Danofloxacin Treatment Alters the Diversity and Resistome Profile of Gut Microbiota in Calves
Abstract
:1. Introduction
2. Results
2.1. Results of 16S rRNA Gene Analysis
2.1.1. Bacterial Phyla by Sampling Days
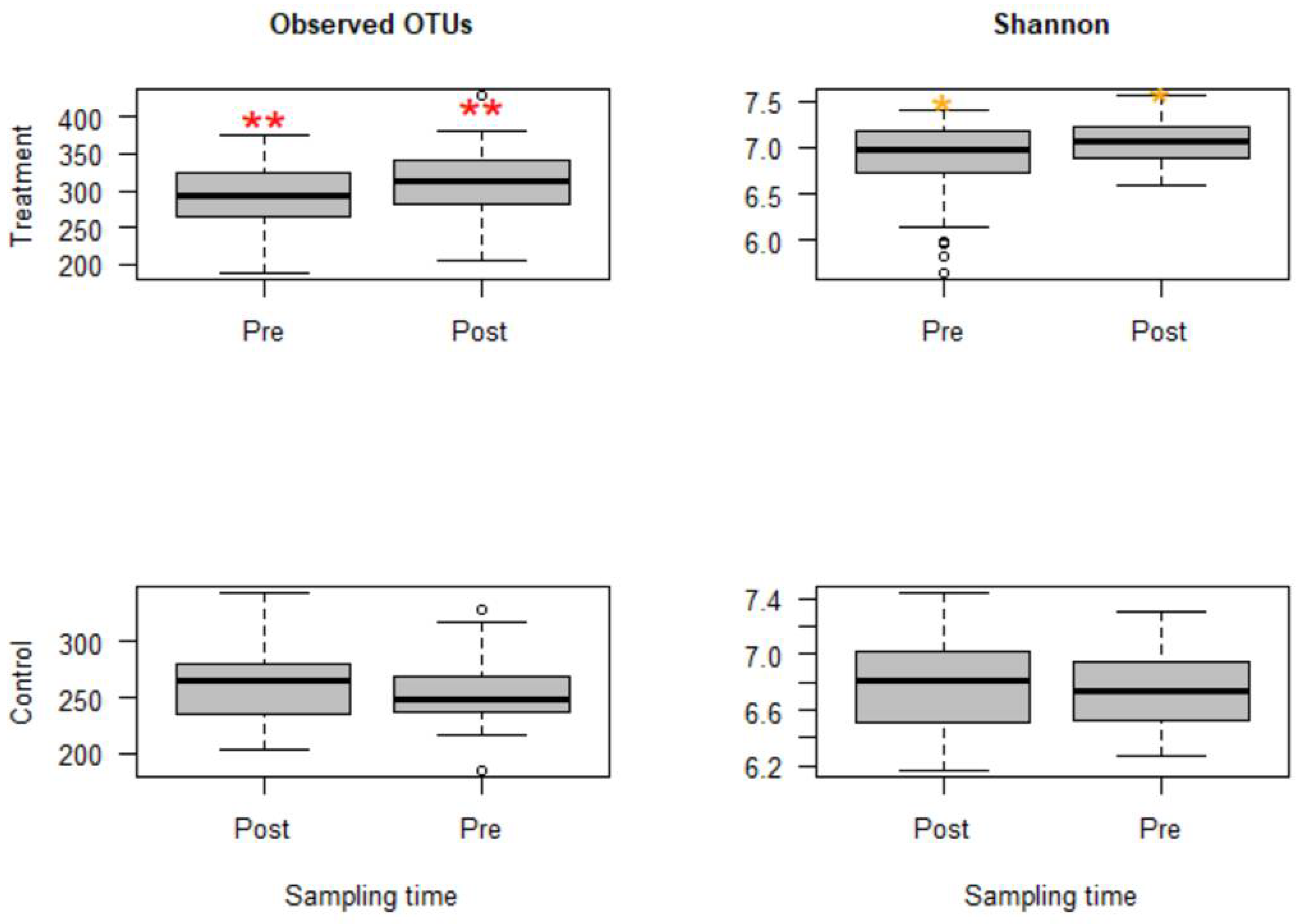
2.1.2. Alpha Diversities
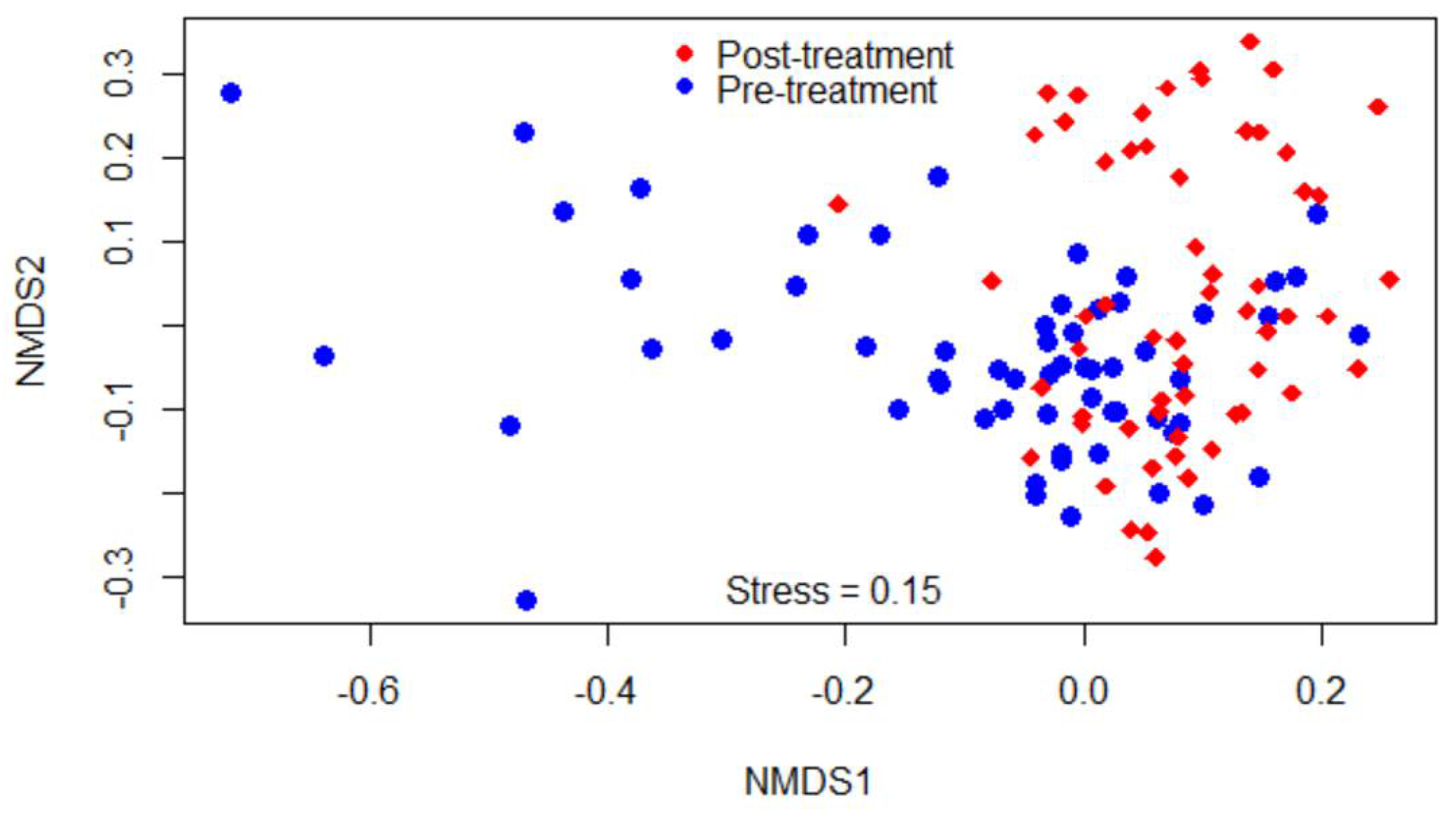
2.1.3. Beta Diversities
2.1.4. Comparisons of Compositions of Bacterial Classes among Study Calf Groups
2.1.5. Comparisons of Relative Abundance of Bacterial Genera between Pre- and Post-Treatment Communities
2.2. The Compositions of Campylobacter
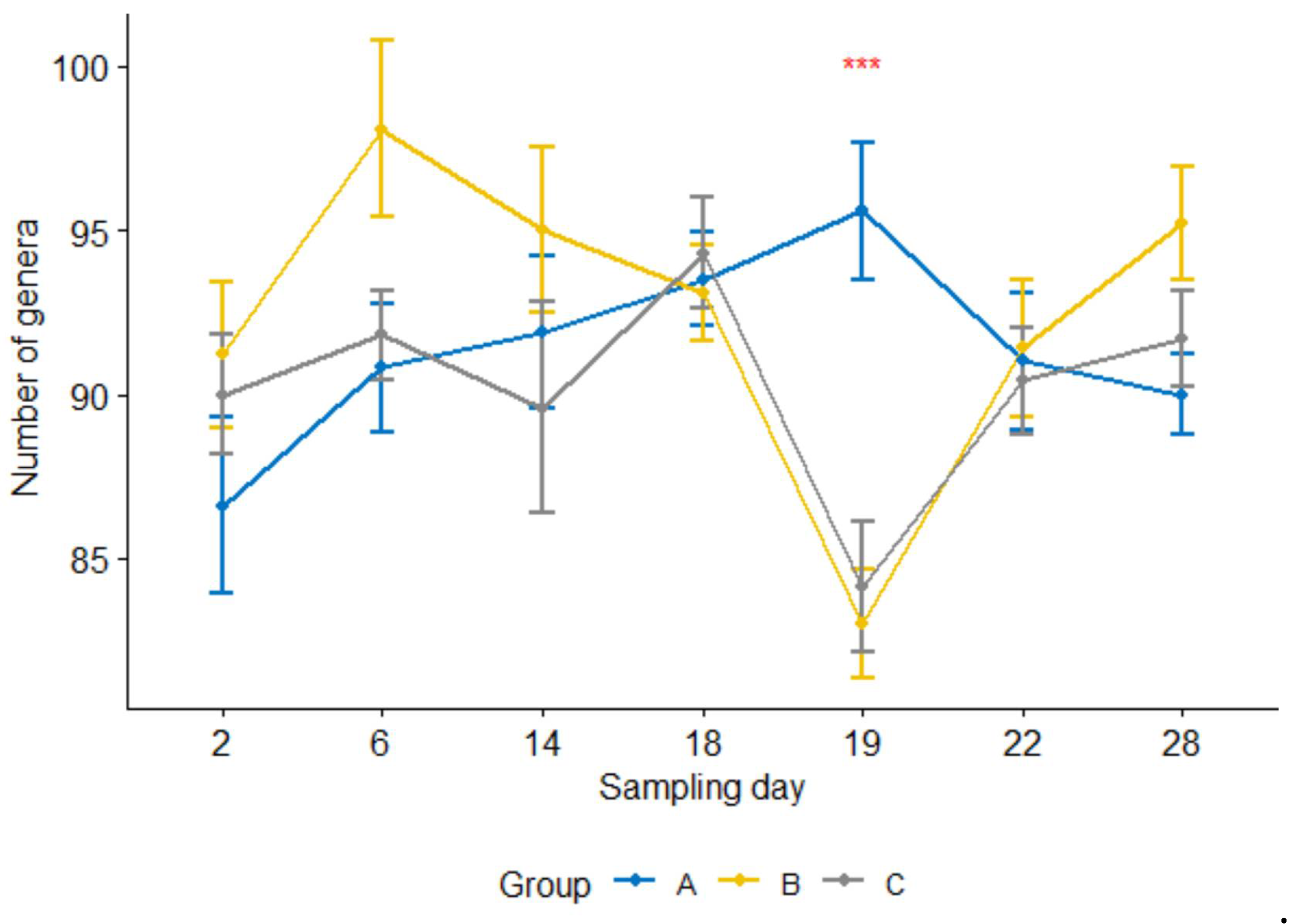
2.2.1. Comparison by Sampling Days
2.2.2. Correlation with Other Genera
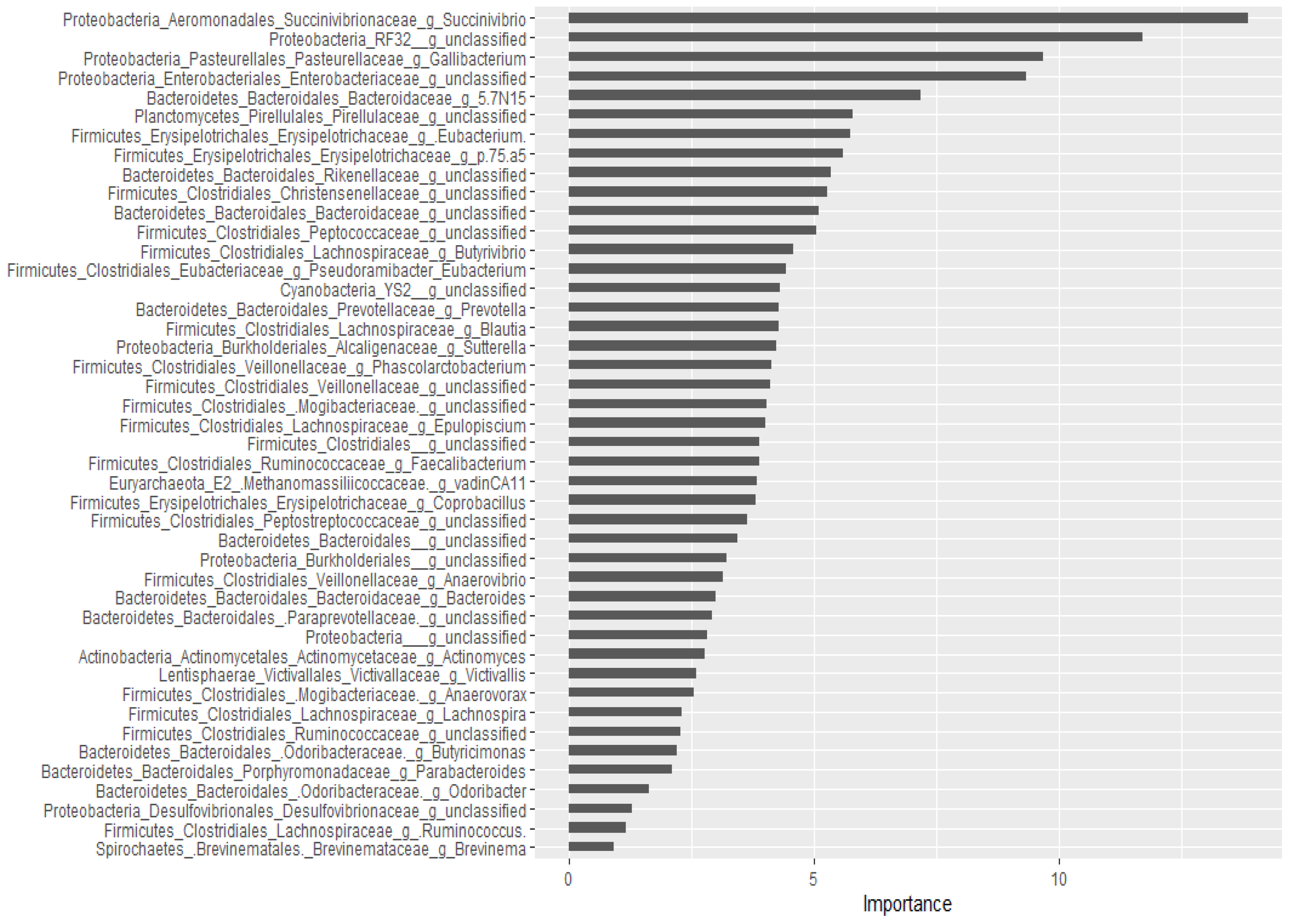
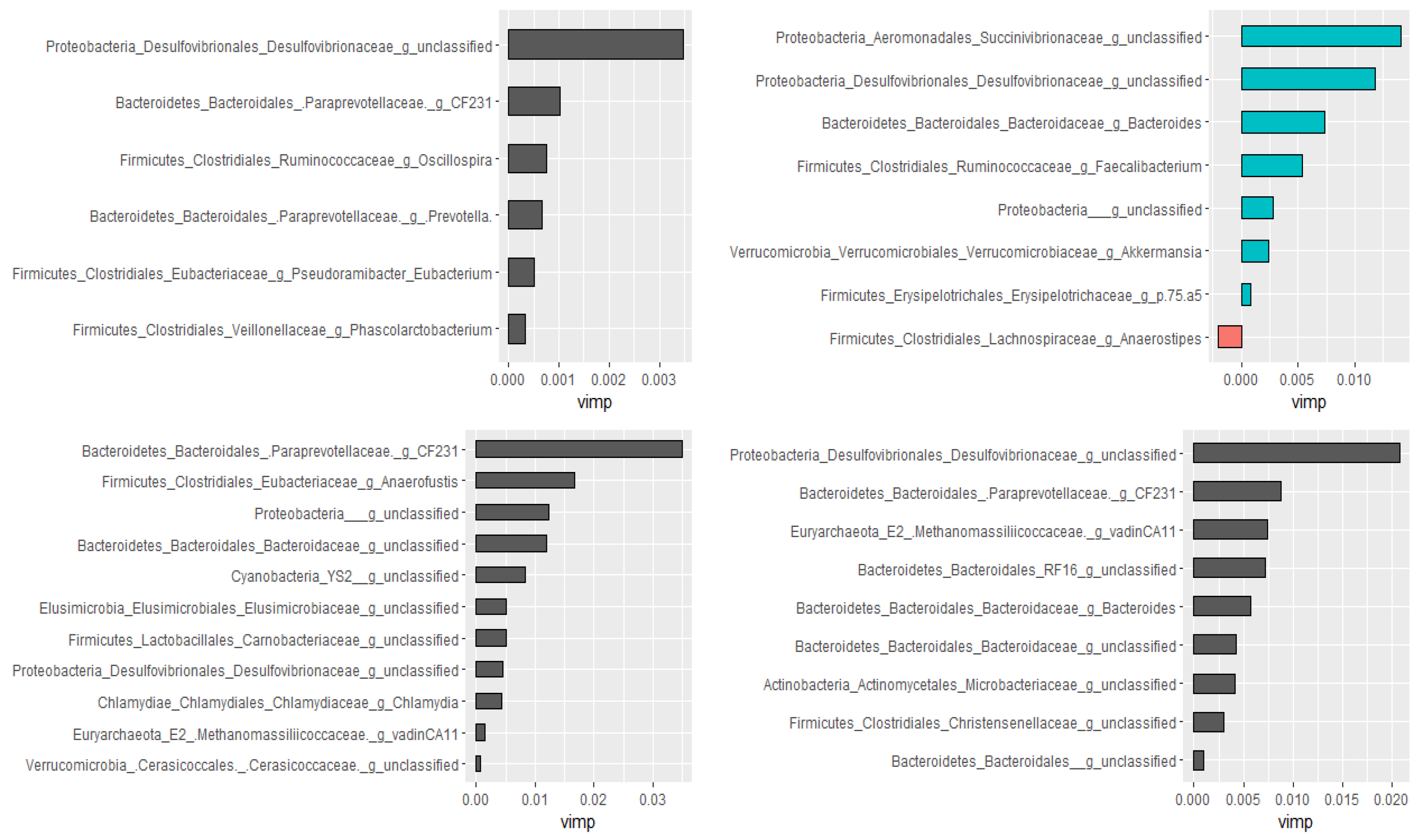
