Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures
Abstract
:1. Introduction
2. Results and Discussion
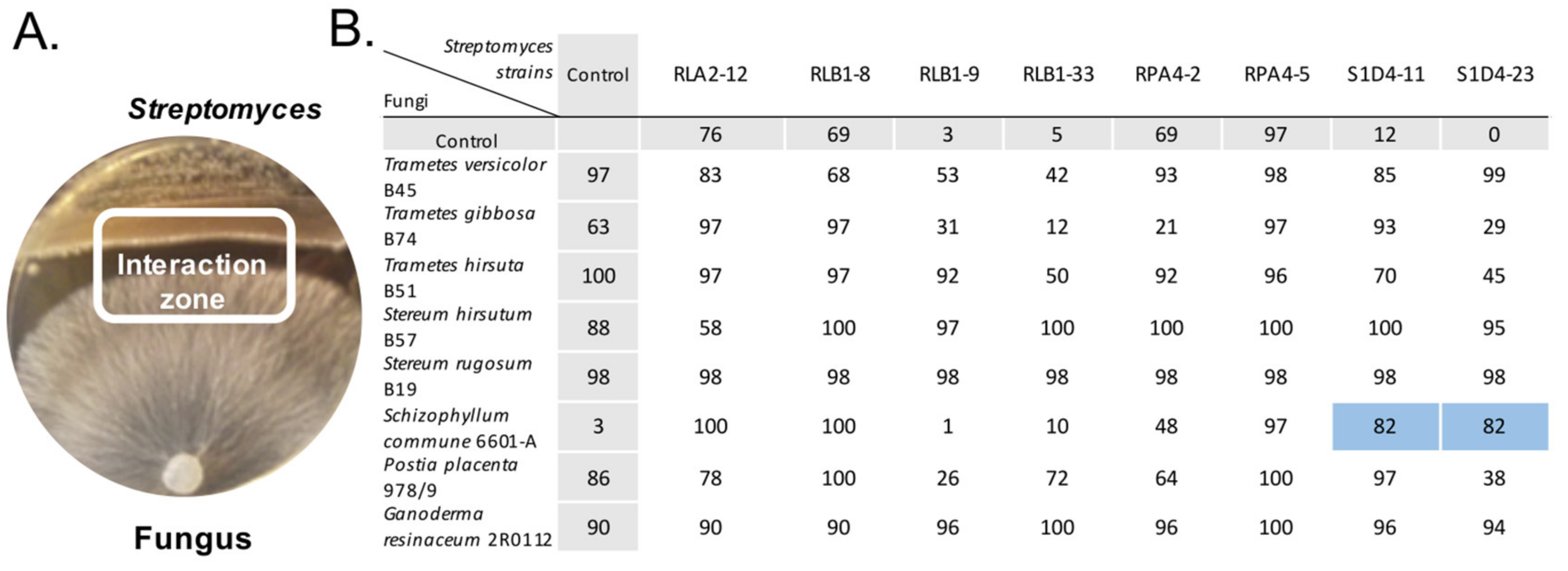
2.1. Induction of Antibacterial Activities by Streptomyces-Fungus Co-Culture
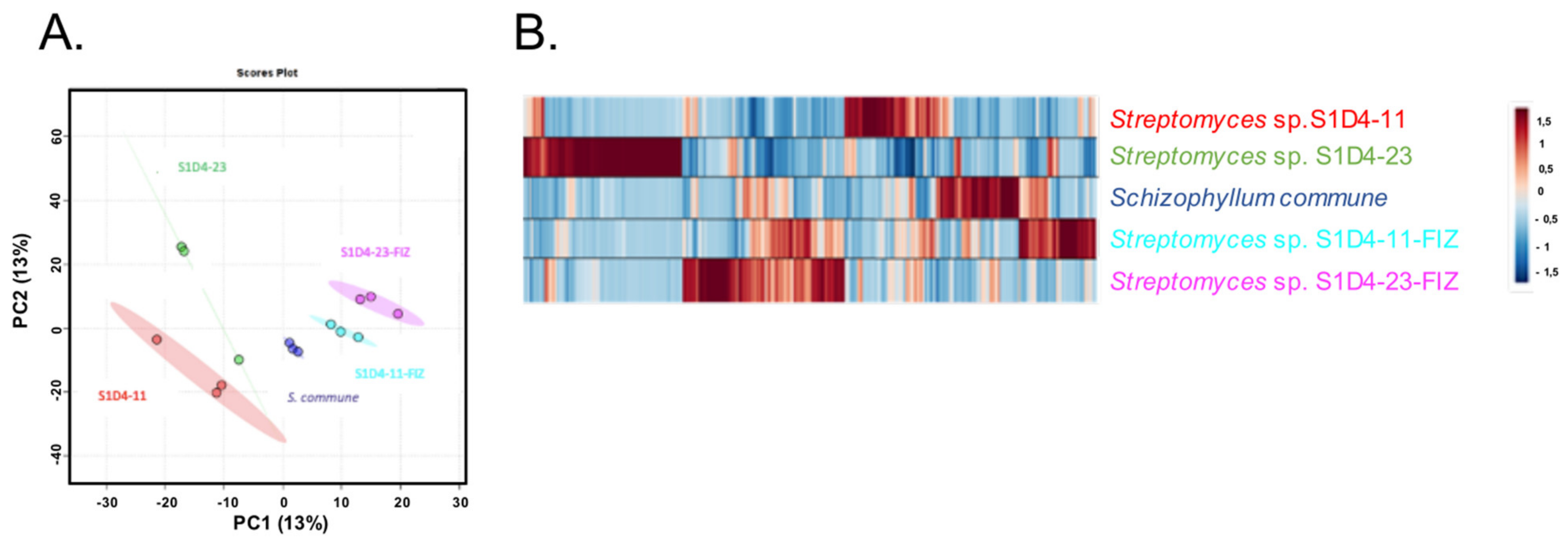
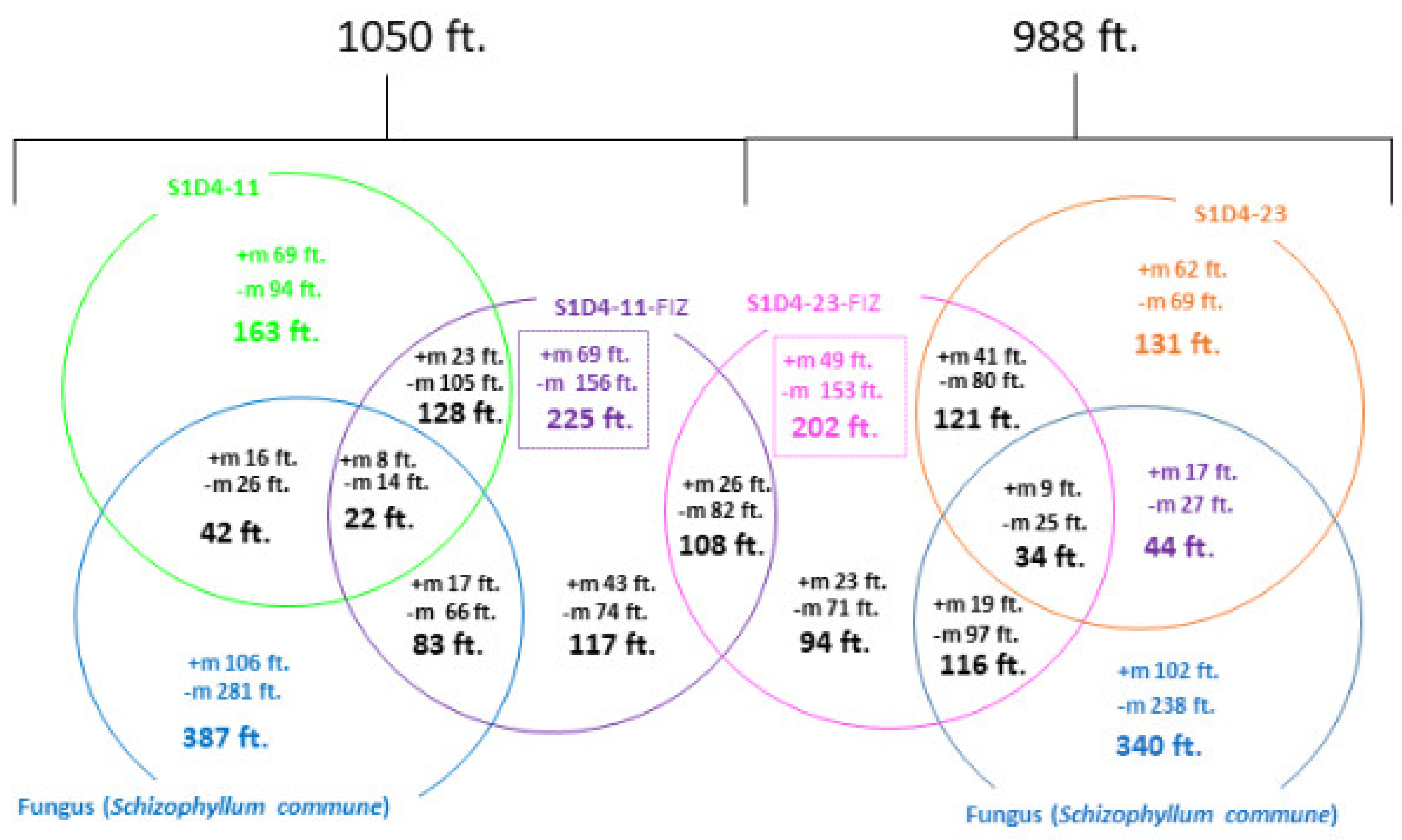
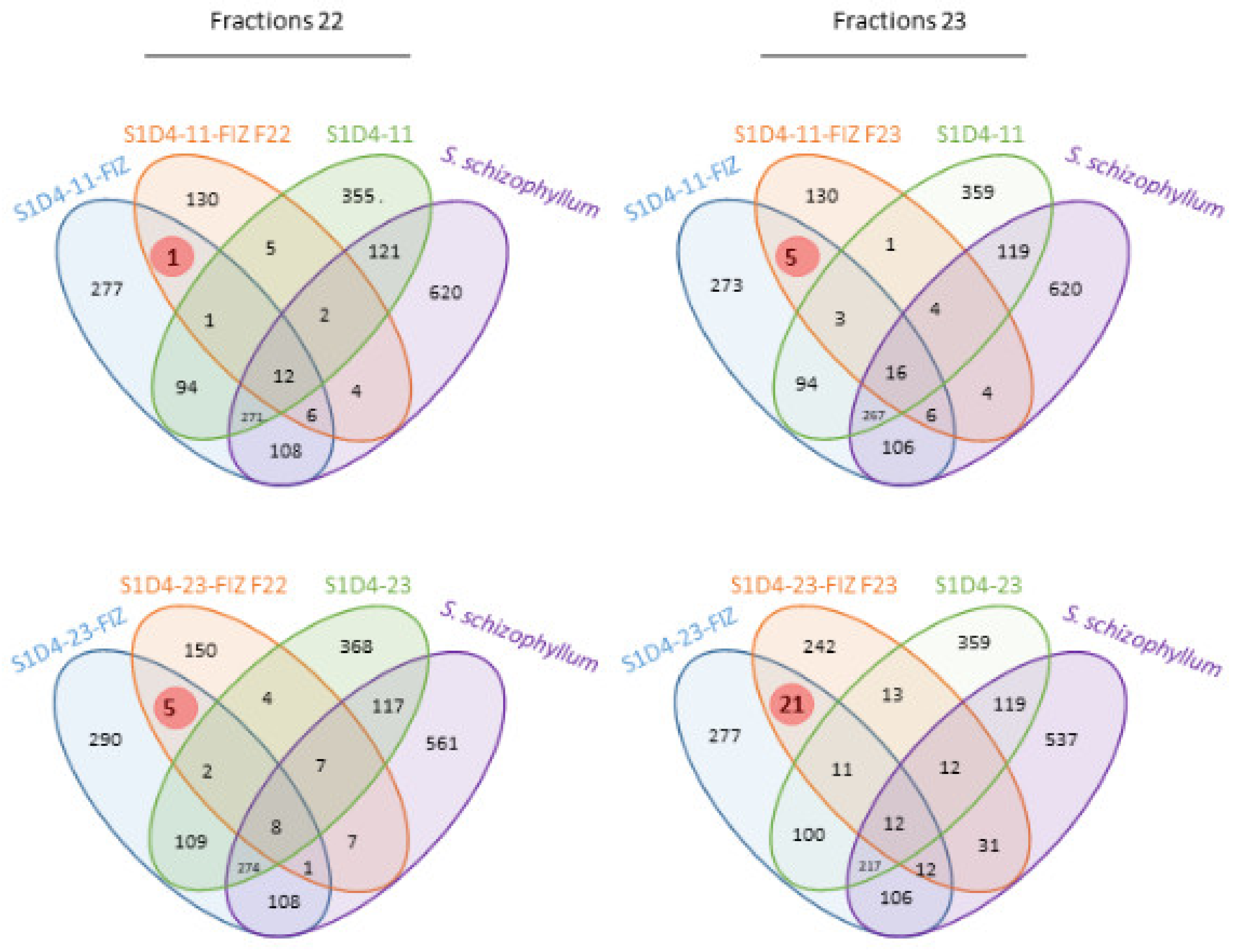
2.2. Impact of Co-Culture on Metabolome
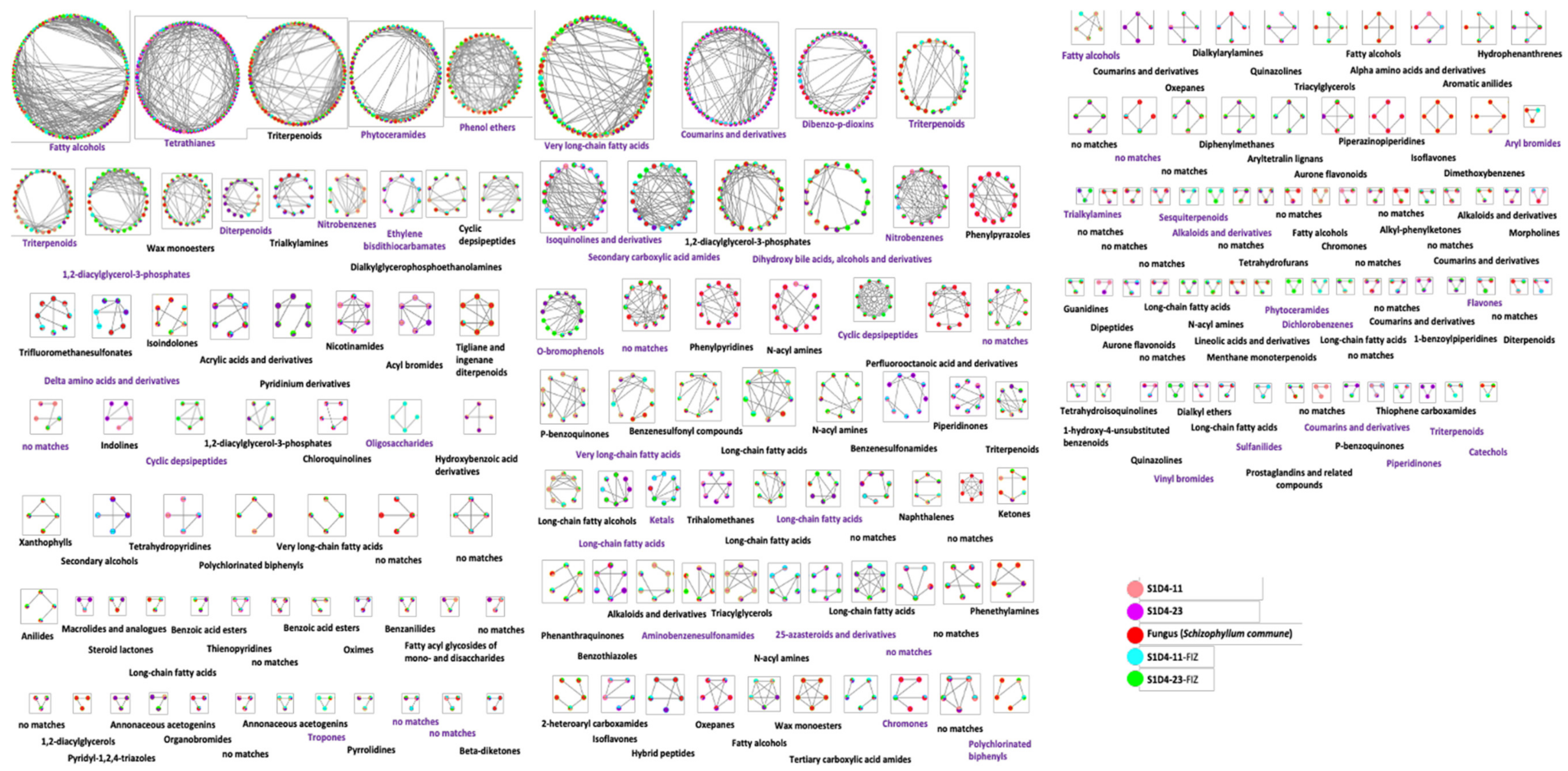
2.3. Towards the Identification of Potential Bioactive Compounds
3. Conclusions
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Metabolite Extraction
4.3. Anti-Bacterial Screening
4.4. GC-MS and LC-MS/MS Metabolite Profiling
4.5. Extract Fractioning
4.6. Molecular Networking
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial–fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Velez, P.; Espinosa-Asuar, L.; Figueroa, M.; Gasca-Pineda, J.; Aguirre-von-Wobeser, E.; Eguiarte, L.E.; Hernandez-Monroy, A.; Souza, V. Nutrient dependent cross-kingdom interactions: Fungi and bacteria from an oligotrophic desert oasis. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef] [Green Version]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Liras, P. Harnessing microbiota interactions to produce bioactive metabolites: Communication signals and receptor proteins. Curr. Opin. Pharmacol. 2019, 48, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites: A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Nicault, M.; Tidjani, A.-R.; Gauthier, A.; Dumarcay, S.; Gelhaye, E.; Bontemps, C.; Leblond, P. Mining the biosynthetic potential for specialized metabolism of a Streptomyces soil community. Antibiotics 2020, 9, 271. [Google Scholar] [CrossRef]
