Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials
Abstract
:1. Introduction
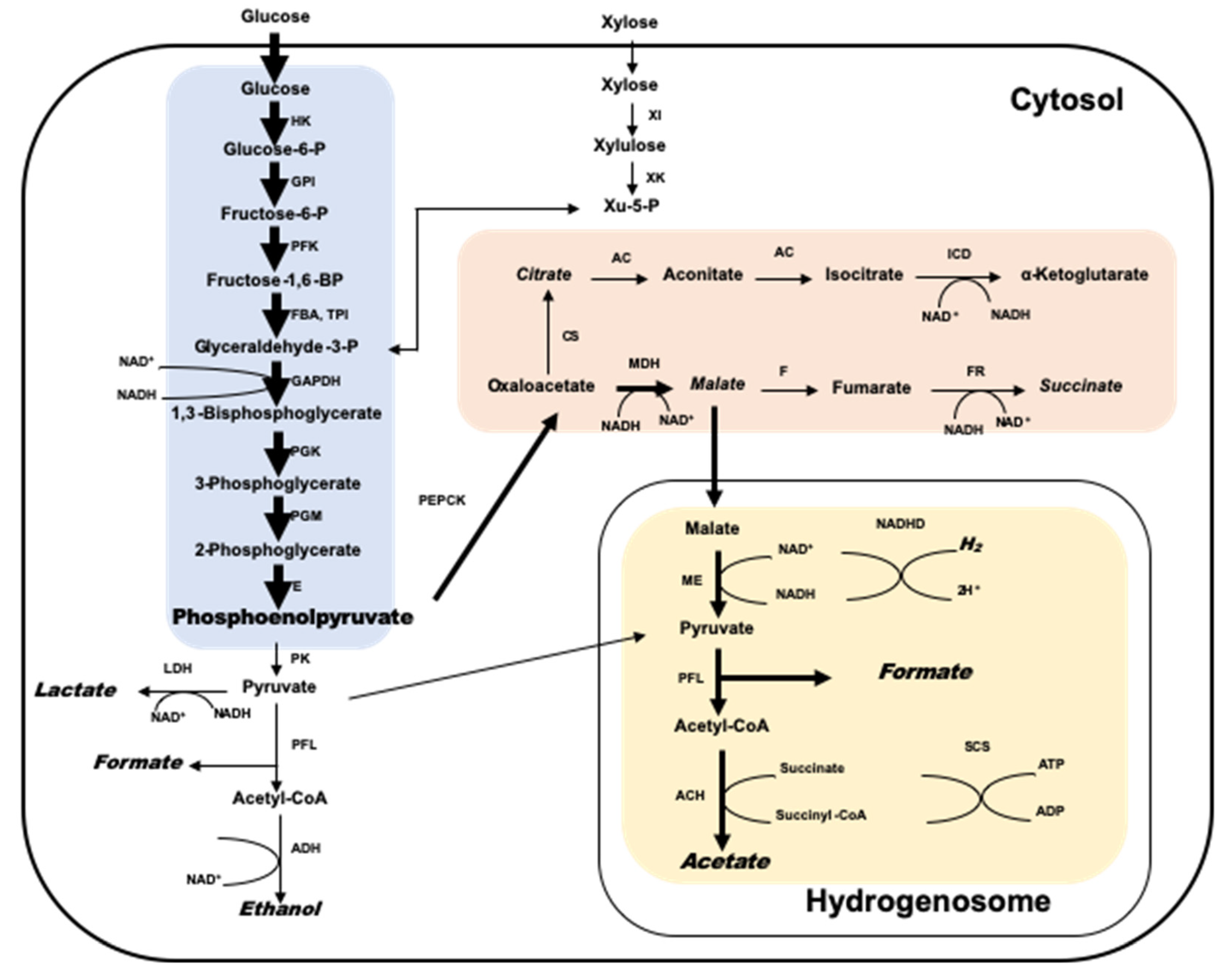
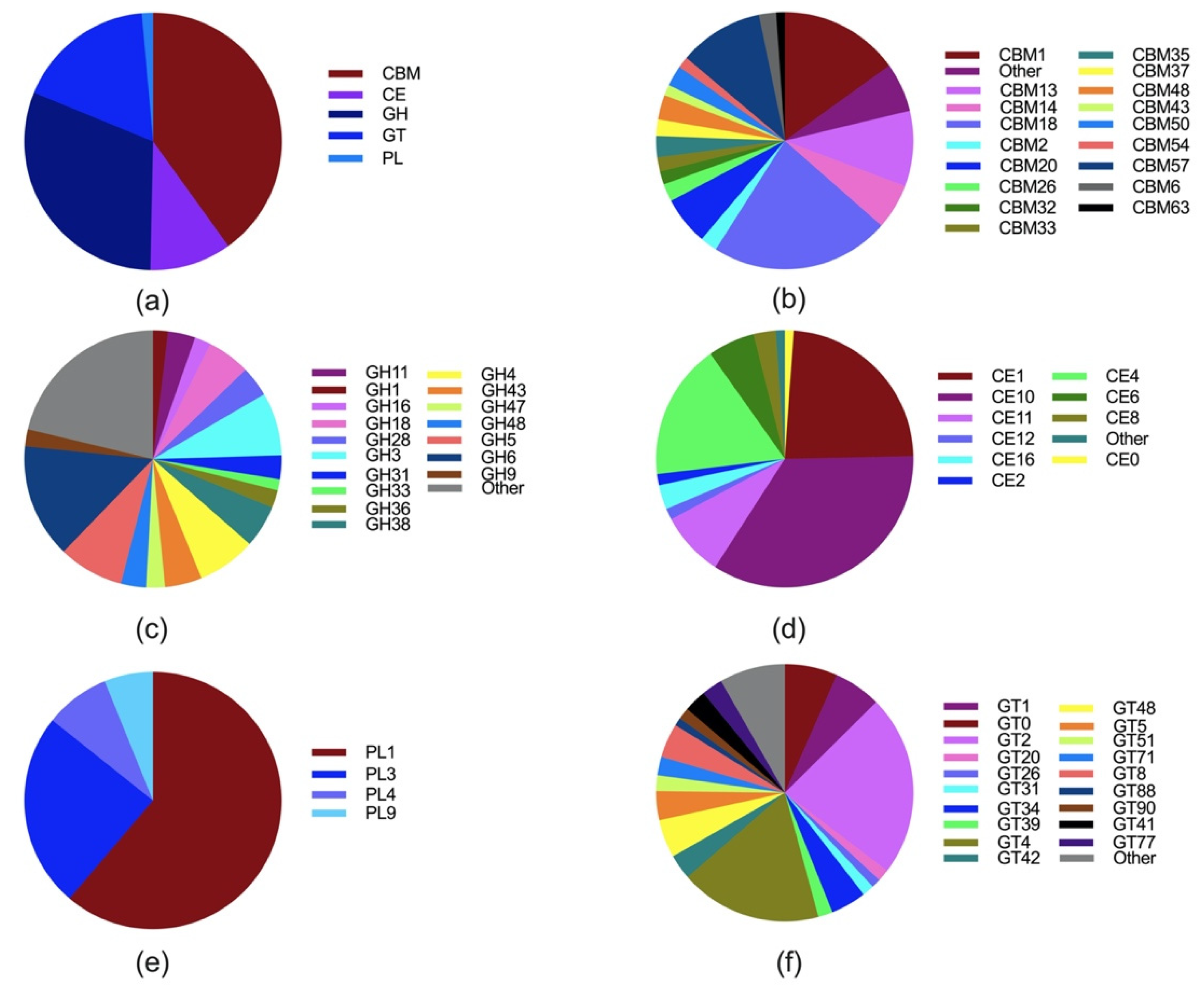
2. Taxonomy, Distribution, Metabolism, and Fiber Degrading Enzymes of Anaerobic Fungi
3. Taxonomy, Distribution, and Metabolism of Methanogens
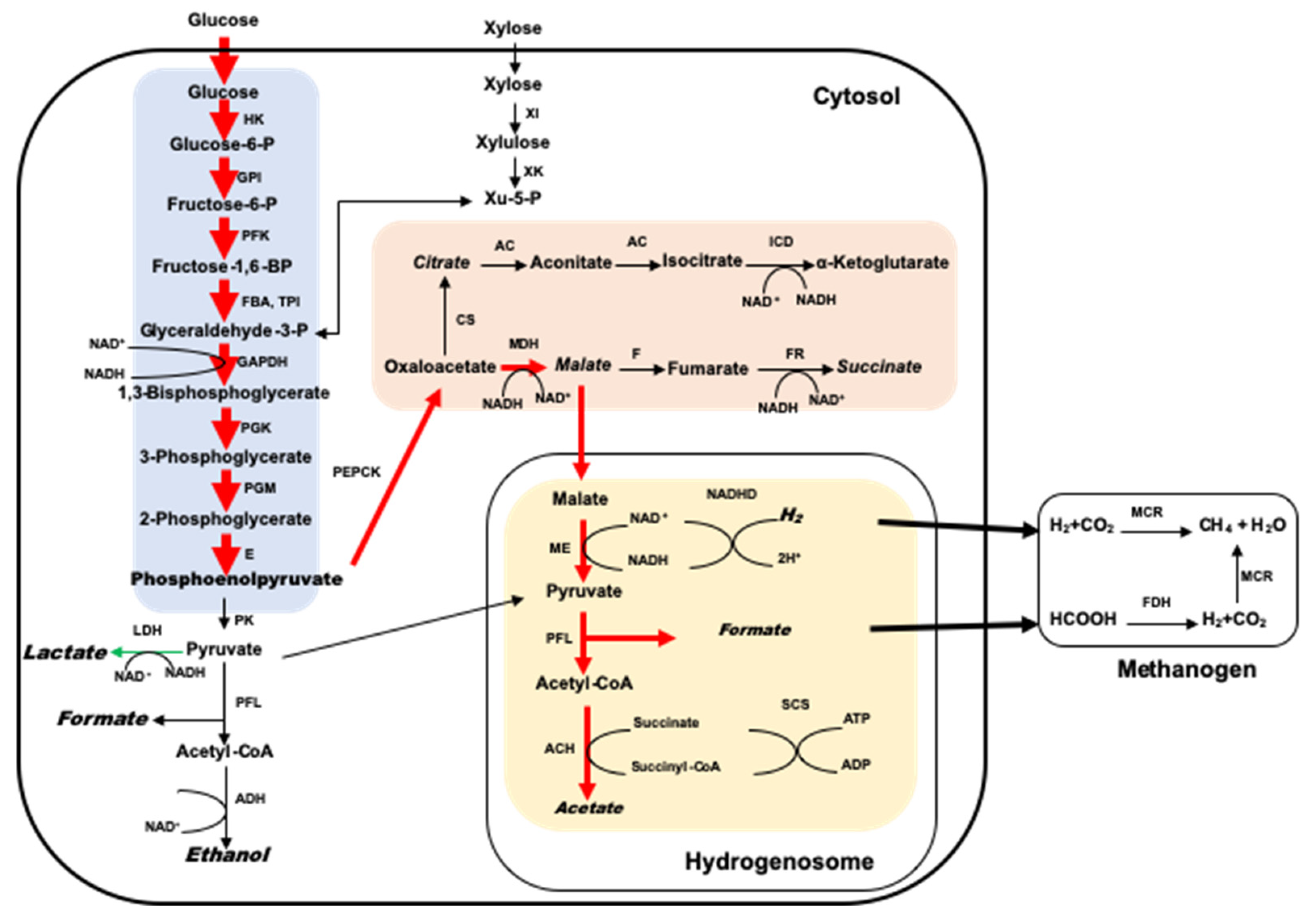
4. Interaction of Anaerobic Fungi and Methanogens in the Rumen
5. The Biotechnological Potential for Biogas Production from Lignocellulosic Materials
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Zhang, G.; Li, Y.; Zhang, Y. Effects of High Forage/Concentrate Diet on Volatile Fatty Acid Production and the Microorganisms Involved in VFA Production in Cow Rumen. Animals 2020, 10, 223. [Google Scholar] [CrossRef] [Green Version]
- Demeyer, D.I. Rumen microbes and digestion of plant cell walls. Agric. Environ. 1981, 6, 295–337. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Akin, D.E.; Borneman, W.S.; Lyon, C.E. Degradation of leaf blades and stems by monocentric and polycentric isolates of ruminal fungi. Anim. Feed Sci. Tech. 1990, 31, 205–221. [Google Scholar] [CrossRef]
- Bauchop, T.; Mountfort, D.O. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl. Environ. Microb. 1981, 42, 1103–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.F.; Edwards, J.E.; Allison, G.G.; Zhu, W.-Y.; Theodorou, M.K. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour. Technol. 2009, 100, 4821–4828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orpin, C.G.; Joblin, K.N. The rumen anaerobic fungi. In The Rumen Microbial Ecosystem; Springer: Dordrecht, The Netherlands, 1997. [Google Scholar]
- Jin, W.; Cheng, Y.F.; Mao, S.Y.; Zhu, W.Y. Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour. Technol. 2011, 102, 7925–7931. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Shi, Q.C.; Sun, R.L.; Liang, D.; Li, Y.F.; Li, Y.Q.; Jin, W.; Zhu, W.Y. The biotechnological potential of anaerobic fungi on fiber degradation and methane production. World J. Microbiol. Biotechnol. 2018, 34, 8. [Google Scholar] [CrossRef]
- Moss, A.R.; Jouany, J.P.; Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 2000, 49, 231–253. [Google Scholar] [CrossRef] [Green Version]
- Millen, D.D.; Mario, D.B.A.; Pacheco, R.D.L. Rumenology; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Nakashimada, Y.; Srinivasan, K.; Murakami, M.; Nishio, N. Direct conversion of cellulose to methane by anaerobic fungus Neocallimastix frontalis and defined methanogens. Biotechnol. Lett. 2000, 22, 223–227. [Google Scholar] [CrossRef]
- Li, Y.; Jin, W.; Cheng, Y.; Zhu, W. Effect of the Associated Methanogen Methanobrevibacter thaueri on the Dynamic Profile of End and Intermediate Metabolites of Anaerobic Fungus Piromyces sp F1. Curr. Microbiol. 2016, 73, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Yildirim, E.; Ince, O.; Ince, B. Rumen anaerobic fungi create new opportunities for enhanced methane production from microalgae biomass. Algal Res. 2017, 23, 150–160. [Google Scholar] [CrossRef]
- Yildirim, E.; Ince, O.; Aydin, S.; Ince, B. Improvement of biogas potential of anaerobic digesters using rumen fungi. Renew. Energy 2017, 109, 346–353. [Google Scholar] [CrossRef]
- Ferraro, A.; Dottorini, G.; Massini, G.; Miritana, V.M.; Signorini, A.; Lembo, G.; Fabbricino, M. Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour. Technol. 2018, 260, 364–373. [Google Scholar] [CrossRef]