2.2.3. Prediction of Important Genera by Random Forests
2.3. Results of Metagenomic Hi-C (ProxiMeta)
2.4. Results of qPCR
3. Discussion
4. Materials and Methods
4.1. Study Design and Animals
4.2. DNA Extraction, Library Preparation, and Sequencing
- 16S rRNA: To determine the effects of danofloxacin on gut microbiota, 16S rRNA analysis was conducted. DNA extractions were performed following ZymoBIOMICS™ protocol from 210 fecal samples (10 calves per group and seven sampling time points). The V4-V5 hypervariable regions of the bacterial 16S rRNA gene were amplified using a universal 16S forward primer (515F: GTGYCAGCMGCCGCGGTAA) and a reverse primer (926R: CCGYCAATTYMTTTRAGTTT). Briefly, the fecal samples were thawed at room temperature for approximately 30 min. From each sample, 200 mg of feces was transferred to a 2 mL ZR BashingBead™ lysis tube and mixed with 250 µL deionized sterile water, 750 µL lysis solution, and 50 µL proteinase K. The samples were processed by a bead beater for 10 min followed by incubation for at least 30 min in a water bath at 55 °C. Then, the lysis tubes were centrifuged in a microcentrifuge at 10,000 × g for 3 min. The supernatant was harvested to columns and then washed with DNA Wash Buffer 1 and 2. The final product was eluted with DNase/RNase free water, and the concentration of eluted DNA was measured first by NanoDrop 3300 Fluorospectrophotometer (Nanodrop technologies, USA) and confirmed by Qubit fluorometer (Invitrogen). Following