- Waksman, S.A.; Harris, D.A.; Kupferberg, A.B.; Singher, H.O.; Styles, H. Streptocin, antibiotic isolated from mycelium of Streptomyces griseus, active against Trichomonas vaginalis, and certain bacteria. Exp. Biol. Med. 1949, 70, 308–312. [Google Scholar] [CrossRef]
- Ikeda, H.; Nonomiya, T.; Usami, M.; Ohta, T.; Omura, S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 1999, 96, 9509–9514. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, Y.; Ishikawa, J.; Hara, H.; Suzuki, H.; Ikenoya, M.; Ikeda, H.; Yamashita, A.; Hattori, M.; Horinouchi, S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008, 190, 4050–4060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ōmura, S.; Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Takahashi, C.; Shinose, M.; Takahashi, Y.; Horikawa, H.; Nakazawa, H.; Osonoe, T.; et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 2001, 98, 12215–12220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Borodina, I. Genome-scale analysis of Streptomyces coelicolor A3 (2) metabolism. Genome Res. 2005, 15, 820–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef]
- Xia, H.; Li, X.; Li, Z.; Zhan, X.; Mao, X.; Li, Y. The application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Laureti, L.; Song, L.; Huang, S.; Corre, C.; Leblond, P.; Challis, G.L.; Aigle, B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 2011, 108, 6258–6263. [Google Scholar] [CrossRef] [Green Version]