- Akyol, C.; Ince, O.; Bozan, M.; Ozbayram, E.G.; Ince, B. Fungal bioaugmentation of anaerobic digesters fed with lignocellulosic biomass: What to expect from anaerobic fungus Orpinomyces sp. Bioresour. Technol. 2019, 277, 1–10. [Google Scholar] [CrossRef]
- Shi, Q.; Li, Y.; Li, Y.; Cheng, Y.; Zhu, W. Effects of steam explosion on lignocellulosic degradation of, and methane production from, corn stover by a co-cultured anaerobic fungus and methanogen. Bioresour. Technol. 2019, 290. [Google Scholar] [CrossRef]
- Ferdes, M.; Dinca, M.N.; Moiceanu, G.; Zabava, B.S.; Paraschiv, G. Microorganisms and Enzymes Used in the Biological Pretreatment of the Substrate to Enhance Biogas Production: A Review. Sustainability 2020, 12, 7205. [Google Scholar] [CrossRef]
- Orpin, C.G. Studies on the rumen flagellate Neocallimastix frontalis. J. Gen. Microbiol. 1975, 91, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Orpin, C.G. The occurrence of chitin in the cell walls of the rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J. Gen. Microbiol. 1977, 99, 215–218. [Google Scholar] [CrossRef] [Green Version]
- Barr, D.J.S. An outline for the reclassification of the Chytridiales, and for a new order, the Spizellomycetales. Can. J. Bot. 2011, 58, 2380–2394. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerova, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef] [PubMed]
- Gruninger, R.J.; Puniya, A.K.; Callaghan, T.M.; Edwards, J.E.; Youssef, N.; Dagar, S.S.; Fliegerova, K.; Griffith, G.W.; Forster, R.; Tsang, A.; et al. Anaerobic fungi (phylum Neocallimastigomycota): Advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. Fems Microbiol. Ecol. 2014, 90, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, S.-Y.; Huo, W.-J.; Zhu, W.-Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R.; Theodorou, M.K.; Lawrence, M.I.G.; Trinci, A.P.J. Distribution of anaerobic fungi in the digestive-tract of cattle and their survival in feces. J. Gen. Microbiol. 1993, 139, 1395–1400. [Google Scholar] [CrossRef] [Green Version]
- Heath, I.B.; Bauchop, T.; Skipp, R.A. Assignment of the rumen anaerobe Neocallimastix frontalis to the Spizellomycetales (Chytridiomycetes) on the basis of its polyflagellate zoospore ultrastructure. Can. J. Bot. 1983, 61, 295–307. [Google Scholar] [CrossRef]
- Gold, J.J.; Heath, I.B.; Bauchop, T. Ultrastructural description of a new chytrid genus of cecum anaerobe, caecomyces-equi gen-nov, sp-nov, assigned to the neocallimasticaceae. Biosystems 1988, 21, 403–415. [Google Scholar] [CrossRef]
- Barr, D.J.S.; Kudo, H.; Jakober, K.D.; Cheng, K.J. Morphology and development of rumen fungi—Neocallimastix sp piromyces communis, and orpinomyces-bovis gen-nov sp-nov. Can. J. Bot. 1989, 67, 2815–2824. [Google Scholar] [CrossRef]
- Breton, A.; Bernalier, A.; Dusser, M.; Fonty, G.; Gaillardmartinie, B.; Guillot, J. Anaeromyces-mucronatus nov-gen, nov-sp - a new strictly anaerobic rumen fungus with polycentric thallus. Fems Microbiol. Lett. 1990, 70, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Ozkose, E.; Thomas, B.J.; Davies, D.R.; Griffith, G.W.; Theodorou, M.K. Cyllamyces aberensis gen.nov sp.nov., a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Can. J. Bot. 2001, 79, 666–673. [Google Scholar]
- Callaghan, T.M.; Podmirseg, S.M.; Hohlweck, D.; Edwards, J.E.; Puniya, A.K.; Dagar, S.S.; Griffith, G.W. Buwchfawromyces eastonii gen. nov., sp nov.: A new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. Mycokeys 2015, 9, 11–28. [Google Scholar]
