Vinification without Saccharomyces: Interacting Osmotolerant and “Spoilage” Yeast Communities in Fermenting and Ageing Botrytised High-Sugar Wines (Tokaj Essence)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Media and Reference Yeast Strain
2.2. Sample Collection and Analysis
2.3. Yeast Isolation and Phenotypic Categorisation
2.4. Molecular Taxonomy
2.5. Electrophoretic Karyotyping and Microsatellite-Primed PCR (MSP-PCR) Fingerprinting
2.6. Extraction and Restriction Fragment Length Polymorphism (RFLP) Analysis of Mitochondrial DNA
2.7. Cluster Analysis of Molecular Patterns
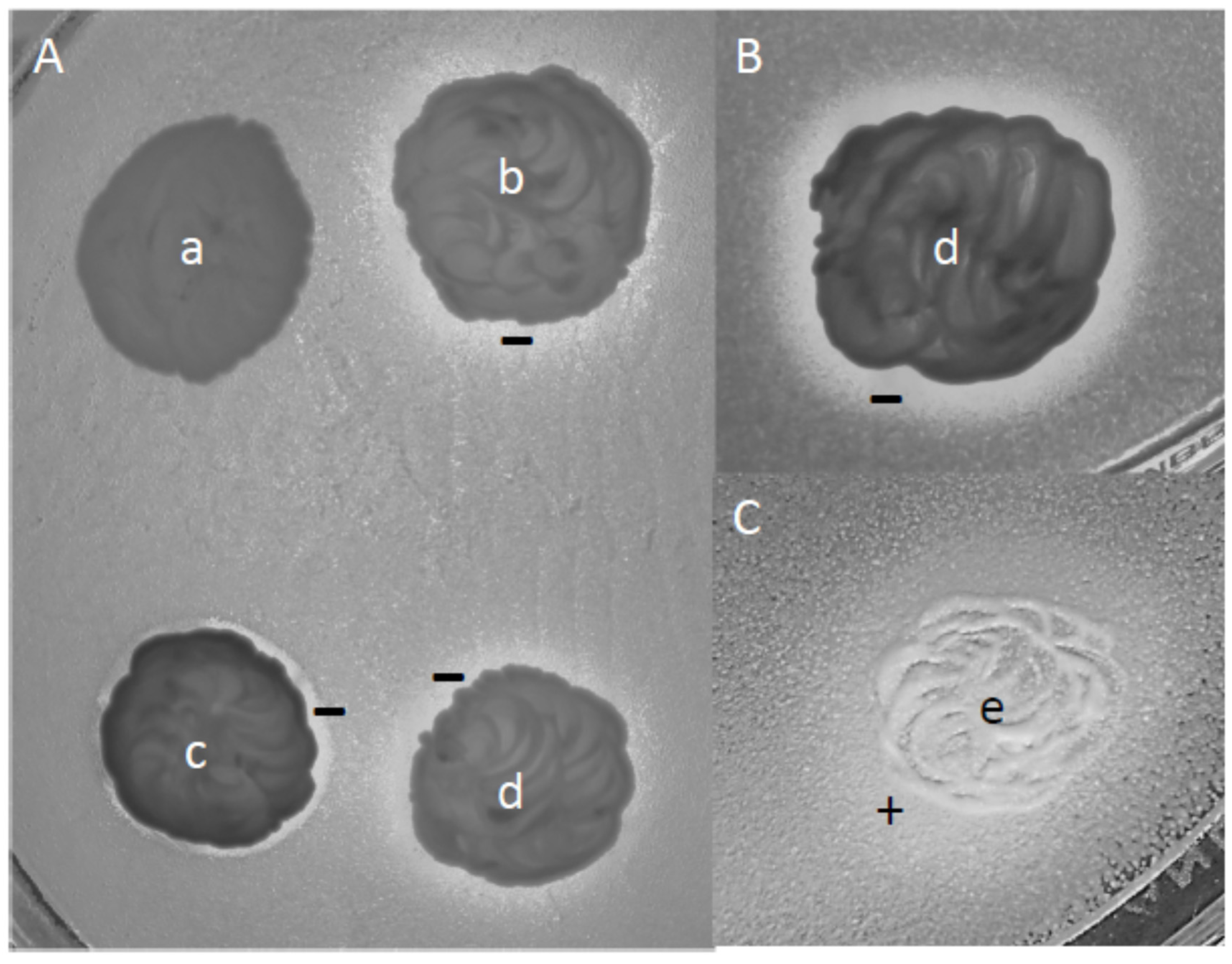
2.8. Phenotypic Characterisation via Drop Tests
2.8.1. Determination of the MIC (Minimal Inhibitory Concentration) of Glucose, Ethanol and Potassium Bisulfite (K2S2O5)
2.8.2. Growth at Various Temperatures
2.8.3. H2S Production
2.8.4. Organic Acid Production
2.9. Examination of Biofilm Formation
2.10. Growth Assay with Microplates
2.11. Interaction and Growth Competition Tests
3. Results
3.1. Sampling and Sample Characterisation
3.2. Isolation and Phenotypic Categorisation of Yeasts
3.3. Taxonomic Identification
3.4. Diverse Osmotolerance of the Species
3.5. Intraspecies Phenotypic Diversity
3.6. Intraspecies Molecular Diversity in Z. rouxii
3.7. Interactions and Competitions of Isolates
4. Discussion
4.1. Dominant and Associated Yeasts
4.2. Why These Yeasts Populate Essence Wines
4.3. Intraspecies Clonal Diversity and Segregation
5. Conclusions
- The fermentation of high-sugar wine can take place in the absence of Saccharomyces.
- Instead of Saccharomyces, osmotolerant “spoilage” yeasts can ferment when the sugar concentration is extremely high.
- In botrytised Tokaj Essence wines of sugar concentrations ranging from 365 to 752 g∙L−1, Zygosaccharomyces rouxii, Candida lactis-condensi and C. zemplinina were the dominating species.
- The minor species were either other “spoilage” yeasts or less osmotolerant biofilm-producing yeasts.
- The high phenotypical and molecular (karyotype, mtDNA-RFLP and MSP-PCR) diversity of the conspecific strains indicates that diverse clones of the species coexisted in the wines.
