Antimicrobial Activities of Marine Sponge-Associated Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sponge Collection, Processing, and Identification
2.2. Isolation of Bacteria
2.3. Morphological Identification of Bacteria
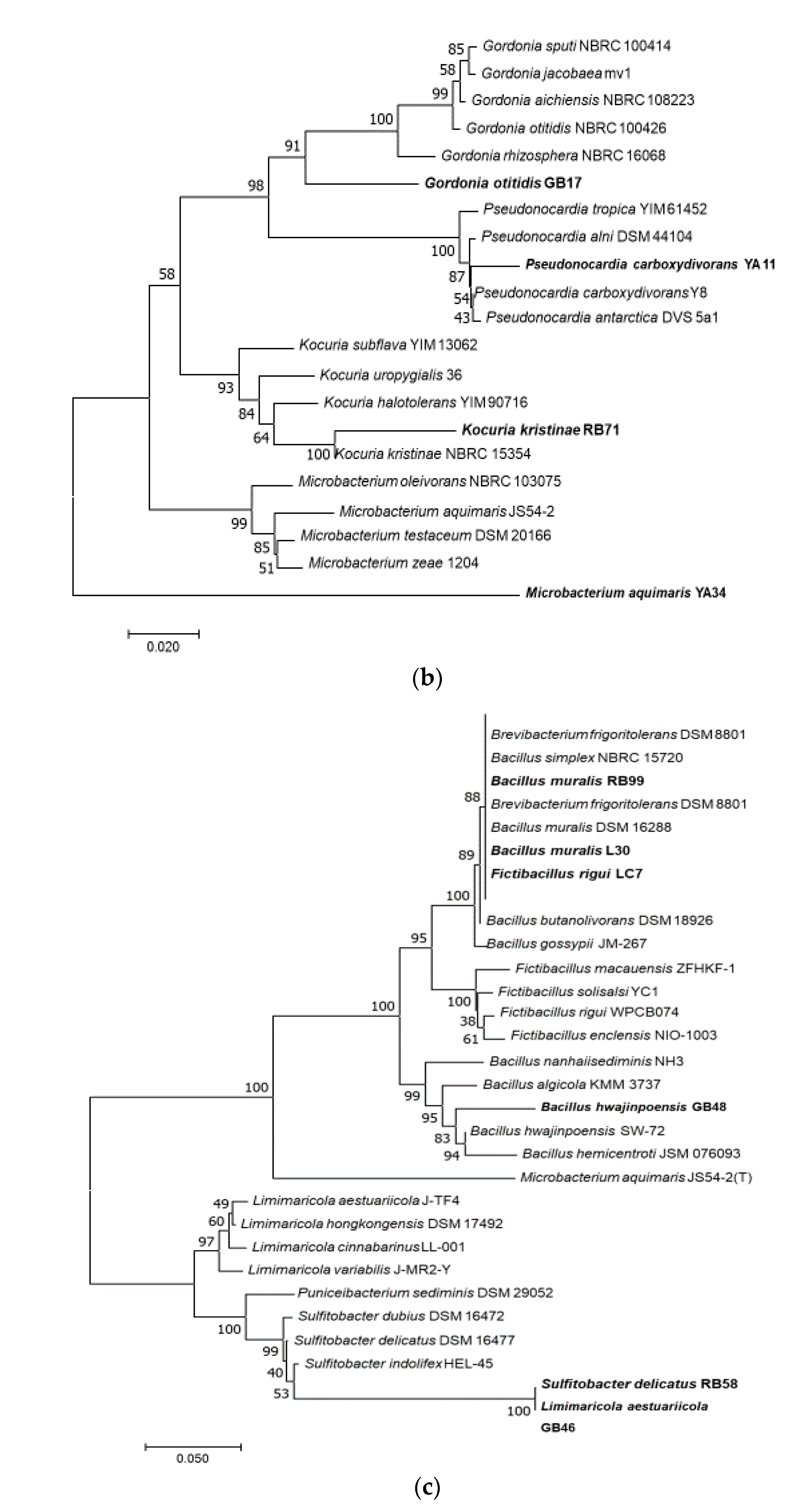
2.4. Genotypic Identification of Bacteria
2.5. Test Organisms
2.6. Agar Plugs Antimicrobial Sceening
2.6.1. Antibacterial and Anticandida Screening
2.6.2. Antifungal Screening
2.7. Evaluation of Different Media for Antimicrobial Production
2.8. Liquid State Antimicrobial Production, Screening and Large Scale Production
2.9. Bioautogram of Extract from RB27
2.10. Purfication of Extract from RB27
2.11. Nuclear Magnetic Resonance Spectroscopy and Accurate Mass Analysis of Extract from RB27
2.12. MIC and MBC of the Compound
3. Results
3.1. Sponge Taxonomy
3.2. Bacterial Profile and Test Strains
3.3. Antimicrobial Activities
3.4. Role of Media for Antimicrobial Production
3.5. Antimicrobial Production from Strain RB27, Preliminary Investigation and Purification
3.6. Bioautogram of the Purified Compounds and Their Identification
3.7. Minimum Inhibitory Concentration and Mnimum Bactericidal Concentration of the Compound
4. Discussion
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, R.S.; Finch, R.; Wegener, H.C.; Bywater, R.; Walters, J.; Lipsitch, M. Antibiotic resistance—The interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 2003, 3, 47–51. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Makvandi, P.; Zare, E.N.; Tay, F.R.; Niu, L. Advances in Antimicrobial Organic and Inorganic Nanocompounds in Biomedicine. Adv. Ther. 2020, 3, 2000024. [Google Scholar] [CrossRef]
- Makvandi, P.; Wang, C.; Zare, E.N.; Borzacchiello, A.; Niu, L.; Tay, F.R. Metal-Based Nanomaterials in Biomedical Applications: Antimicrobial Activity and Cytotoxicity Aspects. Adv. Funct. Mater. 2020, 30, 1910021. [Google Scholar] [CrossRef]
- Webster, N.S.; Taylor, M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2012, 14, 335–346. [Google Scholar] [CrossRef]
- Marques, D.N.; De Almeida, A.S.; Sousa, A.R.D.O.; Pereira, R.; Andrade, A.L.; Chaves, R.P.; Carneiro, R.F.; Vasconcelos, M.A.; Neto, L.G.D.N.; Pinheiro, U.; et al. Antibacterial activity of a new lectin isolated from the marine sponge Chondrilla caribensis. Int. J. Biol. Macromol. 2018, 109, 1292–1301. [Google Scholar] [CrossRef]
- Beesoo, R.; Bhagooli, R.; Neergheen, V.S.; Li, W.-W.; Kagansky, A.; Bahorun, T. Antibacterial and antibiotic potentiating activities of tropical marine sponge extracts. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 196, 81–90. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Hu, W.-P.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2007, 24, 31–86. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Lei, J.; Franco, C.M.M.; Zhang, W. Marine Sponge Derived Natural Products between 2001 and 2010: Trends and Opportunities for Discovery of Bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [Green Version]