- Dagar, S.S.; Kumar, S.; Griffith, G.W.; Edwards, J.E.; Callaghan, T.M.; Singh, R.; Nagpal, A.K.; Puniya, A.K. A new anaerobic fungus (Oontomyces anksri gen. nov., sp nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol. 2015, 119, 731–737. [Google Scholar] [CrossRef] [Green Version]
- Hanafy, R.A.; Elshahed, M.S.; Liggenstoffer, A.S.; Griffith, G.W.; Youssef, N.H. Pecoramyces ruminantium, gen. nov., sp nov., an anaerobic gut fungus from the feces of cattle and sheep. Mycologia 2017, 109, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, R.A.; Elshahed, M.S.; Youssef, N.H. Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild Barbary sheep and fallow deer. Mycologia 2018, 110, 1. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Lanjekar, V.B.; Dhakephalkar, P.K.; Callaghan, T.M.; Griffith, G.W.; Dagar, S.S. Liebetanzomyces polymorphus gen. et sp nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. Mycokeys 2018, 40, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Elshahed, M.S.; Youssef, N.H. Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia 2020, 112, 1212–1239. [Google Scholar]
- Orpin, C.G. Isolation of Cellulolytic Phycomycete Fungi from the Caecum of the Horse. J. Gen. Microbiol. 1981, 123, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Milne, A.; Theodorou, M.K.; Jordan, M.G.C.; Kingspooner, C.; Trinci, A.P.J. Survival of anaerobic fungi in feces, in saliva, and in pure culture. Exp. Mycol. 1989, 13, 27–37. [Google Scholar] [CrossRef]
- Brookman, J.L.; Mennim, G.; Trinci, A.P.J.; Theodorou, M.K.; Tuckwell, D.S. Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS1 and 18S rRNA. Microbiology 2000, 146, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Teunissen, M.J.; Dencamp, H.; Orpin, C.G.; Veld, J.; Vogels, G.D. Comparison of growth-characteristics of anaerobic fungi isolated from ruminant and non-ruminant herbivores during cultivation in a defined medium. J. Gen. Microbiol. 1991, 137, 1401–1408. [Google Scholar] [CrossRef] [Green Version]
- Liggenstoffer, A.S.; Youssef, N.H.; Couger, M.B.; Elshahed, M.S. Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J. 2010, 4, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Lowe, S.E.; Theodorou, M.K.; Trinci, A.P.J. Growth and fermentation of an anaerobic rumen fungus on various carbon-sources and effect of temperature on development. Appl. Environ. Microb. 1987, 53, 1210–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockhart, R.J.; Van Dyke, M.I.; Beadle, I.R.; Humphreys, P.; McCarthy, A.J. Molecular biological detection of anaerobic gut fungi (Neocallimastigales) from landfill sites. Appl. Environ. Microb. 2006, 72, 5659–5661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belivanova, V.; Astrom, M.E. Anaerobic consortia of fungi and sulfate reducing bacteria in deep granite fractures. Nat. Commun. 2017, 8, 1–9. [Google Scholar]
- Bauchop, T. Rumen anaerobic fungi of cattle and sheep. Appl. Environ. Microb. 1979, 38, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Kessler, D.; Leibrecht, I.; Knappe, J. Pyruvate-formate-lyase-deactivase and acetyl-coa reductase activities of escherichia-coli reside on a polymeric protein particle encoded by adhe. FEBS Lett. 1991, 281, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.E.; Kingston-Smith, A.H.; Jimenez, H.R.; Huws, S.A.; Skot, K.P.; Griffith, G.W.; Mcewan, N.R.; Theodorou, M.K. Dynamics of initial colonization of nonconserved perennial ryegrass by anaerobic fungi in the bovine rumen. Fems Microbiol. Ecol. 2008, 66, 537–545. [Google Scholar] [CrossRef]