- Genetic segregation of certain clones and interaction of the species (antagonism and crossfeeding) could also shape the fermenting yeast biota.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valera, C.; Borneman, A. Yeasts found in vineyards and wineries. Yeast 2017, 34, 111–128. [Google Scholar]
- Morrison-Whittle, P.; Goddard, M.R. From vineyard to winery: A source map of microbial diversity driving wine fermentation. Environ. Microbiol. 2018, 20, 75–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: Role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef]
- Heard, G.M.; Fleet, G.H. The effects of temperature and pH on the growth of yeast species during the fermentation of grape juice. J. Appl. Bacteriol. 1988, 65, 23–28. [Google Scholar] [CrossRef]
- Tofalo, R.; Chaves-López, C.; Di Fabio, F.; Schirone, M.; Felis, G.E.; Torriani, S.; Paparella, A.; Suzzi, G. Molecular identification and osmotolerant profile of wine yeasts that ferment a high sugar grape must. Int. J. Food Microbiol. 2009, 130, 179–187. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Spiropoulos, A.E.; Nychas, G.J.E. Yeast community structures and dynamics in healthy and Botrytis-affected grape must fermentations. Appl. Environ. Microbiol. 2007, 73, 6705–6713. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Aponte, M.; Piombino, P.; Lisanti, M.T.; Moio, L.; Ercolini, D.; Blaiotta, G. Influence of microbial communities on the chemical and sensory features of Falanghina sweet passito wines. Food Res. Int. 2019, 120, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Barata, A.; Santos, S.C.; Malfeito-Ferreira, M.; Loureiro, V. New insights into the ecological interaction between grape berry microorganisms and Drosophila flies during the development of sour rot. Microb. Ecol. 2012, 64, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Mills, D.A.; Johannsen, E.A.; Cocolin, L. Yeast diversity and persistence in Botrytis-affected wine fermentations. Appl. Environ. Microbiol. 2002, 68, 4884–4893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and future of non-Saccharomyces yeasts: From spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malfeito-Ferreira, M.; Silva, A.C. Spoilage yeasts in wine production. In Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G.H., Eds.; Springer Nature: New York, NY, USA, 2019; pp. 375–394. [Google Scholar]
- Escott, C.; Loira, I.; Morata, A.; Bañuelos, M.; Suárez-Lepe, J. Wine spoilage yeasts: Control strategy. In Yeast-Industrial Applications; Morata, A., Loira, I., Eds.; InTech: Rijeka, Croatia, 2017; pp. 89–116. [Google Scholar]
- Ivit, N.N.; Longo, R.; Kemp, B. The effect of non-Saccharomyces and Saccharomyces non-cerevisiae yeasts on ethanol and glycerol levels in wine. Fermentation 2020, 6, 77. [Google Scholar] [CrossRef]
- Morata, A.; Escott, C.; Bañuelos, M.A.; Loira, I.; Fresno, J.M.D.; González, C.; Suárez-Lepe, J.A. Contribution of non-Saccharomyces yeasts to wine freshness. A review. Biomolecules 2019, 10, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtin, C.D.; Varela, C.; Borneman, A. Harnessing improved understanding of Brettanomyces bruxellensis biology to mitigate the risk of wine spoilage. Aust. J. Grape Wine Res. 2015, 21, 680–692. [Google Scholar] [CrossRef]
- Agnolucci, M.; Tirelli, A.; Cocolin, L.; Toffanin, A. Brettanomyces bruxellensis yeasts: Impact on wine and winemaking. World. J. Microbiol. Biotechnol. 2017, 33, 180. [Google Scholar] [CrossRef]
- Jermini, M.F.G.; Geiges, O.; Schmidt-Lorenz, W. Detection, isolation and identification of osmotolerant yeasts from high-sugar products. J. Food Prot. 1987, 50, 468–472. [Google Scholar] [CrossRef]
- Gordon, J.L.; Wolfe, K.H. Recent allopolyploid origin of Zygosaccharomyces rouxii strain ATCC 42981. Yeast 2008, 25, 449–456. [Google Scholar] [CrossRef] [Green Version]
- Solieri, L.; Cassanelli, S.; Croce, M.A.; Giudici, P. Genome size and ploidy level: New insights for elucidating relationships in Zygosaccharomyces species. Fungal Genet. Biol. 2008, 45, 1582–1590. [Google Scholar] [CrossRef]
- Wrent, P.; Rivas, E.M.; Peinado, J.M.; de Silóniz, M.I. Zygosaccharomyces rouxii strains CECT 11923 and Z. rouxii CECT 10425: Two new putative hybrids? Int. J. Food Microbiol. 2017, 241, 7–14. [Google Scholar] [CrossRef]
- Braun-Galleani, S.; Ortiz-Merino, R.A.; Wu, Q.; Xu, Y.; Wolfe, K.H. Zygosaccharomyces pseudobailii, another yeast interspecies hybrid that regained fertility by damaging one of its MAT loci. Fems Yeast Res. 2018, 18, foy079. [Google Scholar] [CrossRef] [Green Version]
- Csoma, H.; Sipiczki, M. Taxonomic reclassification of Candida stellata strains reveals frequent occurrence of Candida zemplinina in wine fermentation. Fems Yeast Res. 2008, 8, 328–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sipiczki, M. Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int. J. System. Evol. Microbiol. 2003, 53, 2079–2083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granchi, L.; Ganucci, D.; Messini, A.; Rosellini, D.; Vincenzini, M. Oenological properties of Hanseniaspora osmophila and Kloeckera cortices from wines produced by spontaneous fermentations of normal and dried grapes. FEMS Yeast Res. 2002, 2, 403–407. [Google Scholar] [PubMed] [Green Version]