- Gogineni, V.; Hamann, M.T. Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 81–196. [Google Scholar] [CrossRef]
- Newman, D.J. Natural Products as Leads to Potential Drugs: An Old Process or the New Hope for Drug Discovery? J. Med. Chem. 2008, 51, 2589–2599. [Google Scholar] [CrossRef] [PubMed]
- Meesala, S.; Singh, R.; Watve, M.G.; Naik, D.G. A new antibacterial imidazole from the marine sponge Iricinia fusca. Indian J. Nat. Prod. Resour. 2018, 9, 75–78. [Google Scholar]
- Raslan, A.E.; Radwan, M.M.; Ahmed, S.A.; Nafady, A.M.; Zaki, M.A.; Wanas, A.S.; Abou-Karam, M.; Shier, T.W.; Hassanean, H.A.; ElSohly, M.A. Monanchoramides A–D, ceramides from the marine sponge Monanchora clathrata with cytotoxic activity. Phytochem. Lett. 2018, 23, 83–89. [Google Scholar] [CrossRef]
- Liu, H.-B.; Lauro, G.; O’Connor, R.D.; Lohith, K.; Kelly, M.; Colin, P.; Bifulco, G.; Bewley, C.A. Tulongicin, an Antibacterial Tri-Indole Alkaloid from a Deep-Water Topsentia sp. Sponge. J. Nat. Prod. 2017, 80, 2556–2560. [Google Scholar] [CrossRef]
- Visamsetti, A.; Ramachandran, S.S.; Kandasamy, D. Penicillium chrysogenum DSOA associated with marine sponge (Tedania anhelans) exhibit antimycobacterial activity. Microbiol. Res. 2016, 185, 55–60. [Google Scholar] [CrossRef]
- Maloof, A.C.; Rose, C.V.; Beach, R.; Samuels, B.M.; Calmet, C.C.; Erwin, U.H.; Poirier, G.R.; Yao, N.; Simons, F.J. Possible animal-body fossils in pre-Marinoan limestones from South Australia. Nat. Geosci. 2010, 3, 653–659. [Google Scholar] [CrossRef]
- Van Soest, R.; Boury-Esnault, N.; Hooper, J.; Rützler, K.; de Voogd, N.; de Glasby, B.A.; Hajdu, E.; Pisera, A.; Manconi, R.; Schoenberg, C.; et al. World Porifera Database. 2011. Available online: http://www.marinespecies.org/ (accessed on 16 December 2012).
- Webster, N.S.; Wilson, K.J.; Blackall, L.L.; Hill, R.T.; Toledo-Arana, A.; Valle, J.; Solano, C.; Arrizubieta, M.J.; Cucarella, C.; Lamata, M.; et al. Phylogenetic Diversity of Bacteria Associated with the Marine Sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 2001, 67, 4538–4545. [Google Scholar] [CrossRef] [Green Version]
- Mearns-Spragg, A.; Bregu, M.; Boyd, K.; Burgess, J. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett. Appl. Microbiol. 1998, 27, 142–146. [Google Scholar] [CrossRef]
- Selvin, J.; Kennedy, J.; Lejon, D.P.H.; Kiran, G.S.; Dobson, A.D. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb. Cell Factories 2012, 11, 72. [Google Scholar] [CrossRef] [Green Version]