- Nicholson, M.J.; McSweeney, C.S.; Mackie, R.I.; Brookman, J.L.; Theodorou, M.K. Diversity of anaerobic gut fungal populations analysed using ribosomal ITS1 sequences in faeces of wild and domesticated herbivores. Anaerobe 2010, 16, 66–73. [Google Scholar] [CrossRef]
- Kittelmann, S.; Seedorf, H.; Walters, W.A.; Clemente, J.C.; Knight, R.; Gordon, J.I.; Janssen, P.H. Simultaneous Amplicon Sequencing to Explore Co-Occurrence Patterns of Bacterial, Archaeal and Eukaryotic Microorganisms in Rumen Microbial Communities. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Solomon, K.V.; Henske, J.K.; Theodorou, M.K.; O’Malley, M.A. Anaerobic Gut Fungi: Advances in Isolation, Culture, and Cellulolytic Enzyme Discovery for Biofuel Production. Biotechnol. Bioeng. 2014, 111, 1471–1482. [Google Scholar] [CrossRef]
- Mountfort, D.O.; Asher, R.A. Production and regulation of cellulase by 2 strains of the rumen anaerobic fungus neocallimastix-frontalis. Appl. Environ. Microb. 1985, 49, 1314–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellemond, J.J.; Ricard, G.; Huynen, M.; Tielens, A.G.M.; Hackstein, J.H.P. The anaerobic chytridiomycete fungus Piromyces sp E2 produces ethanol via pyruvate: Formate lyase and an alcohol dehydrogenase E. Mol. Microbiol. 2004, 51, 1389–1399. [Google Scholar]
- Borneman, W.S.; Akin, D.E.; Ljungdahl, L.G. Fermentation products and plant-cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl. Environ. Microb. 1989, 55, 1066–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhmanova, A.; Voncken, F.G.J.; Hosea, K.M.; Harhangi, H.; Keltjens, J.T.; den Camp, H.; Vogels, G.D.; Hackstein, J.H.P. A hydrogenosome with pyruvate formate-lyase: Anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol. Microbiol. 1999, 32, 1103–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, R.R.; Pedezzi, R.; Souto, T.B. Exploring the bioprospecting and biotechnological potential of white-rot and anaerobic Neocallimastigomycota fungi: Peptidases, esterases, and lignocellulolytic enzymes. Appl. Microbiol. Biotechnol. 2017, 101, 3089–3101. [Google Scholar] [CrossRef]
- Janssen, P.H.; Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microb. 2008, 74, 3619–3625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youssef, N.H.; Couger, M.B.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Prade, R.A.; Najar, F.Z.; Atiyeh, H.K.; Wilkins, M.R.; Elshahed, M.S. The Genome of the Anaerobic Fungus Orpinomyces sp Strain C1A Reveals the Unique Evolutionary History of a Remarkable Plant Biomass Degrader. Appl. Environ. Microb. 2013, 79, 4620–4634. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.-Y.; Chen, H.-L.; Lu, M.-Y.J.; Chen, Y.-C.; Sung, H.-M.; Mao, C.-T.; Cho, H.-Y.; Ke, H.-M.; Hwa, T.-Y.; Ruan, S.-K.; et al. Functional characterization of cellulases identified from the cow rumen fungus Neocallimastix patriciarum W5 by transcriptomic and secretomic analyses. Biotechnol. Biofuels 2011, 4. [Google Scholar] [CrossRef] [Green Version]
- Couger, M.B.; Youssef, N.H.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Elshahed, M.S. Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp strain C1A. Biotechnol. Biofuels 2015, 8. [Google Scholar] [CrossRef] [Green Version]
- Ferry, J.G. Methanogenesis || Diversity and Taxonomy of Methanogens; Springer: Boston, MA, USA, 1993; Chapter 2; pp. 35–80. [Google Scholar] [CrossRef]
- Hackstein, J.H.P. Methanogens in the Gastrointestinal Tract of Animals; Springer: Cham, Switzerland, 2018; pp. 121–152. [Google Scholar]