- Myronovskyi, M.; Luzhetskyy, A. heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 2019, 36, 1281–1294. [Google Scholar] [CrossRef]
- Ochi, K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017, 70, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Freitas, P.R.; Araújo, A.C.J.; Alencar Menezes, I.R.; Santos, E.L.; Tintino, S.R.; Moura, T.F.; Filho, J.R.; Ferreira, V.A.; Silva, A.C.A.; et al. GC-MS profile and enhancement of antibiotic activity by the essential oil of Ocotea odorífera and safrole: Inhibition of Staphylococcus aureus efflux pumps. Antibiotics 2020, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef] [PubMed]
- Netzker, T.; Flak, M.; Krespach, M.K.; Stroe, M.C.; Weber, J.; Schroeckh, V.; Brakhage, A.A. Microbial interactions trigger the production of antibiotics. Curr. Opin. Microbiol. 2018, 45, 117–123. [Google Scholar] [CrossRef] [PubMed]
- El Ariebi, N.; Hiscox, J.; Scriven, S.A.; Müller, C.T.; Boddy, L. Production and effects of volatile organic compounds during interspecific interactions. Fungal Ecol. 2016, 20, 144–154. [Google Scholar] [CrossRef]
- Hiscox, J.; O’Leary, J.; Boddy, L. Fungus wars: Basidiomycete battles in wood decay. Stud. Mycol. 2018, 89, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Christofides, S.R.; Hiscox, J.; Savoury, M.; Boddy, L.; Weightman, A.J. Fungal control of early-stage bacterial community development in decomposing wood. Fungal Ecol. 2019, 42, 100868. [Google Scholar] [CrossRef]
- Hervé, V.; Le Roux, X.; Uroz, S.; Gelhaye, E.; Frey-Klett, P. Diversity and structure of bacterial communities associated with Phanerochaete chrysosporium during wood decay. Environ. Microbiol. 2014, 16, 2238–2252. [Google Scholar] [CrossRef]