- Indraningrat, A.A.G.; Smidt, H.; Sipkema, D. Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, T.R.A.; Kavlekar, D.P.; Bharathi, P.A.L. Marine Drugs from Sponge-Microbe Association—A Review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laport, M.S.; Santos, O.C.S.; Muricy, G. Marine Sponges: Potential Sources of New Antimicrobial Drugs. Curr. Pharm. Biotechnol. 2009, 10, 86–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 2006, 33, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Santos-Gandelman, J.F.; Giambiagi-Demarval, M.; Oelemann, W.M.R.; Laport, M.S. Biotechnological Potential of Sponge-Associated Bacteria. Curr. Pharm. Biotechnol. 2014, 15, 143–155. [Google Scholar] [CrossRef]
- Graça, A.P.; Viana, F.; Bondoso, J.; Correia, M.I.; Gomes, L.; Humanes, M.; Reis, A.; Xavier, J.R.; Gaspar, H.; Lage, O.M. The antimicrobial activity of heterotrophic bacteria isolated from the marine sponge Erylus deficiens (Astrophorida, Geodiidae). Front. Microbiol. 2015, 6, 389. [Google Scholar] [CrossRef] [Green Version]
- Hoppers, A.; Stoudenmire, J.; Wu, S.; Lopanik, N.B. Antibiotic activity and microbial community of the temperate sponge, Haliclona sp. J. Appl. Microbiol. 2015, 118, 419–430. [Google Scholar] [CrossRef]
- Tawfike, A.F.; Attia, E.Z.; Desoukey, S.Y.; Hajjar, D.; Makki, A.A.; Schupp, P.J.; Edrada-Ebel, R.; Abdelmohsen, U.R. New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive Investigation of Marine Actinobacteria Associated with the Sponge Halichondria panicea. Appl. Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef] [Green Version]
- Bibi, F.; Faheem, M.; Azhar, E.; Yasir, M.; Alvi, S.; Kamal, M.; Ullah, I.; Naseer, M. Bacteria From Marine Sponges: A Source of New Drugs. Curr. Drug Metab. 2017, 18, 11–15. [Google Scholar] [CrossRef]
- Kennedy, J.; Baker, P.; Piper, C.; Cotter, P.D.; Walsh, M.; Mooij, M.J.; Bourke, M.B.; Rea, M.C.; O’Connor, P.M.; Ross, R.P.; et al. Isolation and Analysis of Bacteria with Antimicrobial Activities from the Marine Sponge Haliclona simulans Collected from Irish Waters. Mar. Biotechnol. 2008, 11, 384–396. [Google Scholar] [CrossRef]
- Kiran, G.S.; Priyadharsini, S.; Sajayan, A.; Ravindran, A.; Selvin, J. An antibiotic agent pyrrolo[1,2-a]pyrazine-1,4-dione,hexahydro isolated from a marine bacteria Bacillus tequilensis MSI45 effectively controls multi-drug resistant Staphylococcus aureus. RSC Adv. 2018, 8, 17837–17846. [Google Scholar] [CrossRef] [Green Version]
- Reimer, A.; Blohm, A.; Quack, T.; Grevelding, C.G.; Kozjak-Pavlovic, V.; Rudel, T.; Hentschel, U.; Abdelmohsen, U.R. Inhibitory activities of the marine streptomycete-derived compound SF2446A2 against Chlamydia trachomatis and Schistosoma mansoni. J. Antibiot. 2015, 68, 674–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomo, S.; González, I.; De La Cruz, M.; Martín, J.; Tormo, R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Gendy, M.M.A.; El-Bondkly, A.M.A. Production and genetic improvement of a novel antimycotic agent, Saadamycin, against Dermatophytes and other clinical fungi from Endophytic Streptomyces sp. Hedaya48. J. Ind. Microbiol. Biotechnol. 