- Garcia, J.L.; Patel, B.K.C.; Ollivier, B. Taxonomic phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 2000, 6, 205–226. [Google Scholar] [CrossRef]
- Borrel, G.; Parisot, N.; Harris, H.M.B.; Peyretaillade, E.; Gaci, N.; Tottey, W.; Bardot, O.; Raymann, K.; Gribaldo, S.; Peyret, P.; et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genom. 2014, 15, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St-Pierre, B.; Wright, A.-D.G. Molecular analysis of methanogenic archaea in the forestomach of the alpaca (Vicugna pacos). BMC Microbiol. 2012, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, E.E.; Smith, R.P.; St-Pierre, B.; Wright, A.-D.G. Differences in the Rumen Methanogen Populations of Lactating Jersey and Holstein Dairy Cows under the Same Diet Regimen. Appl. Environ. Microb. 2011, 77, 5682–5687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seedorf, H.; Kittelmann, S.; Janssen, P.H. Few Highly Abundant Operational Taxonomic Units Dominate within Rumen Methanogenic Archaeal Species in New Zealand Sheep and Cattle. Appl. Environ. Microb. 2015, 81, 986–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.D.; Tan, H.Y.; Long, R.; Liang, J.B.; Wright, A.-D.G. Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai-Tibetan plateau, China. BMC Microbiol. 2012, 12, 237. [Google Scholar] [CrossRef] [Green Version]
- Seedorf, H.; Kittelmann, S.; Henderson, G.; Janssen, P.H. RIM-DB: A taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ 2014, 2, e424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.S.; Tian, J.Q.; Jiang, N.; Guo, X.P.; Wang, Y.F.; Dong, X.Z. Methanogen community in Zoige wetland of Tibetan plateau and phenotypic characterization of a dominant uncultured methanogen cluster ZC-I. Environ. Microbiol. 2008, 10, 1850–1860. [Google Scholar] [CrossRef]
- Conrad, R.; Erkel, C.; Liesack, W. Rice Cluster I methanogens, an important group of Archaea producing greenhouse gas in soil. Curr. Opin. Biotechnol. 2006, 17, 262–267. [Google Scholar] [CrossRef]
- Newberry, C.J.; Webster, G.; Cragg, B.A.; Parkes, R.J.; Weightman, A.J.; Fry, J.C. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 2004, 6, 274–287. [Google Scholar] [CrossRef] [Green Version]
- Ferry, J.G.; Lessner, D.J. Methanogenesis in Marine Sediments. Ann. N. Y. Acad. Sci. 2008, 1125, 147–157. [Google Scholar] [CrossRef]
- Lu, Y.H.; Conrad, R. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 2005, 309, 1088–1090. [Google Scholar] [CrossRef] [PubMed]
- Strapoc, D.; Mastalerz, M.; Dawson, K.; Macalady, J.; Callaghan, A.V.; Wawrik, B.; Turich, C.; Ashby, M. Biogeochemistry of Microbial Coal-Bed Methane: Annual Review of Earth and Planetary Sciences. Annu. Rev. Earth Planet. Sci. 2011, 39, 617–656. [Google Scholar] [CrossRef]
- Simankova, M.V.; Parshina, S.N.; Tourova, T.P.; Kolganova, T.V.; Zehnder, A.J.B.; Nozhevnikova, A.N. Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments. Syst. Appl. Microbiol. 2001, 24, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Karakashev, D.; Batstone, D.J.; Angelidaki, I. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl. Environ. Microb. 2005, 71, 331–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angel, R.; Claus, P.; Conrad, R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 2012, 6, 847–862. [Google Scholar] [CrossRef] [Green Version]
- Huber, R.; Kurr, M.; Jannasch, H.W.; Stetter, K.O. A novel group of abyssal methanogenic archaebacteria (methanopyrus) growing at 110-degrees-c. Nature 1989, 342, 833–834. [Google Scholar] [CrossRef]