- Passoth, V.; Fredlund, E.; Druvefors, U.Ä.; Schnürer, J. Biotechnology, physiology and genetics of the yeast Pichia anomala. FEMS Yeast Res. 2006, 6, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, Y.; Watanabe, M. Effects of yeast strains and environmental conditions on formation of organic acids in must during fermentation. J. Fermen. Technol. 1981, 59, 27–32. [Google Scholar]
- Sponholz, W.R.; Dittrich, H.H. Die Bildung von SO2 bindenden Gärungsnebenprodukten, höheren Alkoholen und Estern bei einigen Reinzuchthefestämmen und bei einigen für die Weinbereitung wichtigen “wilden” Hefen. Wein-Wiss 1974, 29, 301–314. [Google Scholar]
- Kántor, A.; Petrová, J.; Hleba, L.; Kluz, M.; Kačániová, M. Determination of spoilage yeasts in different red and white wines. Sci. Pap. Anim. Sci. Biotechnol. 2016, 49, 57–65. [Google Scholar]
- Sánchez-Rubio, M.; Guerrouj, K.; Taboada-Rodríguez, A.; López-Gómez, A.; Marín-Iniesta, F. Control of native spoilage yeast on dealcoholized red wine by preservatives alone and in binary mixtures. J. Food Sci. 2017, 82, 2128–2133. [Google Scholar] [CrossRef]
- Romano, P.; Suzzi, G. Origin and production of acetoin during wine yeast fermentation. Appl. Environ. Microbiol. 1996, 62, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Chandra, M.; Oro, I.; Ferreira-Dias, S.; Malfeito Ferreira, M. Effect of ethanol, sulfur dioxide and glucose on the growth of wine spoilage yeasts using response surface methodology. PLoS ONE 2015, 10, e0128702. [Google Scholar] [CrossRef] [Green Version]
- Scarr, M.P.; Rose, D. Study of osmophilic yeasts producing invertase. J. Gen. Microbiol. 1966, 45, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Tokuoka, K.; Ishitani, T. Minimum water activities for the growth of yeasts isolated from high-sugar foods. J. Gen. Appl. Microbiol. 1991, 37, 111–119. [Google Scholar] [CrossRef]
- Stratford, M.; Steels, H.; Novodvorska, M.; Archer, D.B.; Avery, S.V. Extreme osmotolerance and halotolerance in food-relevant yeasts and the role of glycerol-dependent cell individuality. Front. Microbiol. 2019, 9, 3238. [Google Scholar] [CrossRef] [PubMed]
- Combina, M.; Daguerre, C.; Massera, A.; Mercado, L.; Sturm, M.E.; Ganga, A.; Martinez, C. Yeast identification in grape juice concentrates from Argentina. Lett. Appl. Microbiol. 2008, 46, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Rojo, M.; Arroyo Lopez, F.; Lerena, M.; Mercado, L.; Torres, A.; Combina, M. Effects of pH and sugar concentration in ‘Zygosaccharomyces rouxii’ growth and time for spoilage in concentrated grape juice at isothermal and nonisothermal conditions. Food Microbiol. 2013, 38, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Hu, Z.Q.; Long, F.Y.; Niu, C.; Yuan, Y.H.; Yue, T.L. Characterization of osmotolerant yeasts and yeast-like molds from apple orchards and apple juice processing plants in China and investigation of their spoilage potential. J. Food Sci. 2015, 80, M1850–M1860. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, V.; Malfeito-Ferreira, M. Spoilage yeasts in the wine industry (review). Int. J. Food Microbiol. 2003, 86, 23–50. [Google Scholar] [CrossRef]
- Thompson, S. Microbiological spoilage of high-sugar products. In Compendium of the Microbiological Spoilage of Foods and Beverages: Food Microbiology and Food Safety; Sperber, W.H., Doyle, M.P., Eds.; Springer: New York, NY, USA, 2009; pp. 301–324. [Google Scholar]
- Magyar, I. Botrytized wines. Adv Food Nutr. Res. 2011, 63, 147–206. [Google Scholar]
- Sipiczki, M. Overwintering of vineyard yeasts: Survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 2016, 7, 212. [Google Scholar] [CrossRef]
- Donèche, B.J. Botrytized wines. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Chur, New York, NY, USA, 1993; pp. 327–353. [Google Scholar]
- Sipiczki, M. Yeasts in botrytised wine making. In Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G.H., Eds.; Springer Nature: New York, NY, USA, 2019; pp. 229–261. [Google Scholar]
- Greger, M. Notes on the Pure or Natural Wines of Hungary, Their Properties and Uses; Jas. Truscott & Sons: London, UK, 1879. [Google Scholar]
- Allen, H.W. The Romance of Tokay; Berry Bross: London, UK, 1928. [Google Scholar]
- Jackson, R.S. Wine Science; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- Eftimová, Z.; Eftimová, J.; Balážová, Ľ. Antioxidant activity of Tokaj Essence. Slov. J. Food Sci. 2018, 12, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Csoma, H.; Sipiczki, M. Taxonomic investigation of the yeast biota of botrytized grapes and “Essence” in the Tokaj wine region. In Book of Abstracts, 8th International Enology Symposium; Vigne et Vin Publications Internationales: Villenave d’Ornon, France, 2007; p. 174. [Google Scholar]