2010, 37, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Tanner, J.E.; Barnett, S.J.; Bekker, J.; Franco, C.M.M.; Zhang, W. A controlled aquarium system and approach to study the role of sponge-bacteria interactions using Aplysilla rosea and Vibrio natriegens. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Van Soest, R.W.M.; Boury-Esnault, N.; Vacelet, J.; Dohrmann, M.; Erpenbeck, D.; De Voogd, N.J.; Santodomingo, N.; Vanhoorne, B.; Kelly, M.; Hooper, J.N.A. Global Diversity of Sponges (Porifera). PLoS ONE 2012, 7, e35105. [Google Scholar] [CrossRef]
- Sorokin, S.; Fromont, J.; Currie, D. Demosponge Biodiversity in the Benthic Protection Zone of the Great Australian Bight. Trans. R. Soc. South Aust. 2007, 131, 192–204. [Google Scholar] [CrossRef]
- Sorokin, S.J.; Currie, D.R. The distribution and diversity of sponges in Spencer Gulf. In Report to Nature Foundation SA Inc; F2008/001153-1; SARDI Aquatic Sciences Publication: Adelaide, Australia, 2008. [Google Scholar]
- Hooper, J.N.A.; Van Soest, R. Systema Porifera. A Guide to the Classification of Sponges; Plenum Publishing: New York, NY, USA, 2002; Volume 18, pp. 1–7. [Google Scholar]
- Morrow, C.; Cárdenas, P. Proposal for a revised classification of the Demospongiae (Porifera). Front. Zoöl. 2015, 12, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Hooper, J.N. Sponguide: Guide to Sponge Collection and Identification; Queensland Museum: South Brisbane, Australia, 2000. [Google Scholar]
- Yang, Q.; Franco, C.M.M.; Sorokin, S.J.; Zhang, W. Development of a multilocus-based approach for sponge (phylum Porifera) identification: Refinement and limitations. Sci. Rep. 2017, 7, 41422. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.W.; Schupp, P.J.; Dahllöf, I.; Kjelleberg, S.; Steinberg, P.D. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 2003, 6, 121–130. [Google Scholar] [CrossRef]
- Yang, Q.; Franco, C.M.M.; Zhang, W. Sponge-associated actinobacterial diversity: Validation of the methods of actinobacterial DNA extraction and optimization of 16S rRNA gene amplification. Appl. Microbiol. Biotechnol. 2015, 99, 8731–8740. [Google Scholar] [CrossRef] [PubMed]
- Mincer, T.J.; Fenical, W.; Jensen, P.R. Culture-Dependent and Culture-Independent Diversity within the Obligate Marine Actinomycete Genus Salinispora. Appl. Environ. Microbiol. 2005, 71, 7019–7028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Lee, Y.K.; Zhang, W.; Lee, H.K. Culturable Actinobacteria from the Marine Sponge Hymeniacidon perleve: Isolation and Phylogenetic Diversity by 16S rRNA gene-RFLP Analysis. Antonie van Leeuwenhoek 2006, 90, 159–169. [Google Scholar] [CrossRef]