- Johnston, S.R.; Boddy, L.; Weightman, A.J. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol. Ecol. 2016, 92, fiw179. [Google Scholar] [CrossRef] [Green Version]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Ueda, K.; Kawai, S.; Ogawa, H.-O.; Kiyama, A.; Kubota, T.; Kawanobe, H.; Beppu, T. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J. Antibiot. 2000, 53, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Charusanti, P.; Fong, N.L.; Nagarajan, H.; Pereira, A.R.; Li, H.J.; Abate, E.A.; Su, Y.; Gerwick, W.H.; Palsson, B.O. Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. PLoS ONE 2012, 7, e33727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luti, K.J.K.; Mavituna, F. Elicitation of Streptomyces coelicolor with dead cells of bacillus subtilis and Staphylococcus aureus in a bioreactor increases production of undecylprodigiosin. Appl. Microbiol. Biotechnol. 2011, 90, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, J.; Muñoz-Dorado, J.; Braña, A.F.; Shimkets, L.J.; Sevillano, L.; Santamaría, R.I. Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microbiol. Biotechnol. 2011, 4, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Wakefield, J.; Hassan, H.M.; Jaspars, M.; Ebel, R.; Rateb, M.E. Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeckh, V.; Scherlach, K.; Nutzmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef] [Green Version]