- Duarte, F.L.; Pimentel, N.H.; Teixeira, A.; Fonseca, A. Saccharomyces bacillaris is not a synonym of Candida stellata: Reinstatement as Starmerella bacillaris comb. nov. Antonie Van Leeuwenhoek 2012, 102, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.R.O.; Leon, M.P.; Barros, K.O.; Freitas, L.F.D.; Hughes, A.F.S.; Morais, P.B.; Lachance, M.-A.; Rosa, C.A. Starmerella camargoi f.a., sp. nov., Starmerella ilheusensis f.a., sp. nov., Starmerella litoralis f.a., sp. nov., Starmerella opuntiae f.a., sp. nov., Starmerella roubikii f.a., sp. nov. and Starmerella vitae f.a., sp. nov., isolated from flowers and bees, and transfer of related Candida species to the genus Starmerella as new combinations. Int. J. Syst. Evol. Microbiol. 2018, 68, 1333–1343. [Google Scholar] [PubMed]
- Zara, S.; Mannazu, I. Detection, quantification, and identification of yeast in winemaking. In Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G.H., Eds.; Springer Nature: New York, NY, USA, 2019; pp. 81–115. [Google Scholar]
- Stefanini, I.; Cavalieri, D. Metagenomic approaches to investigate the contribution of the vineyard environment to the quality of wine fermentation: Potentials and difficulties. Front. Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef] [PubMed]
- Hierro, N.; Esteve-Zarzoso, B.; González, Á.; Mas, A.; Guillamón, J.M. Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine. Appl. Environ. Microbiol. 2006, 72, 7148–7155. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Esteve-Zarzoso, B.; Cocolin, L.; Mas, A.; Rantsiou, K. Viable and culturable populations of Saccharomyces cerevisiae, Hanseniaspora uvarum and Starmerella bacillaris (synonym Candida zemplinina) during Barbera must fermentation. Food Res. Int. 2015, 78, 195–200. [Google Scholar] [CrossRef]
- Vendrame, M.; Manzano, M.; Comi, G.; Bertrand, J.; Iacumin, L. Use of propidium monoazide for the enumeration of viable Brettanomyces bruxellensis in wine and beer by quantitative PCR. Food Microbiol. 2014, 42, 196–204. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 87–110. [Google Scholar]
- Wickerham, L.J. Taxonomy of Yeasts; Techn. Bull. 1029; U.S. Deptartment of Agriculture: Washington, DC, USA, 1951.
- Commission Regulation (EEC) No 2676/90 of 17 September 1990 determining Community methods for the analysis of wines, Commission of the European Communities. Official Journal of the European Communities L 272, 03/10/1990 P. 0001–0192. Available online: https://op.europa.eu/en/publication-detail/-/publication/6528497d-1ece-4355-ab08-c73b3242f7ee (accessed on 20 December 2020).
- Nguyen, H.V.; Lepingle, A.; Gaillardin, C.A. Molecular typing demonstrates homogeneity of Saccharomyces uvarum strains and reveals the existence of hybrids between S. uvarum and S. cerevisiae, including the S. bayanus type strain CBS 380. Syst. Appl. Microbiol. 2000, 23, 71–85. [Google Scholar] [CrossRef]
- Baleiras-Couto, M.M.; Hartog, B.J.; Huis in’t Veld, J.H.J.; Hofstra, H.; van der Vossen, J.M.B.M. Identification of spoilage yeasts in a food-production chain by microsatellite polymerase chain reaction fingerprinting. Food Microbiol. 1996, 13, 59–67. [Google Scholar] [CrossRef]
- Pavel, A.B.; Vasile, C.I. PyElph—A software tool for gel images analysis and phylogenetics. BMC Bioinform. 2012, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- Dice, L.R. Measurements of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Garcia-Vallve, S.; Palau, J.; Romeu, A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol. Biol. Evol. 1999, 9, 1125–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tofalo, R.; Schirone, M.; Torriani, S.; Rantsiou, K.; Cocolin, L.; Perpetuini, G.; Suzzi, G. Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol. 2012, 29, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Bader, O. Fungal species identification by MALDI-ToF mass spectrometry. Methods Mol. Biol. 2017, 1508, 323–337. [Google Scholar]
- Sipiczki, M. Metschnikowia pulcherrima and related pulcherrimin-producing yeasts: Fuzzy species boundaries and complex antimicrobial antagonism. Microorganisms 2020, 8, 1029. [Google Scholar] [CrossRef]
- Riedl, R.; Futterer, J.; Goderbauer, P.; Michel, M.; Jacob, F.; Hutzler, M. Combined yeast biofilm screening—Characterization and validation of yeast related biofilms in a brewing environment with combined cultivation and specific real-time PCR screening of selected indicator species. J. Am. Soc. Brew. Chem. 2019, 77, 99–112. [Google Scholar] [CrossRef]
- Zara, G.; Budroni, M.; Mannazzu, I.; Fancello, F.; Zara, S. Yeast biofilm in food realms: Occurrence and control. World J. Microbiol. Biotechnol. 2020, 36, 134. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Academic Press Australia: Sydney, Australia, 1985. [Google Scholar]
- Stratford, M.; Capell, C.J. Soft drinks. Microbiology. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Trugo, L., Finglas, P.M., Eds.; Academic Press: San Diego, CA, USA, 2003; pp. 5358–5366. [Google Scholar]
- Kuanyshev, N.; Adamo, G.M.; Porro, D.; Branduardi, P. The spoilage yeast Zygosaccharomyces bailii: Foe or friend? Yeast 2017, 34, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Perrusquía-Luévano, S.; Cano-Herrera, M.S.; Guigón-López, C.; Avitia-Talamantes, M.D.C.; Torres-Torres, C.; Villalpando, I. Microbiology of high-sugar must fermentation by novel yeasts from the chihuahuan desert. FEMS Yeast Res. 2019, 19, foy099. [Google Scholar] [CrossRef]