- Kaewkla, O.; Franco, C.M.M. Rational Approaches to Improving the Isolation of Endophytic Actinobacteria from Australian Native Trees. Microb. Ecol. 2012, 65, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Matobole, R.M.; Van Zyl, L.J.; Parker-Nance, S.; Davies-Coleman, M.T.; Trindade, M. Antibacterial Activities of Bacteria Isolated from the Marine Sponges Isodictya compressa and Higginsia bidentifera Collected from Algoa Bay, South Africa. Mar. Drugs 2017, 15, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laich, F.; Vaca, I.; Chávez, R. Rhodotorula portillonensis sp. nov., a basidiomycetous yeast isolated from Antarctic shallow-water marine sediment. Int. J. Syst. Evol. Microbiol. 2013, 63, 3884–3891. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Rashad, F.M.; Fathy, H.M.; El-Zayat, A.S.; Elghonaimy, A.M. Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol. Res. 2015, 175, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, Z.; Miao, X.; Zhang, F. The Screening of Antimicrobial Bacteria with Diverse Novel Nonribosomal Peptide Synthetase (NRPS) Genes from South China Sea Sponges. Mar. Biotechnol. 2008, 11, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Akbar, A.; Shah, A.; Abbas, G.; Hussain, M.; Khan, T. Isolation optimization and characterization of antimicrobial peptide producing bacteria from soil. J. Anim. Plant. Sci. 2015, 25, 1107–1113. [Google Scholar]
- Ahmed, N.; Uzair, B.; Ayaz, S.; Ahmed, V.U. Antibacterial activity of marine bacteria from Arabian Sea of Pakistan. Internet J. Microbiol. 2008, 4, 1–5. [Google Scholar] [CrossRef]
- Dona, A.C.; Kyriakides, M.; Scott, F.; Shephard, E.A.; Varshavi, D.; Veselkov, K.; Everett, J.R. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput. Struct. Biotechnol. J. 2016, 14, 135–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Franco, C.M.M.; Zhang, W. Uncovering the hidden marine sponge microbiome by applying a multi-primer approach. Sci. Rep. 2019, 9, 6214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Franco, C.M.M.; Lin, H.-W.; Zhang, W. Untapped sponge microbiomes: Structure specificity at host order and family levels. FEMS Microbiol. Ecol. 2019, 95, 95. [Google Scholar] [CrossRef] [PubMed]
- Santos, O.C.S.; Pontes, P.V.; Santos, J.F.; Muricy, G.; Giambiagi-Demarval, M.; Laport, M.S. Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res. Microbiol. 2010, 161, 604–612. [Google Scholar] [CrossRef]
- Lafi, F.F.; Garson, M.J.; Fuerst, J.A. Culturable Bacterial Symbionts Isolated from Two Distinct Sponge Species (Pseudoceratina clavata and Rhabdastrella globostellata) from the Great Barrier Reef Display Similar Phylogenetic Diversity. Microb. Ecol. 2005, 50, 213–220. [Google Scholar] [CrossRef]
- Margassery, L.M.; Kennedy, J.; O’Gara, F.; Dobson, A.D.W.; Morrissey, J.P. Diversity and antibacterial activity of bacteria isolated from the coastal marine sponges Amphilectus fucorum and Eurypon major. Lett. Appl. Microbiol. 2012, 55, 2–8. [Google Scholar] [CrossRef]
- Bibi, F.; Alvi, S.A.; Al-Sofyani, A.; Yasir, M.; Kensarah, E.A.; Azhar, E.I. Research Article Two marine sponges-associated cultivable bacteria: Diversity and biological activities. Genet. Mol. Res. 2018, 17, 16039910. [Google Scholar] [CrossRef]