- Pace, N.R. A molecular view of microbial diversity and the biosphere. Science 1997, 276, 734–740. [Google Scholar] [CrossRef]
- Krishnan, S.; Singh, L.; Sakinah, M.; Thakur, S.; Wahid, Z.A.; Alkasrawi, M. Process enhancement of hydrogen and methane production from palm oil mill effluent using two-stage thermophilic and mesophilic fermentation. Int. J. Hydrog. Energy 2016, 41, 12888–12898. [Google Scholar] [CrossRef] [Green Version]
- Mountfort, D.O.; Asher, R.A.; Bauchop, T. Fermentation of Cellulose to Methane and Carbon Dioxide by a Rumen Anaerobic Fungus in a Triculture with Methanobrevibacter sp. Strain RA1 and Methanosarcina barkeri. Appl. Environ. Microb. 1982, 44, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Yuanfei, L.I.; Meizhou, S.; Yanfen, C.; Weiyun, Z. Effects of Associated Methanogen on Organic Acid Profile of Metabolism by Anaerobic Fungus Revealed Using High Performance Liquid Chromatography. Chin. J. Anim. Nutr. 2017, 29, 1198–1204. [Google Scholar]
- Jin, W.; Liu, J.; Li, Y.; Cheng, Y.; Zhu, W. Effect of methanogens on carbon metabolism of anaerobic fungi. Weishengwu Xuebao 2017, 57, 1106–1111. [Google Scholar]
- Swift, C.L.; Brown, J.L.; Seppala, S.; O’Malley, M.A. Co-cultivation of the anaerobic fungus Anaeromyces robustus with Methanobacterium bryantii enhances transcription of carbohydrate active enzymes. J. Ind. Microbiol. Biotechnol. 2019, 46, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Bernalier, A.; Fonty, G.; Gouet, P. Cellulose degradation by 2 rumen anaerobic fungi in monoculture or in coculture with rumen bacteria. Anim. Feed Sci. Tech. 1991, 32, 131–136. [Google Scholar] [CrossRef]
- Leis, S.; Dresch, P.; Peintner, U.; Fliegerova, K.; Sandbichler, A.M.; Insam, H.; Podmirseg, S.M. Finding a robust strain for biomethanation: Anaerobic fungi (Neocallimastigomycota) from the Alpine ibex (Capra ibex) and their associated methanogens. Anaerobe 2014, 29, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Joblin, K.N.; Naylor, G.E.; Williams, A.G. Effect of methanobrevibacter-smithii on xylanolytic activity of anaerobic ruminal fungi. Appl. Environ. Microb. 1990, 56, 2287–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dollhofer, V.; Podmirseg, S.M.; Callaghan, T.M.; Griffith, G.W.; Fliegerova, K. Anaerobic Fungi and Their Potential for Biogas Production. In Biogas Science and Technology; Guebitz, G.M., Bauer, A., Bochmann, G., Gronauer, A., Weiss, S., Eds.; Springer: Cham, Switzerland, 2015; Volume 151, pp. 41–61. [Google Scholar]
- Boxma, B.; Voncken, F.; Jannink, S.; van Alen, T.; Akhmanova, A.; van Weelden, S.W.H.; van Croce, S.; Wei, Q.; D’Imporzano, G.; Dong, R.; et al. Anaerobic digestion of straw and corn stover: The effect of biological process optimization and pre-treatment on total bio-methane yield and energy performance. Biotechnol. Adv. 2016, 34, 1289–1304. [Google Scholar]
- Kumar, S.; Indugu, N.; Vecchiarelli, B.; Pitta, D.W. Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows. Front. Microbiol. 2015, 6, 781. [Google Scholar] [CrossRef] [Green Version]
- Flad, V.; Young, D.; Seppl, S.; Hooker, C.; Theodorou, M.K. The Biotechnological Potential of Anaerobic Gut Fungi. In Genetics and Biotechnology; Springer: Cham, Switzerland, 2020; pp. 413–437. [Google Scholar]
- Li, Y.; Hou, Z.; Shi, Q.; Cheng, Y.; Zhu, W. Methane Production From Different Parts of Corn Stover via a Simple Co-culture of an Anaerobic Fungus and Methanogen. Front. Bioeng. Biotechnol. 2020, 8, 8. [Google Scholar] [CrossRef]