- Fleet, G.H.; Lafon-Lafourcade, S.; Ribéreau-Gayon, P. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl. Environ. Microbiol. 1984, 48, 1034–1038. [Google Scholar] [CrossRef] [Green Version]
- Divol, B.; Lonvaud-Funel, A. Evidence for viable nonculturable yeasts in Botrytis-affected wine. J. Appl. Microbiol. 2005, 99, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Mateus, D.; Sousa, S.; Coimbra, C.; Rogerson, F.S.; Simões, J. Identification and characterization of non-Saccharomyces species isolated from port wine spontaneous fermentations. Foods 2020, 9, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minarik, E.; Jungova, E.; Emeriaud, M. Fruktophile Hefen und deren Einfluss auf süsse Naturweine. Wein-Wiss 1978, 33, 42–47. [Google Scholar]
- Domizio, P.; Romani, C.; Lencioni, L.; Comitini, F.; Gobbi, M.; Mannazzu, I.; Ciani, M. Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 2011, 147, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Romano, P.; Suzzi, G. Potential use for Zygosaccharomyces species in winemaking. J. Wine Res. 1993, 4, 87–94. [Google Scholar] [CrossRef]
- Garavaglia, J.; Schneider, R.C.S.; Mendes, S.D.C.; Welke, J.E.; Zini, C.A.; Caramão, E.B.; Valente, P. Evaluation of Zygosaccharomyces bailii BCV 08 as a co-starter in wine fermentation for the improvement of ethyl esters production. Microbiol. Res. 2015, 173, 59–65. [Google Scholar] [CrossRef]
- Suh, S.O.; Gujjari, P.; Beres, C.; Beck, B.; Zhou, J. Proposal of Zygosaccharomyces parabailii sp. nov. and Zygosaccharomyces pseudobailii sp. nov., novel species closely related to Zygosaccharomyces bailii. Int. J. Syst. Evol. Microbiol. 2013, 63, 1922–1929. [Google Scholar] [CrossRef] [Green Version]
- Barata, A.; Seborro, F.; Belloch, C.; Malfeito-Ferreira, M.; Loureiro, V. Ascomycetous yeast species recovered from grapes damaged by honeydew and sour rot. J. Appl. Microbiol. 2008, 104, 1182–1191. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.; Ruiz, J.; Belda, I.; Benito-Vázquez, I.; Marquina, D.; Calderón, F.; Santos, A.; Benito, S. The genus Metschnikowia in enology. Microorganisms 2020, 8, 1038. [Google Scholar] [CrossRef]
- Steels, H.; James, S.A.; Roberts, I.N.; Stratford, M. Zygosaccharomyces lentus: A significant new osmophilic, preservative-resistant spoilage yeast, capable of growth at low temperature. J. Appl. Microbiol. 1999, 87, 520–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Hwang, C.F.; Liu, S.; Boundy-Mills, K.L.; Mills, D.A. Profiling the yeast communities of wine fermentations using terminal restriction fragment length polymorphism analysis. Am. J. Enol. Vitic. 2012, 63, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Antunovics, Z.; Csoma, H.; Sipiczki, M. Molecular and genetic analysis of the yeast flora of botrytized Tokaj wines. Bull. De L’oiv (Off. Int. De La Vigne Et Du Vin Paris) 2003, 76, 380–397. [Google Scholar]
- Csoma, H.; Sipiczki, M. Investigation of the yeast microflora of “Tokaj essence”. In Proceedings of the Abstract Book, 1st FEMS Congress of European Microbiologists, Ljubljana, Slovenia, 29 June–3 July 2003; p. 213. [Google Scholar]
- Magyar, I.; Bene, Z. Morphological and taxonomic study on mycobiota of noble rotted grapes in the Tokaj wine district. Acta Aliment. 2006, 35, 237–246. [Google Scholar] [CrossRef]
- Masneuf-Pomarede, I.; Juquin, E.; Miot-Sertier, C.; Renault, P.; Laizet, Y.; Salin, F.; Alexandre, H.; Capozzi, V.; Cocolin, L.; Colonna-Ceccaldi, B.; et al. The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res. 2015, 15, fov045. [Google Scholar] [CrossRef] [Green Version]
- Urso, R.; Rantsiou, K.; Dolci, P.; Rolle, L.; Comi, G.; Cocolin, L. Yeast biodiversity and dynamics during sweet wine production as determined by molecular methods. FEMS Yeast Res. 2008, 8, 1053–1062. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hu, W.; Huang, X.; Xu, Y. Investigation of yeast population diversity and dynamics in spontaneous fermentation of Vidal blanc icewine by traditional culture-dependent and high-throughput sequencing methods. Food Res. Int. 2018, 112, 66–77. [Google Scholar] [CrossRef]
- Englezos, V.; Giacosa, S.; Rantsiou, K.; Rolle, L.; Cocolin, L. Starmerella bacillaris in winemaking: Opportunities and risks. Curr. Opin. Food Sci. 2017, 17, 30–35. [Google Scholar] [CrossRef]
- Pawlikowska, E.; James, S.A.; Breierova, E.; Antolak, H.; Kregiel, D. Biocontrol capability of local Metschnikowia sp. isolates. Antonie Van Leeuwenhoek 2019, 112, 1425–1445. [Google Scholar] [CrossRef] [Green Version]
- Sipiczki, M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef] [Green Version]
- Morales, M.L.; Fierro-Risco, J.; Ríos-Reina, R.; Ubeda, C.; Paneque, P. Influence of Saccharomyces cerevisiae and Lachancea thermotolerans co-inoculation on volatile profile in fermentations of a must with a high sugar content. Food Chem. 2019, 276, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Pozo-Bayón, M.; Moreno-Arribas, M.V. Sherry wines. Adv. Food Nutr. Res. 2011, 63, 17–40. [Google Scholar]