- Hervé, V.; Ketter, E.; Pierrat, J.-C.; Gelhaye, E.; Frey-Klett, P. Impact of Phanerochaete chrysosporium on the functional diversity of bacterial communities associated with decaying wood. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Johnston, S.R.; Hiscox, J.; Savoury, M.; Boddy, L.; Weightman, A.J. Highly competitive fungi manipulate bacterial communities in decomposing beech wood (Fagus sylvatica). FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef] [Green Version]
- Aqueveque, P.; Céspedes, C.L.; Becerra, J.; Aranda, M.; Sterner, O. Antifungal activities of secondary metabolites isolated from liquid fermentations of Stereum hirsutum (Sh134-11) against Botrytis cinerea (grey mould agent). Food Chem. Toxicol. 2017, 109, 1048–1054. [Google Scholar] [CrossRef]
- Cilerdzic, J.; Stajic, M.; Vukojevic, J. Potential of submergedly cultivated mycelia of Ganoderma spp. as antioxidant and antimicrobial agents. Curr. Pharm. Biotechnol. 2016, 17, 275–282. [Google Scholar] [CrossRef]
- Han, J.-J.; Bao, L.; He, L.-W.; Zhang, X.-Q.; Yang, X.-L.; Li, S.-J.; Yao, Y.-J.; Liu, H. Phaeolschidins A–E, five hispidin derivatives with antioxidant activity from the fruiting body of Phaeolus schweinitzii collected in the Tibetan Plateau. J. Nat. Prod. 2013, 76, 1448–1453. [Google Scholar] [CrossRef] [PubMed]
- Knežević, A.; Stajić, M.; Sofrenić, I.; Stanojković, T.; Milovanović, I.; Tešević, V.; Vukojević, J. Antioxidative, antifungal, cytotoxic and antineurodegenerative activity of selected trametes species from Serbia. PLoS ONE 2018, 13, e0203064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef] [Green Version]
