach, G.; Landschoot, S.; Audenaert, K.; Echodu, R.; Haesaert, G. Genetic Characterization of Fungal Biodiversity in Storage Grains: Towards Enhancing Food Safety in Northern Uganda. Microorganisms 2021, 9, 383. https://doi.org/10.3390/microorganisms9020383
Wokorach G, Landschoot S, Audenaert K, Echodu R, Haesaert G. Genetic Characterization of Fungal Biodiversity in Storage Grains: Towards Enhancing Food Safety in Northern Uganda. Microorganisms. 2021; 9(2):383. https://doi.org/10.3390/microorganisms9020383
Chicago/Turabian StyleWokorach, Godfrey, Sofie Landschoot, Kris Audenaert, Richard Echodu, and Geert Haesaert. 2021. "Genetic Characterization of Fungal Biodiversity in Storage Grains: Towards Enhancing Food Safety in Northern Uganda" Microorganisms 9, no. 2: 383. https://doi.org/10.3390/microorganisms9020383
APA StyleWokorach, G., Landschoot, S., Audenaert, K., Echodu, R., & Haesaert, G. (2021). Genetic Characterization of Fungal Biodiversity in Storage Grains: Towards Enhancing Food Safety in Northern Uganda. Microorganisms, 9(2), 383. https://doi.org/10.3390/microorganisms9020383