- Miklos, I.; Sipiczki, M.; Benko, Z. Osmotolerant yeasts isolated from Tokaj wines. J. Basic. Microbiol. 1994, 6, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Benito, S. The impacts of Lachancea thermotolerans yeast strains on winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 6775–6790. [Google Scholar] [CrossRef] [Green Version]
- Bordet, F.; Joran, A.; Klein, G.; Roullier-Gall, C.; Alexandre, H. Yeast-yeast interactions: Mechanisms, methodologies and impact on composition. Microorganisms 2020, 8, 600. [Google Scholar] [CrossRef] [Green Version]
- Radler, F.; Herzberger, S.; Schönig, I.; Schwarz, P. Investigation of a killer strain of Zygosaccharomyces bailii. J. Gen. Microbiol. 1993, 139, 495–500. [Google Scholar] [CrossRef] [Green Version]
- Weiler, F.; Schmitt, M.J. Zygocin, a secreted antifungal toxin of the yeast Zygosaccharomyces bailii, and its effect on sensitive fungal cells. FEMS Yeast Res. 2003, 3, 69–76. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Alonso, A.; Marquina, D.; Santos, A. The biology of Pichia membranifaciens killer toxins. Toxins 2017, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Alonso, A.; Belda, I.; Santos, A.; Navascués, E.; Marquina, D. Advances in the control of the spoilage caused by Zygosaccharomyces species on sweet wines and concentrated grape musts. Food Control 2015, 51, 129–134. [Google Scholar] [CrossRef]
- Voordeckers, K.; De Maeyer, D.; van der Zande, E.; Vinces, M.D.; Meert, W.; Cloots, L.; Ryan, O.; Marchal, K.; Verstrepen, K.J. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 2012, 86, 225–239. [Google Scholar] [CrossRef] [Green Version]
- James, S.; Stratford, M. Zygosaccharomyces Barker (1901). In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 937–947. [Google Scholar]
- Csoma, H.; Acs-Szabo, L.; Papp, L.A.; Sipiczki, M. Application of different markers and data-analysis tools to the examination of biodiversity can lead to different results: A case study with Starmerella bacillaris (synonym Candida zemplinina) strains. Fems Yeast Res. 2018, 18, foy021. [Google Scholar] [CrossRef] [PubMed]
- Longo, E.; Vezinhet, F. Chromosomal rearrangements during vegetative growth of a wild strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1993, 59, 322–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütz, M.; Gafner, J. Analysis of yeast diversity during spontaneous and induced alcoholic fermentations. J. Appl. Bacteriol. 1993, 75, 551–558. [Google Scholar] [CrossRef]
- Nadal, D.; Carro, D.; Fernández, L.J.; Pina, B. Analysis and dynamics of the chromosomal complement of wild sparkling-wine yeast strains. Appl. Environ. Microbiol. 1999, 65, 1688–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, M.; Vinagre, A.; Ambrona, J.; Molina, F.; Maqueda, M.; Robello, J.E. Genetic instability of heterozygous, hybrid, natural wine yeasts. Appl. Environ. Microbiol. 2004, 70, 4686–4691. [Google Scholar] [CrossRef] [Green Version]
- Schütz, M.; Gafner, J. Dynamics of the yeast strain population during spontaneous determined by CHEF gel electrophoresis. Lett. Appl. Microbiol. 1994, 19, 253–259. [Google Scholar] [CrossRef]
- Povhe-Jemec, K.; Cadez, N.; Zagorc, T.; Bubic, V.; Zupec, A.; Raspor, P. Yeast population dynamics in five spontaneous fermentations of Malvasia must. Food Microbiol. 2001, 18, 247–259. [Google Scholar] [CrossRef]
- Sipiczki, M. Diversity, variability and fast adaptive evolution of the wine yeast (Saccharomyces cerevisiae) genome—A review. Ann. Microbiol. 2011, 61, 85–93. [Google Scholar] [CrossRef]
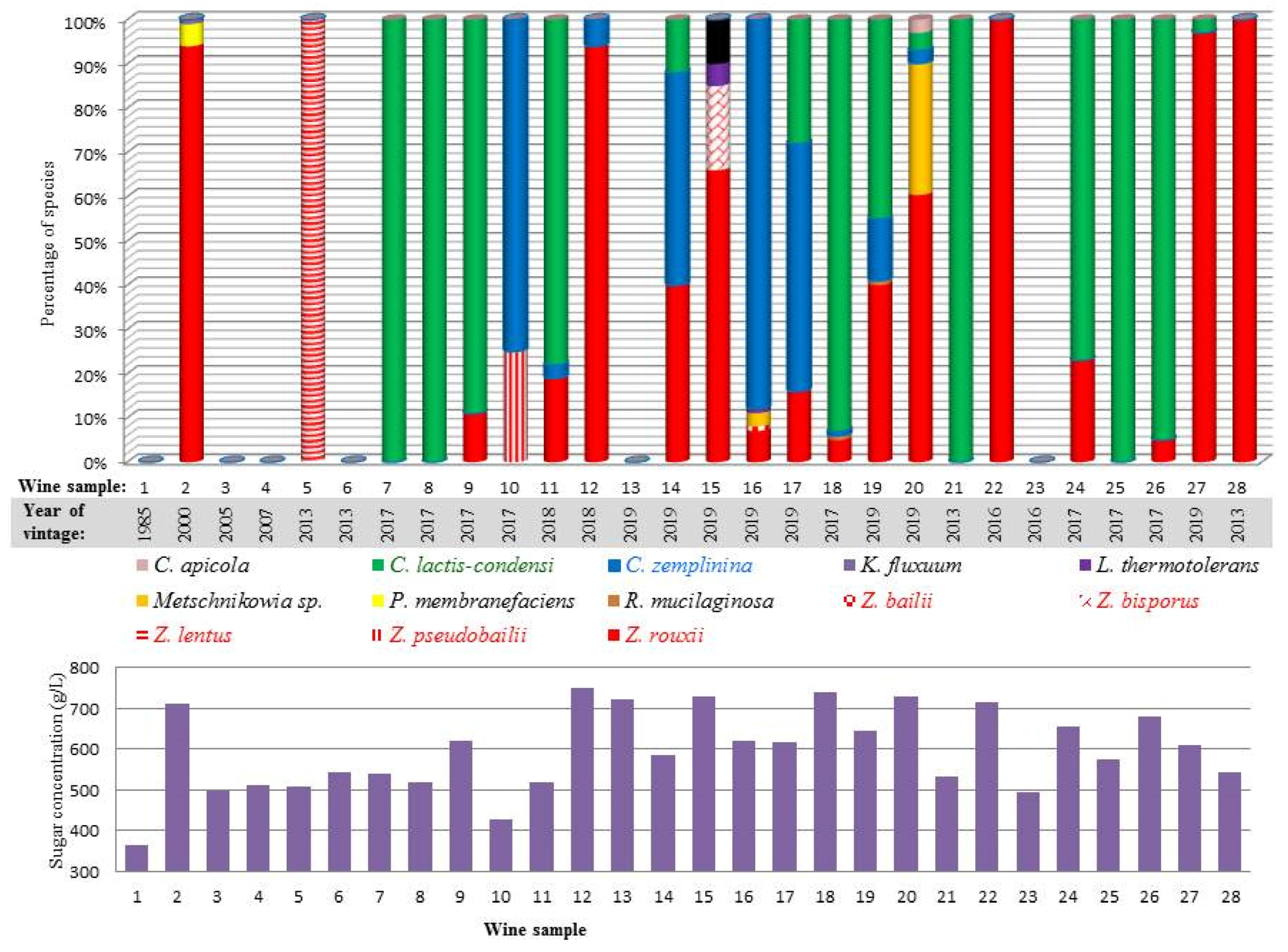
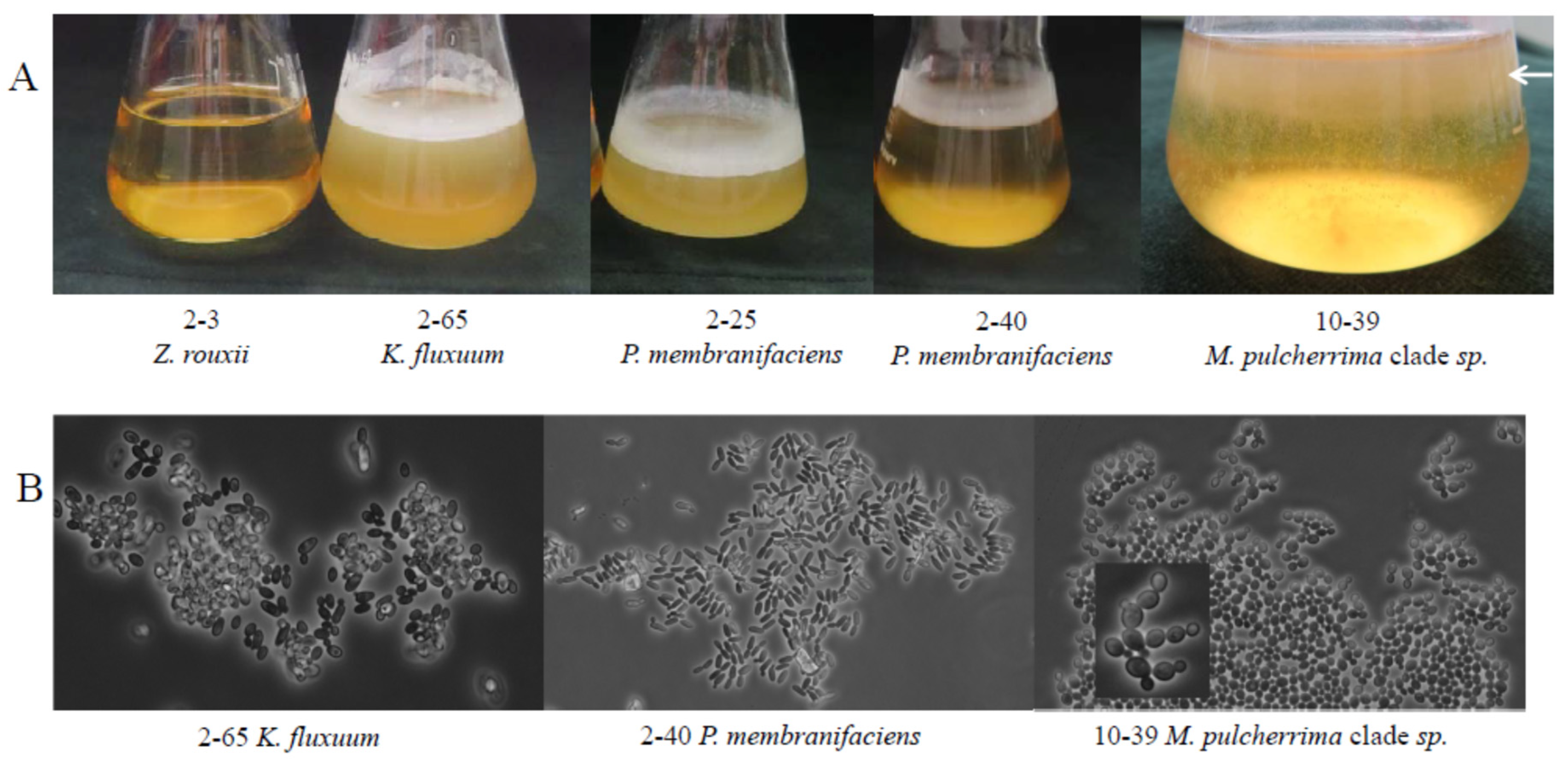
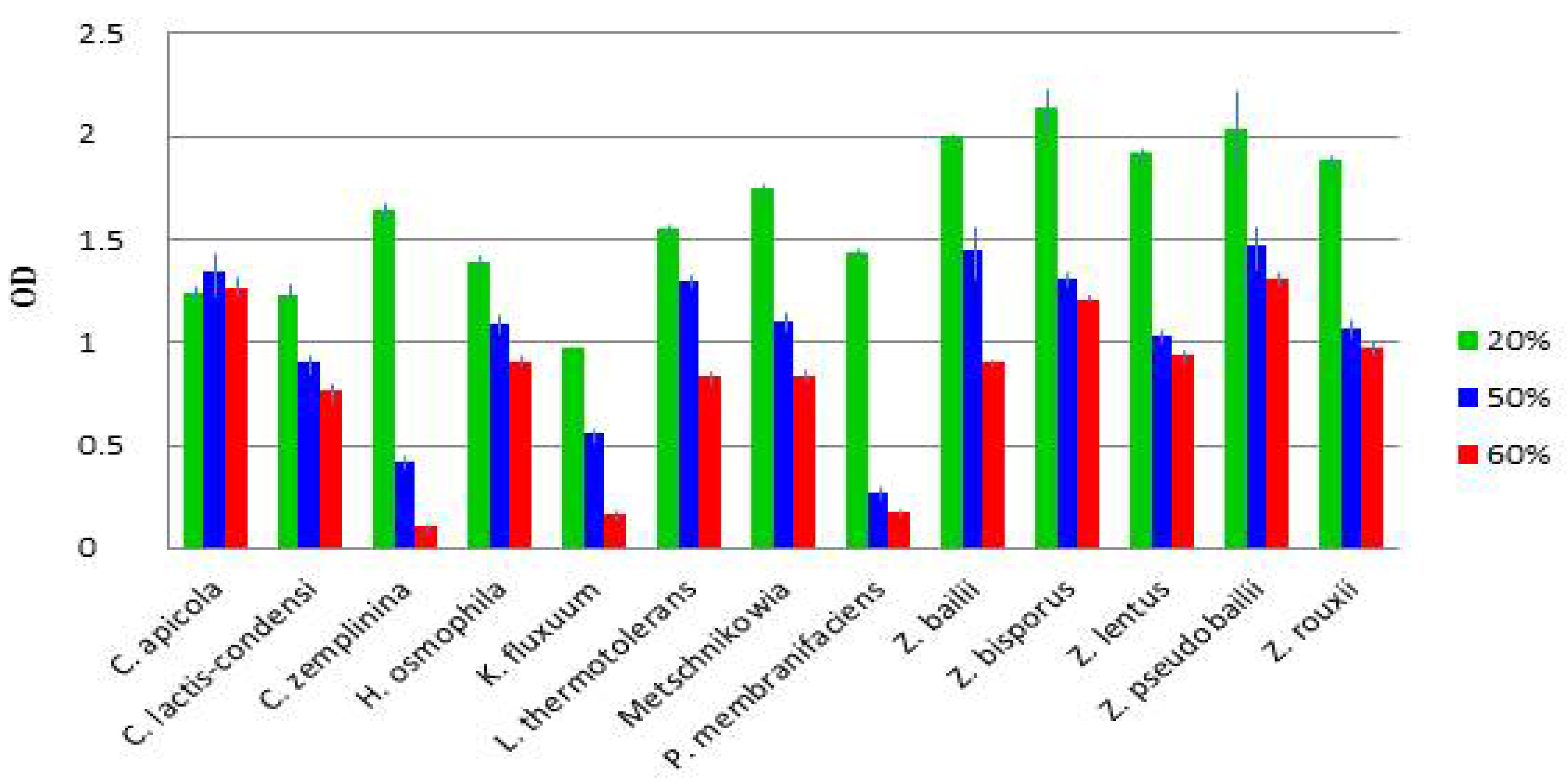


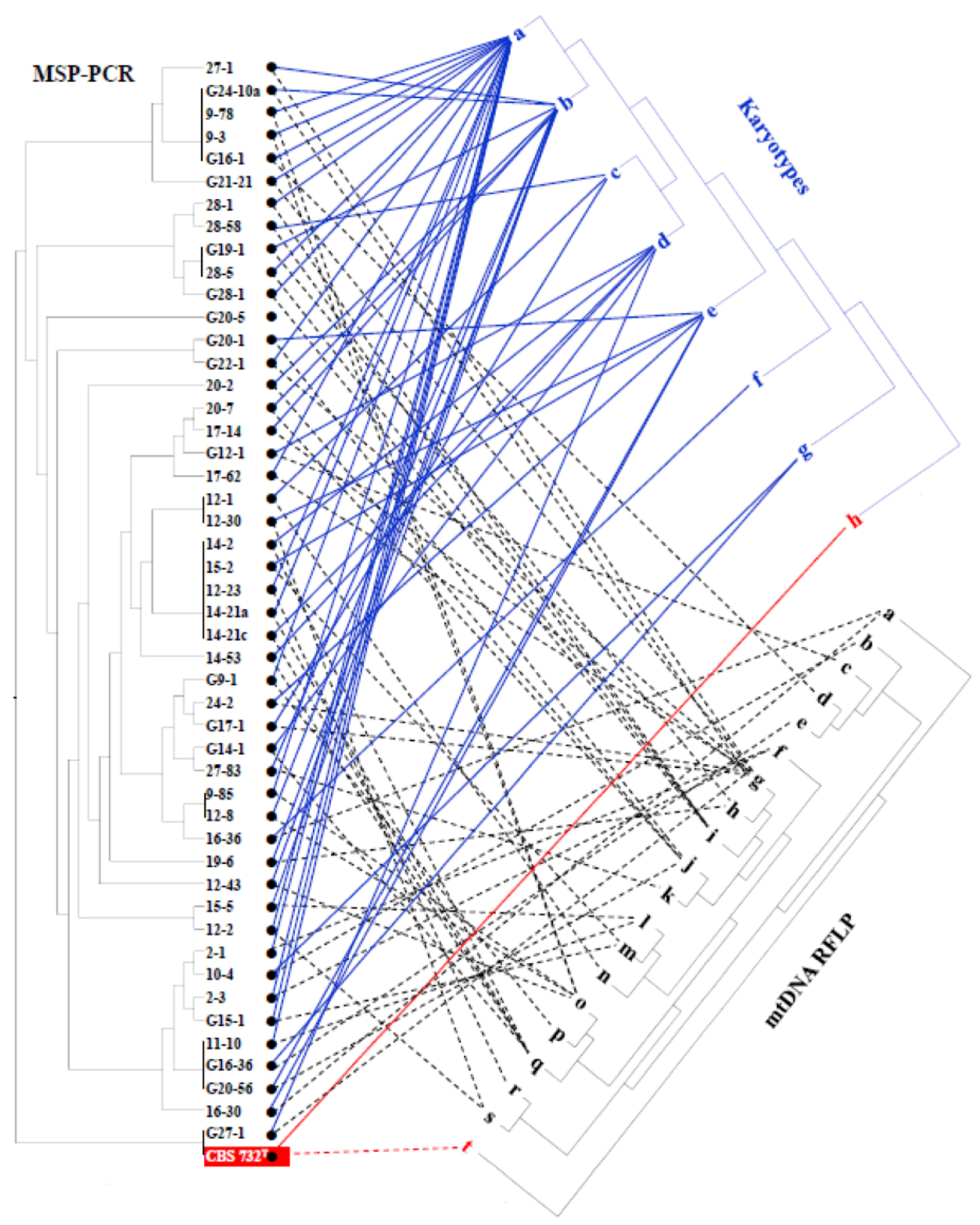
| Sample | Location of Winery | Vintage | Stored in 1 | Reducing Sugar g/L | Alcohol% | Extract | pH | Acid | SO2 2 | CFU 3 (108/mL) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugar-free (g/L) | Sum (g/L) | Titr. (g/L) | Vol. (g/L) | Free (mg/L) | All (mg/L) | ||||||||
| 1 | Vámosújfalu | 1985 | B | 365.00 | 5.53 | 94.00 | 459.00 | 3.3 | 12.12 | 0.93 | 10 | 130 | 0 |
| 2 | Tolcsva | 2000 | GB | 711.60 | 1.12 | 99.60 | 811.20 | 3.26 | 13.96 | 0.39 | 8 | 24 | 46.5 |
| 3 | Vámosújfalu | 2005 | B | 498.10 | 3.64 | 60.90 | 559.00 | 3.12 | 12.12 | 0.90 | 10 | 248 | 0 |
| 4 | Unspecified | 2007 | B | 510.80 | 3.46 | 81.50 | 592.30 | 3.44 | 8.48 | 0.84 | 6 | 236 | 0 |
| 5 | Hercegkút | 2013 | B | 509.00 | 1.95 | 69.20 | 578.20 | 2.99 | 10.50 | 0.87 | 8 | 250 | 0.00045 |
| 6 | Tokaj | 2013 | B | 544.00 | 2.49 | 60.50 | 604.50 | 3.19 | 11.58 | 0.84 | 10 | 306 | 0 |
| 7 | Unspecified | 2017 | GB | 538.70 | 5.11 | 79.60 | 618.30 | 3.44 | 13.60 | 1.08 | 12 | 384 | 1 |
| 8 | Unspecified | 2017 | OB | 518.40 | 3.87 | 77.80 | 596.20 | 3.17 | 12.68 | 1.20 | 8 | 300 | 1 |
| 9 | Mád | 2017 | B | 620.10 | 2.76 | 55.60 | 675.70 | 3.32 | 12.45 | 0.05 | 6 | 18 | 6 |
| 10 | Tolcsva | 2017 | B | 426.90 | 4.76 | 43.80 | 470.70 | 3.39 | 9.96 | 0.69 | 6 | 16 | 0.0004 |
| 11 | Tolcsva | 2018 | T | 517.90 | 3.80 | 154.70 | 672.60 | 3.52 | 10.19 | 1.08 | 12 | 210 | 0.05 |