- Flemer, B.; Kennedy, J.; Margassery, L.M.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D. Diversity and antimicrobial activities of microbes from two Irish marine sponges, Suberites carnosus and Leucosolenia sp. J. Appl. Microbiol. 2011, 112, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.; Yang, Y.-T.; Lu, M.-C.; Wong, T.-Y.; Sung, P.-J.; Huang, Y.-S. Antimicrobial activity and diversity of bacteria associated with Taiwanese marine sponge Theonella swinhoei. Ann. Microbiol. 2019, 69, 253–265. [Google Scholar] [CrossRef]
- Liu, T.; Wu, S.; Zhang, R.; Wang, D.; Chen, J.; Zhao, J. Diversity and antimicrobial potential of Actinobacteria isolated from diverse marine sponges along the Beibu Gulf of the South China Sea. FEMS Microbiol. Ecol. 2019, 95, 95. [Google Scholar] [CrossRef] [Green Version]
- Baig, U.; Dahanukar, N.; Shintre, N.; Holkar, K.; Pund, A.; Lele, U.; Gujarathi, T.; Patel, K.; Jakati, A.; Singh, R.; et al. Phylogenetic diversity and activity screening of cultivable actinobacteria isolated from marine sponges and associated environments from the western coast of India. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bibi, F.; Yasir, M.; Al-Sofyani, A.; Naseer, M.I.; Azhar, E.I. Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. EA348. Saudi J. Biol. Sci. 2020, 27, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Cita, Y.P.; Suhermanto, A.; Radjasa, O.K.; Sudharmono, P. Antibacterial activity of marine bacteria isolated from sponge Xestospongia testudinaria from Sorong, Papua. Asian Pac. J. Trop. Biomed. 2017, 7, 450–454. [Google Scholar] [CrossRef]
- Graça, A.P.; Bondoso, J.; Gaspar, H.; Xavier, J.R.; Monteiro, M.C.; De La Cruz, M.; Oves-Costales, D.; Vicente, F.; Lage, O.M. Antimicrobial Activity of Heterotrophic Bacterial Communities from the Marine Sponge Erylus discophorus (Astrophorida, Geodiidae). PLoS ONE 2013, 8, e78992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, U.R.; Pimentel-Elardo, S.M.; Hanora, A.; Radwan, M.; Abou-El-Ela, S.H.; Ahmed, S.; Hentschel, U. Isolation, Phylogenetic Analysis and Anti-infective Activity Screening of Marine Sponge-Associated Actinomycetes. Mar. Drugs 2010, 8, 399–412. [Google Scholar] [CrossRef]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Rajasabapathy, R.; Ghadi, S.C.; Manikandan, B.; Mohandass, C.; Surendran, A.; Dastager, S.G.; Meena, R.M.; Rathinam, A.J. Antimicrobial profiling of coral reef and sponge associated bacteria from southeast coast of India. Microb. Pathog. 2020, 141, 103972. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Stach, J.E. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Anand, T.P.; Bhat, A.W.; Shouche, Y.S.; Roy, U.; Siddharth, J.; Sarma, S.P. Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of South East India. Microbiol. Res. 2006, 161, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C.K.M. Strategies for Fermentation Medium Optimization: An In-Depth Review. Front. Microbiol. 2017, 7, 2087. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.H.; Yuan, W.; Li, Z.Y.; Li, J.; Li, L.; Sun, J.B.; Gui, Y.H.; Wang, J.; Ye, B.P.; Lin, H.W. Anti-MRSA actinomycins D1-D4 from the marine sponge-associated Streptomyces sp. LHW52447. Tetrahedron 2018, 74, 5914–5919. [Google Scholar] [CrossRef]


| Sample Code | Sample ID | SYP | AP | NS | HV | MA | NA | TS | Total |
|---|---|---|---|---|---|---|---|---|---|
| GB_SP_01 | Geodia sp. | 4 (1) * | 5 (2) | - | 3 (1) | 1 (1) | 2 (1) | 1 (1) | 16 (7) |
| GB_SP_08 | Chondrosida sp. | 7 (2) | 14 (4) | 4 (2) | 6 (2) | 2 (1) | 2 (1) | 1 (1) | 36 (14) |
| GB_SP_ 21 | Chondrosida sp. | - | 4 (1) | 3 (1) | - | 3 (1) | 3 (1) | 2 (1) | 15 (5) |
| GB_SP_23 | Chondrosida sp. | 5 (2) | 7 (3) | - | 5 (2) | 6 (2) | 6 (2) | - | 29 (11) |
| RB_SP_01 | Ircinia sp. | - | 8 (3) | - | 6 (3) | - | 5 (2) | 7 (3) | 26 (11) |
| RB_SP_02 | Poecilosclerida sp. | 2 (1) | 4 (2) | - | 2 (1) | 5 (2) | - | - | 13 (6) |
| RB_SP_03 | Crella sp. | - | - | - | - | 4 (2) | 4 (2) | 4 (2) | 12 (6) |
| RB_SP_11 | Sarcotragus sp. | 6 (3) | 4 (2) | - | 3 (1) | 5 (2) | 2 (1) | - | 20 (9) |
| RB_SP_12 | Carteriospongia foliascens | - | 4 (2) | - | 3 (1) | - | - | 4 (2) | 11 (5) |
| RB_SP_16 | Aplysilla sulfurea | 17 (8) | 35 (16) | 6 (3) | 28 (14) | 11 (5) | 12 (6) | 8 (4) | 117 (56) |
| RB_SP_17 | Dendrilla sp. | 3 (1) | 10 (4) | - | 5 (2) | 6 (3) | 4 (2) | 2 (1) | 30 (13) |
| RB_SP_18 | Tedania tubulifera | 9 (4) | 15 (6) | 2 (1) | 10 (5) | 7 (3) | 7 (3) | 8 (4) | 58 (26) |
| Strains | Phylum | Positive * | Negative | Total Screened |
|---|---|---|---|---|
| Streptomyces | Actinobacteria | 58 | 14 | 72 |
| Gordonia | Actinobacteria | 1 | 2 | 3 |
| Bacillus | Firmicutes | 3 | 23 | 26 |
| Fictibacillus | Firmicutes | 1 | 4 | 5 |
| Sulfitobacter | Proteobacteria | 1 | 7 | 8 |
| Kocuria | Actinobacteria | 2 | 7 | 9 |
| Pseudonocardia | Actinobacteria | 1 | 1 | 2 |
| Microbacterium | Actinobacteria | 1 | 6 | 7 |
| Limimaricola | Proteobacteria | 1 | 3 | 4 |
| Janibacter | Actinobacteria | 0 | 1 | 1 |
| Muricauda | Bacteroidetes | 0 | 1 | 1 |
| Staphylococcus | Firmicutes | 0 | 2 | 2 |
| Micrococcus | Actinobacteria | 1 | 5 | 6 |
| Falsibacillus | Firmicutes | 0 | 5 | 5 |
| Rhodovulum | Proteobacteria | 0 | 2 | 2 |
| Mycolicibacterium | Actinobacteria | 0 | 1 | 1 |
| Rhodococcus | Actinobacteria | 0 | 8 | 8 |
| Pseudomonas | Proteobacteria | 0 | 3 | 3 |
| Leisingera | Proteobacteria | 0 | 1 | 1 |
| Isoptericola | Actinobacteria | 0 | 2 | 2 |
| Pseudoalteromonas | Proteobacteria | 0 | 1 | 1 |
| Total | 70 | 99 | 169 |
| Strains | YP * | PD | SYP | MS | ISP2 |
|---|---|---|---|---|---|
| RB27 | - | 14 | - | 26 | 26 |