- Drake, H.; Ivarsson, M.; Bengtson, S.; Heim, C. Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose. Biotechnol. Bioeng. 2018, 115, 874–884. [Google Scholar]
- Al-Addous, M.; Alnaief, M.; Class, C.; Nsair, A.; Kuchta, K.; Alkasrawi, M. Technical Possibilities of Biogas Production from Olive and Date Waste in Jordan. BioResources 2017, 12, 9383–9395. [Google Scholar]
- Almomani, F.; Shawaqfah, M.; Bhosale, R.R.; Kumar, A.; Khraisheh, M.A.M. Intermediate ozonation to enhance biogas production in batch and continuous systems using animal dung and agricultural waste. Int. Biodeterior. Biodegrad. 2017, 119, 176–187. [Google Scholar] [CrossRef]
- Almomani, F. Prediction of biogas production from chemically treated co-digested agricultural waste using artificial neural network. Fuel 2020, 280, 118573. [Google Scholar] [CrossRef]
- Joblin, K.N.; Matsui, H.; Naylor, G.E.; Ushida, K. Degradation of fresh ryegrass by methanogenic co-cultures of ruminal fungi grown in the presence or absence of Fibrobacter succinogenes. Curr. Microbiol. 2002, 45, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-Q.; Long, R.-J.; Yang, H.; Yang, H.-J.; Shen, X.-H.; Shi, R.-F.; Wang, Z.-Y.; Du, J.-G.; Qi, X.-J.; Ye, Q.-H. Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe 2016, 39, 158–164. [Google Scholar] [CrossRef]
| Genus | Morphology | Isolation Source | References |
|---|---|---|---|
| Neocallimastix | Monocentric, Polyflagellate, Filamentous | Sheep rumen contents | [28] |
| Caecomyces | Monocentric, Uniflagellate, Bulbous | Horse caecum | [29] |
| Orpinomyces | Polycentric, Polyflagellate, Filamentous | Holstein steer rumen | [30] |
| Piromyces | Monocentric, Uniflagellate, Filamentous | Holstein steer rumen | [30] |
| Anaeromyces | Polycentric, Uniflagellate, Filamentous | Cow rumen | [31] |
| Cyllamyces | Polycentric, Uniflagellate, Bulbous | Cow feces | [32] |
| Buwchfawromyces | Monocentric, Uniflagellate, Filamentous | Buffalo feces | [33] |
| Oontomyces | Monocentric, Uniflagellate, Filamentous | Indian camel stomach | [34] |
| Pecoramyces | Monocentric, Uniflagellate, Filamentous | Sheep feces | [35] |
| Feramyces | Monocentric, Polyflagellate, Filamentous | Barbary sheep | [36] |
| Liebetanzomyces | Monocentric, Uniflagellate, Filamentous | Goats rumen samples | [37] |
| Agriosomyces | Monocentric, Uniflagellate, Filamentous | Mouflon sheep feces | [38] |
| Aklioshbomyces | Monocentric, Uniflagellate, Filamentous | White-tailed deer feces | [38] |
| Capellomyces | Monocentric, Uniflagellate, Filamentous | Boer goat feces | [38] |
| Ghazallomyces | Monocentric, Polyflagellate, Filamentous | Axis deer feces | [38] |
| Joblinomyces | Monocentric, Uniflagellate, Filamentous | Goat feces | [38] |
| Khoyollomyces | Monocentric, Uniflagellate, Filamentous | Grevy’s zebra feces | [38] |
| Tahromyces | Monocentric, Uniflagellate, Filamentous | Nilgiri tahr feces | [38] |
| Order | Family | Genus | Number of Valid Published Species |
|---|---|---|---|
| Methanobacteriales | Methanobacteriaceae | Methanobacterium | 24 |
| Methanobrevibacter | 15 | ||
| Methanosphaera | 2 | ||
| Methanothermobacter | 8 | ||
| Methanothermaceae | Methanothermus | 2 | |
| Methanococcales | Methanocaldococcaceae | Methanocaldococcus | 7 |
| Methanotorris | 2 | ||
| Methanococcaceae | Methanococcus | 4 | |
| Methanothermococcus | 2 | ||
| Methanocellales | Methanocellaceae | Methanocella | 3 |
| Methanomicrobiales | Methanocalculaceae | Methanocalculus | 6 |
| Methanocorpusculaceae | Methanocorpusculum | 4 | |