| 12 | Mád | 2018 | B | 752.20 | 2.60 | 41.80 | 794.00 | 3.24 | 11.68 | 0.54 | 10 | 20 | 1 |
| 13 | Tolcsva | 2019 | GB | 724.30 | 0.00 | 59.60 | 783.90 | 3.53 | 9.10 | 0.90 | 42 | 360 | 0 |
| 14 | Tolcsva | 2019 | C | 584.00 | 4.01 | 30.10 | 614.10 | 3.4 | 11.27 | 0.93 | 6 | 16 | 0.00035 |
| 15 | Unspecified | 2019 | T | 729.30 | 0.00 | 50.40 | 779.70 | 3.28 | 12.64 | 0.90 | 42 | 340 | 0.00005 |
| 16 | Hercegkút | 2019 | T | 620.10 | 1.68 | 46.00 | 666.10 | 3.71 | 9.56 | 0.78 | 6 | 30 | 102 |
| 17 | Tolcsva | 2019 | GB | 617.50 | 1.70 | 47.80 | 665.30 | 3.41 | 11.48 | 0.99 | 6 | 116 | 66 |
| 18 | Tarcal | 2017 | GB | 740.00 | 0.39 | 34.30 | 774.30 | 3.48 | 9.94 | 0.50 | 8 | 16 | 3 |
| 19 | Tarcal | 2019 | GB | 645.50 | 3.21 | 27.90 | 673.40 | 3.25 | 11.64 | 0.00 | n.d. | n.d. | 16 |
| 20 | Mád | 2019 | GB | 731.90 | 2.85 | 9.10 | 741.00 | 3.59 | 8.07 | 0.00 | n.d. | n.d. | 60 |
| 21 | Bodrogolaszi | 2013 | B | 533.70 | 3.24 | 90.30 | 624.00 | 3.42 | 9.37 | 0.35 | n.d. | n.d. | 0.00012 |
| 22 | Bodrogolaszi | 2016 | GB | 716.60 | 2.97 | 45.20 | 761.80 | 3.26 | 22.14 | 0.00 | n.d. | n.d. | 0.00052 |
| 23 | Tolcsva | 2016 | T | 493.00 | 4.15 | 141.40 | 634.40 | 3.3 | 19.98 | 0.53 | n.d. | n.d. | 0 |
| 24 | Bodrogolaszi | 2017 | GB | 655.60 | 3.01 | 33.40 | 689.00 | 3.74 | 10.92 | 0.05 | n.d. | n.d. | 40 |
| 25 | Bodrogolaszi | 2017 | T | 574.30 | 3.47 | 71.80 | 646.10 | 3.74 | 12.63 | 0.47 | n.d. | n.d. | 0.5 |
| 26 | Mixed | 2017 | T | 681.10 | 2.97 | 49.50 | 730.60 | 3.32 | 16.22 | 0.00 | n.d. | n.d. | 0.00025 |
| 27 | Bodrogolaszi | 2019 | T | 609.90 | 3.00 | 76.80 | 686.70 | 3.57 | 13.43 | 0.00 | n.d. | n.d. | 22.3 |
| 28 | Mád | 2013 | GB | 543.80 | 3.19 | 81.20 | 625.00 | 3.51 | 10.63 | 0.40 | n.d. | n.d. | 12 |
| Isolate | Species | MIC (% Glucose) |
|---|---|---|
| 15-1 | H. osmophila | >70 |
| 2-25 | K. fluxuum | 40 |
| 15-11, 16-33, G20-16 | L. thermotolerans | >70 |
| 16-39, 20-1, G20-6 | M. pulcherrima clade sp. | ≤70 |
| 2-40, 2-65 | P. membranifaciens | 60 |
| G10-1, 11-25, 12-63, 14-4, 20-18 | C. zemplinina | >70 |
| G8-1, 11-33 | C. lactis-condensi | >70 |
| G9-4, | 60 | |
| 9-1, 19-1 | 50 | |
| 16-30 | Z. bailii | >70 |
| 5-2, 5-43, G5-1 | Z. lentus | >70 |
| 10-4 | Z. pseudobailii | >70 |
| All isolates | Z. rouxii | >70 |
| Isolate | Colony Morphology | Growth on/at 1 | MIC | Acid Prod (mm) | BiGGY | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mannitol | Galactose | Glycerol | Maltose | Lysine | 37 °C | Glucose (%) | Ethanol (%) | Sulphur (mg∙L−1) | ||||
| 9-3 | Rough white | + | + | + | + | + | + | >70 | 14 | 300 | 3 | 3 |
| 9-78 | Dull white | + | + | + | + | + | + | >70 | 14 | 300 | 3 | 3 |
| 9-85 | Rough white | + | + | + | + | + | + | >70 | 14 | 300 | 3 | 3 |
| G9-1 | Dull white | + | + | + | + | + | + | >70 | 14 | 300 | 3.5 | 4 |
| 12-8 | Dull white | + | + | + | + | + | + | >70 | 12 | 700 | 3 | 4 |
| G12-1 | Dull white | + | + | + | + | + | + | >70 | 12 | 700 | 3.5 | 4 |
| 20-2 | Rough white | + | + | + | + | + | + | >70 | 14 | 700 | 2.5 | 4 |
| 12-1 | Rough white | + | + | + | w | + | + | >70 | 14 | 400 | 3 | 3 |
| 15-2 | Dull white | + | + | + | w | + | + | >70 | 12 | 800 | 2.5 | 3 |
| 15-5 | Dull white | + | + | + | w | + | + | >70 | 14 | 800 | 2 | 3 |
| G17-1 | Dull white | + | + | + | w | + | + | >70 | 14 | 300 | 2 | 3 |
| 11-10 | Dull white | + | + | + | − | + | + | >70 | 14 | 800 | 1.75 | 2 |
| 12-2 | Dull white | + | + | + | − | + | + | >70 | 14 | 300 | 3 | 3 |
| 12-23 | Dull white | + | + | + | − | + | + | >70 | 14 | 400 | 2 | 1 |
| 14-2 | Dull white | + | + | + | − | + | + | >70 | 14 | 300 | 3 | 4 |
| 20-7 | Rough white | + | + | + | − | + | + | >70 | 14 | 300 | 3 | 4 |
| 27-1 | Dull white | + | + | + | + | + | w | >70 | 14 | 700 | 3 | 3 |
| G16-1 | Dull white | + | + | + | + | + | w | >70 | 12 | 700 | 2 | 4 |
| G22-1 | Dull white | + | + | + | + | + | − | >70 | 12 | 700 | 3 | 3 |
| 24-2 | Dull white | + | + | + | + | + | − | >70 | 12 | 700 | 2.5 | 3 |
| 12-43 | Rough white | + | + | + | + | w | − | >70 | 14 | 300 | 3 | 3 |
| 14-21c | White sectored | + | + | + | w | + | w | >70 | 12 | 700 | 2 | 4 |
| 17-14 | Dull white | + | + | + | w | + | w | >70 | 12 | 800 | 1.5 | 3 |
| G16-36 | Rough white | + | + | + | w | + | − | >70 | 14 | 300 | 3 | 3 |
| 17-62 | Dull white | + | + | + | w | + | − | >70 | 14 | 800 | 2 | 2 |
| 27-83 | Rough white | + | + | + | w | + | − | >70 | 10 | 400 | 3.5 | 4 |
| G21-21 | Dull white | + | + | + | − | + | w | >70 | 14 | 700 | 3.5 | 3 |
| 2-1 | Dull white | + | + | + | − | + | − | >70 | 14 | 700 | 3.5 | 3 |
| 2-3 | Dull white | + | + | + | − | + | − | >70 | 14 | 700 | 3.5 | 3 |
| 2-42 | Dull white | + | + | + | − | + | − | >70 | 14 | 600 | 3 | 3 |
| G2-1 | Dull white | + | + | + | − | + | − | >70 | 14 | 600 | 3 | 3 |
| 14-21a | Dull white | + | + | + | − | + | − | >70 | 13 | 300 | 3 | 4 |
| 14-53 | Dull white | + | + | + | − | + | − | >70 | 12 | 300 | 3 | 4 |
| G14-1 | Dull white | + | + | + | − | + | − | >70 | 12 | 300 | 3 | 4 |
| G20-1 | Dull white | + | + | + | − | + | − | >70 | 12 | 400 | 3.5 | 4 |