- Moody, S.C. Microbial co-culture: Harnessing intermicrobial signaling for the production of novel antimicrobials. Future Microbiol. 2014, 9, 575–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, M.; Zhao, P.; Li, G.; Zhang, K. In depth natural product discovery from the basidiomycetes Stereum species. Microorganisms 2020, 8, 1049. [Google Scholar] [CrossRef]
- Oppong-Danquah, E.; Budnicka, P.; Blümel, M.; Tasdemir, D. Design of fungal co-cultivation based on comparative metabolomics and bioactivity for discovery of marine fungal agrochemicals. Mar. Drugs 2020, 18, 73. [Google Scholar] [CrossRef] [Green Version]
- Fischer, J.; Müller, S.Y.; Netzker, T.; Jäger, N.; Gacek-Matthews, A.; Scherlach, K.; Stroe, M.C.; García-Altares, M.; Pezzini, F.; Schoeler, H.; et al. Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites. eLife 2018, 7, e40969. [Google Scholar] [CrossRef]
- Khalil, Z.G.; Cruz-Morales, P.; Licona-Cassani, C.; Marcellin, E.; Capon, R.J. Inter-kingdom beach warfare: Microbial chemical communication activates natural chemical defences. ISME J. 2019, 13, 147–158. [Google Scholar] [CrossRef]
- Krause, K.; Jung, E.-M.; Lindner, J.; Hardiman, I.; Poetschner, J.; Madhavan, S.; Matthäus, C.; Kai, M.; Menezes, R.C.; Popp, J.; et al. Response of the wood-decay fungus Schizophyllum commune to co-occurring microorganisms. PLoS ONE 2020, 15. [Google Scholar] [CrossRef] [Green Version]