| Methanomicrobiaceae | Methanoculleus | 11 | |
| Methanofollis | 5 | ||
| Methanogenium | 4 | ||
| Methanolacinia | 2 | ||
| Methanomicrobium | 1 | ||
| Methanoplanus | 2 | ||
| Methanoregulaceae | Methanolinea | 2 | |
| Methanoregula | 2 | ||
| Methanosphaerula | 1 | ||
| Methanospirillaceae | Methanospirillum | 4 | |
| Methanosarcinales | Methanosarcinaceae | Halomethanococcus | 1 |
| Methanimicrococcus | 1 | ||
| Methanococcoides | 4 | ||
| Methanohalobium | 1 | ||
| Methanohalophilus | 4 | ||
| Methanolobus | 7 | ||
| Methanomethylovorans | 3 | ||
| Methanosalum | 2 | ||
| Methanosarcina | 13 | ||
| Methanotrichaceae | Methanothrix | 2 | |
| Methermicoccaceae | Methermicoccus | 1 | |
| Methanopyrales | Methanopyraceae | Methanopyrus | 1 |
| Methanomassiliicoccales | Methanomassiliicoccaceae | Methanomassiliicoccus | 3 |
| Combinations of Anaerobic Fungi and Methanogens | Substrate | Incubation Time | Conversion Rate of Methane | Reference |
|---|---|---|---|---|
| Neocallimastix frontalis PN1 + Methanobrevibacter sp. strain RA1 | Whatman no. 1 filter paper | 3 days | 1.78 mmol methane/g substrate | [29] |
| Whatman no. 1 filter paper | 7 days | 3.35 mmol methane/g substrate | [29] | |
| Sisal twin fiber | 7 days | 2.1 mmol methane/g substrate | [83] | |
| Barley straw leaf | 7 days | 1.7 mmol methane/g substrate | [83] | |
| Neocallimastix frontalis ATCC 76100 + Methanobacterium formicicum DSM 3637 | Cellulose | 7 days | 5.7 mmol methane/g substrate | [12] |
| Neocallimastix frontalis ATCC 76100 + Methanosaeta concilii DSM 6752 | Cellulose | 17 days | 4.3 mmol methane/g substrate | [12] |
| Neocallimastix frontalis PNK2 + Methanobrevibacter smithii PS | Fresh ryegrass stem | 6 days | 8.75 mL methane/g substrate | [99] |
| Fresh ryegrass leaf | 6 days | 8 mL methane/g substrate | [99] | |
| Piromyces + Methanobrevibacter thaueri CW | Rice straw | 4 days | 1.05 mmol methane/g DM | [8] |
| Wheat straw | 4 days | 1.16 mmol methane/g DM | [8] | |
| Maize stem | 4 days | 0.71 mmol methane/g DM | [8] | |
| Corncob | 4 days | 2.17 mmol methane/g DM | [8] | |
| Wheat bran | 4 days | 1.06 mmol methane/g DM | [8] | |
| Piromyces + Methanobrevibacter sp. Z8 | Rice straw | 4 days | 1.06 mmol methane/g DM | [8] |
| Wheat straw | 4 days | 0.96 mmol methane/g DM | [8] | |
| Maize stem | 4 days | 0.58 mmol methane/g DM | [8] | |
| Corncob | 4 days | 1.90 mmol methane/g DM | [8] | |
| Wheat bran | 4 days | 1.23 mmol methane/g DM | [8] | |
| Orpinomyces sp. + Methanobrevibacter sp. | Corn core | 4 days | 1.61 mmol methane/g substrate | [85] |
| Neocallimastix sp + Methanobrevibacter sp. | Corn core | 4 days | 1.96 mmol methane/g substrate | [85] |
| Pecoramyces sp. + Methanobrevibacter sp. | Corn stover leaf blade | 3 days | 56.6 mL/g digested substrate | [94] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Meng, Z.; Xu, Y.; Shi, Q.; Ma, Y.; Aung, M.; Cheng, Y.; Zhu, W. Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials. Microorganisms 2021, 9, 190. https://doi.org/10.3390/microorganisms9010190
Li Y, Meng Z, Xu Y, Shi Q, Ma Y, Aung M, Cheng Y, Zhu W. Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials. Microorganisms. 2021; 9(1):190. https://doi.org/10.3390/microorganisms9010190
Chicago/Turabian StyleLi, Yuqi, Zhenxiang Meng, Yao Xu, Qicheng Shi, Yuping Ma, Min Aung, Yanfen Cheng, and Weiyun Zhu. 2021. "Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials" Microorganisms 9, no. 1: 190. https://doi.org/10.3390/microorganisms9010190