| G20-56 | Rough white | + | + | + | − | + | − | >70 | 12 | 400 | 3.5 | 4 |
| G24-10a | Dull white | + | + | + | − | + | − | >70 | 12 | 600 | 3 | 3 |
| G27-1 | Rough white | + | + | + | − | + | − | >70 | 14 | 700 | 3 | 3 |
| 12-30 | Dull white | + | + | + | − | w | + | >70 | 14 | 400 | 2 | 1 |
| 19-6 | Dull white | + | + | + | − | w | w | >70 | 12 | 800 | 3 | 1 |
| 12-18 | Dull white | + | + | + | − | w | − | >70 | 14 | 400 | 3 | 1 |
| G19-1 | Dull white | + | + | + | − | w | − | >70 | 14 | 800 | 2 | 1 |
| 28-1 | Dull white | w | w | w | w | w | − | >70 | 12 | 400 | 2 | 1 |
| 28-5 | Dull white | w | w | w | w | w | − | >70 | 12 | 400 | 2.5 | 1 |
| 28-58 | Dull white | w | w | w | w | w | − | >70 | 12 | 600 | 1 | 1 |
| G28-1 | Dull white | w | w | w | w | w | − | >70 | 12 | 600 | 1 | 1 |
| Colony | Width of Inhibition Zones (mm) in the Lawn of | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G8-1 | G11-4 | 11-25 | 17-1 | 15-1 | 2-25 | 15-11 | 16-33 | 20-1 | 16-39 | 2-65 | 2-40 | 2-3 | 20-2 | 16-30 | 5-2 | 10-4 | ||
| Isolate | Species | C. lactis-condensi | C. lactis-condensi | C. zemplinina | C. zemplinina | H. osmophila | K. fluxuum | L. thermotolerans | L. thermotolerans | M. p. clade sp. | M. p. clade sp. | P. membranifaciens | P. membranifaciens | Z. rouxii | Z. rouxii | Z. bailii | Z. lentus | Z. pseudobailii |
| G8-1 | C. lactis-condensi |  | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| G11-4 | C. lactis-condensi | − |  | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 11-25 | C. zemplinina | − | 2, 2 |  | − | 3, 3 | − | 2,2 | − | − | − | − | − | − | − | − | − | − |
| 17-1 | C. zemplinina | − | − | − |  | 3, 2 | crossf | 2,2 | − | − | − | − | − | − | − | − | − | − |
| 15-1 | H. osmophila | − | − | − | − |  | − | − | − | − | − | − | − | − | − | − | − | − |
| 2-25 | K. fluxuum | − | − | − | − | − |  | crossf | crossf | − | − | − | − | − | − | − | − | − |
| 15-11 | L. thermotolerans | 2, 1 | 3, 3 | 2, 2 | 1, 1 | 2, 1 | 2, 2 |  | 2, 0 | 2, 2 | 2, 2 | 2, 1 | 1, 1 | 1, 0 | 2, 2 | 2, 2 | 2, 2 | 1, 1 |
| 16-33 | L. thermotolerans | 1, 1 | 2, 2 | 2, 2 | 2, 1 | 1, 1 | 2, 2 | 2, 2 |  | 3, 0 | 2, 2 | − | − | 1, 1 | 2, 2 | 2, 1 | 1, 1 | 1, 1 |
| 20-1 | M. pulcherrima clade sp. | 1, 0 | 1, 1 | − | 1, 0 | − | crossf | crossf | − |  | − | − | − | − | 1, 1 | − | − | − |
| 16-39 | M. pulcherrima clade sp. | − | 1, 1 | − | 1, 1 | − | − | − | crossf | − |  | 0.5, 1 | 0.5, 1 | − | 1, 0 | − | − | − |
| 2-65 | P. membranifaciens | − | 1, 0 | − | − | − | − | − | crossf | − | − |  | − | − | − | − | − | − |
| 2-3 | Z. rouxii | − | 1, 0 | − | − | 1, 0 | 1, 0 | − | − | − | − | 1, 1 | 1, 0.5 |  | − | − | − | − |
| 20-2 | Z. rouxii | − | 3, 3 | − | − | − | 1, 0 | crossf | crossf | − | − | 1, 1 | 1, 1 | − |  | − | − | − |
| 16-30 | Z. bailii | − | 1, 0 | − | − | − | − | crossf | crossf | − | − | 0.5, 0 | − | − | − |  | − | − |
| 5-2 | Z. lentus | − | 1, 0 | − | − | − | − | crossf | crossf | − | − | 1, 0 | 0.5, 0 | − | − | − |  | − |
| 10-4 | Z. pseudobailii | − | − | − | − | − | − | crossf | crossf | − | − | − | − | − | − | − | − |  |
| Mixed Cultures | Proportion of Isolates after 48 h of Incubation at | ||
|---|---|---|---|
| Isolates | Species Combination | 2% Glucose | 30% Glucose |
| 2-3 + 10-4 | Z. rouxii + Z. pseudobailii | 1:1.03 | 1:8 |
| 2-3 + 16-30 | Z. rouxii + Z. bailii | n.d. | 1.18:1 |
| 2-3 + G11-4 | Z. rouxii + C. lactis-condensi | 1:1.19 | 15.25:1 |
| 2-3 + 11-25 | Z. rouxii + C. zemplinina | 1:24 | 2.4:1 |
| 11-25 + G11-4 | C. zemplinina + C. lactis-condensi | 1:1.13 | 1:5.4 |
| 2-3 + 15-1 | Z. rouxii + H osmophila | 1:75 | 1:14.3 |
| 2-3 + 15-11 | Z. rouxii + L. thermotolerans | 1:>57 | n.d |
| 2-3 + 16-39 | Z. rouxii + Metschnikowia | 1:>84 | n.d. |
| 2-3 + 2-65 | Z. rouxii + P. membranifaciens | 1:>28 | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csoma, H.; Kállai, Z.; Antunovics, Z.; Czentye, K.; Sipiczki, M. Vinification without Saccharomyces: Interacting Osmotolerant and “Spoilage” Yeast Communities in Fermenting and Ageing Botrytised High-Sugar Wines (Tokaj Essence). Microorganisms 2021, 9, 19. https://doi.org/10.3390/microorganisms9010019
Csoma H, Kállai Z, Antunovics Z, Czentye K, Sipiczki M. Vinification without Saccharomyces: Interacting Osmotolerant and “Spoilage” Yeast Communities in Fermenting and Ageing Botrytised High-Sugar Wines (Tokaj Essence). Microorganisms. 2021; 9(1):19. https://doi.org/10.3390/microorganisms9010019
Chicago/Turabian StyleCsoma, Hajnalka, Zoltán Kállai, Zsuzsa Antunovics, Kinga Czentye, and Matthias Sipiczki. 2021. "Vinification without Saccharomyces: Interacting Osmotolerant and “Spoilage” Yeast Communities in Fermenting and Ageing Botrytised High-Sugar Wines (Tokaj Essence)" Microorganisms 9, no. 1: 19. https://doi.org/10.3390/microorganisms9010019
APA StyleCsoma, H., Kállai, Z., Antunovics, Z., Czentye, K., & Sipiczki, M. (2021). Vinification without Saccharomyces: Interacting Osmotolerant and “Spoilage” Yeast Communities in Fermenting and Ageing Botrytised High-Sugar Wines (Tokaj Essence). Microorganisms, 9(1), 19. https://doi.org/10.3390/microorganisms9010019