- Tillotson, R.D.; Wösten, H.A.B.; Richter, M.; Willey, J.M. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 1998, 30, 595–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saravana Kumar, P.; Al-Dhabi, N.A.; Duraipandiyan, V.; Balachandran, C.; Praveen Kumar, P.; Ignacimuthu, S. In vitro antimicrobial, antioxidant and cytotoxic properties of Streptomyces lavendulae strain SCA5. BMC Microbiol. 2014, 14, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halket, J.M.; Waterman, D.; Przyborowska, A.; Patel, R.K.P.; Fraser, P.D.; Bramley, P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurudeeban, S.; Ramanathan, T.; Satyavani, K. Antimicrobial and radical scavenging effects of alkaloid extracts from Rhizophora mucronata. Pharm. Chem. J. 2013, 47, 50–53. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicault, M.; Zaiter, A.; Dumarcay, S.; Chaimbault, P.; Gelhaye, E.; Leblond, P.; Bontemps, C. Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures. Microorganisms 2021, 9, 178. https://doi.org/10.3390/microorganisms9010178
Nicault M, Zaiter A, Dumarcay S, Chaimbault P, Gelhaye E, Leblond P, Bontemps C. Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures. Microorganisms. 2021; 9(1):178. https://doi.org/10.3390/microorganisms9010178
Chicago/Turabian StyleNicault, Matthieu, Ali Zaiter, Stéphane Dumarcay, Patrick Chaimbault, Eric Gelhaye, Pierre Leblond, and Cyril Bontemps. 2021. "Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures" Microorganisms 9, no. 1: 178. https://doi.org/10.3390/microorganisms9010178
APA StyleNicault, M., Zaiter, A., Dumarcay, S., Chaimbault, P., Gelhaye, E., Leblond, P., & Bontemps, C. (2021). Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures. Microorganisms, 9(1), 178. https://doi.org/10.3390/microorganisms9010178