| RB53 | - | 16 | - | 29 | 19 |
| RB154 | - | - | 10 | 21 | 26 |
| RBYA1 | - | - | 14 | 18 | 16 |
| RBYA2 | - | 14 | - | 20 | 18 |
| RBYA21 | - | - | 14 | - | 21 |
| RBYA22 | - | - | - | - | 16 |
| RBYA32 | - | 14 | - | 18 | 22 |
| Strains | Media | Extract | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | |||||||||
| SA | MRSA | SA | MRSA | SA | MRSA | SA | MRSA | SA | MRSA | SA | MRSA | SA | MRSA | ||
| RB27 | ISP2 | - | - | 23 | 19 | 27 | 23 | 23 | 20 | 24 | 21 | 23 | 20 | 26 | 19 |
| SM | - | - | 14 | - | 12 | - | 14 | 13 | 17 | 15 | 15 | - | - | - | |
| MS | - | - | 27 | 26 | 27 | 27 | 28 | 29 | 28 | 28 | 28 | 26 | 29 | 28 | |
| GM | - | - | 21 | 15 | 21 | 17 | 23 | 18 | 23 | 17 | 23 | 20 | 24 | 19 | |
| RB53 | ISP2 | - | - | 16 | - | 17 | - | 14 | - | - | - | - | - | 7 | - |
| SM | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| MS | - | - | - | - | 22 | 19 | 22 | 18 | 22 | 21 | 22 | 19 | 22 | 19 | |
| GM | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| RB154 | ISP2 | 13 | 13 | 13 | 13 | 16 | 14 | 16 | 11 | 11 | 8 | 9 | 8 | 18 | 10 |
| SM | 14 | 17 | 17 | 18 | 17 | 19 | 13 | 19 | 13 | 13 | 13 | 16 | 13 | 13 | |
| MS | 14 | 14 | 15 | 17 | 13 | 17 | 12 | 12 | 13 | 17 | - | - | - | - | |
| GM | 16 | 17 | 18 | 20 | 18 | 19 | 19 | 18 | 17 | 19 | 18 | 20 | 18 | 20 | |
| Strains | MIC (mg/mL) | MBC (mg/mL) |
|---|---|---|
| Methicillin-resistant Staphylococcus aureus | 1.25 | 2.5 |
| Staphylococcus aureus | 0.625 | 1.25 |
| Enterococcus faecalis | 0.625 | 1.25 |
| Corynebacterium striatum | 1.25 | 2.5 |
| Streptococcusagalactiae | 1.25 | 2.5 |
| Staphylococcuscapitis | 0.3125 | 1.25 |
| Klebsiella pneumoniae | >10 | >10 |
| Corynebacterium sp. | >10 | >10 |
| Pseudomonas aeruginosa | >10 | >10 |
| Escherichia coli | >10 | >10 |
| Micrococcus sp. | >10 | >10 |
| Bacillus cereus | >10 | >10 |
| Staphylococcus epidermidis | >10 | >10 |
| Fictibacillus arsenicus | >10 | >10 |
| Stenotrophomonas maltophilia | >10 | >10 |
| Streptococcus pyogenes | >10 | >10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anteneh, Y.S.; Yang, Q.; Brown, M.H.; Franco, C.M.M. Antimicrobial Activities of Marine Sponge-Associated Bacteria. Microorganisms 2021, 9, 171. https://doi.org/10.3390/microorganisms9010171
Anteneh YS, Yang Q, Brown MH, Franco CMM. Antimicrobial Activities of Marine Sponge-Associated Bacteria. Microorganisms. 2021; 9(1):171. https://doi.org/10.3390/microorganisms9010171
Chicago/Turabian StyleAnteneh, Yitayal S., Qi Yang, Melissa H. Brown, and Christopher M. M. Franco. 2021. "Antimicrobial Activities of Marine Sponge-Associated Bacteria" Microorganisms 9, no. 1: 171. https://doi.org/10.3390/microorganisms9010171
APA StyleAnteneh, Y. S., Yang, Q., Brown, M. H., & Franco, C. M. M. (2021). Antimicrobial Activities of Marine Sponge-Associated Bacteria. Microorganisms, 9(1), 171. https://doi.org/10.3390/microorganisms9010171