normalization of all the DNA extracts, they were transferred to 96-wells plates, and submitted for sequencing to the DNA Facility of Iowa State University. The Earth Microbiome Project protocol was followed for sequencing on the Illumina MiSeq platform (2 × 250 paired-ends) in a single flow cell lane. For control, two community standards were used.
- Shotgun sequencing: To assess the effects of danofloxacin on gut microbial resistome, shotgun and ProxiMeta Hi-C metagenomics were performed. For shotgun library preparation, four samples (pooled two pre-treatment samples and pooled two post-treatment samples from calves in Group B) were selected based on the results of 16S analysis. Samples collected right before danofloxacin injection (i.e., Day 18) and four days later (i.e., Day 22) were used for this purpose. Like in the case of 16S rRNA gene, whole-genome DNA was extracted from the fecal samples according to ZymoBIOMICS™ instructions. The whole-genome extracts were submitted to the DNA Facility of Iowa State University, where a single flow cell lane Illumina HiSeq platform (2 × 150 bp) was used for sequencing.
- Metagenomic Hi-C ProxiMeta DNA extraction and library preparation: The same samples used for the whole-genome shotgun DNA extraction were used for the Metagenomic ProxiMeta sequencing. The Hi-C library was created using a Phase Genomics (Seattle, WA, USA) ProxiMeta Hi-C Microbiome Kit, which is a commercially available version of the Hi-C protocol. Briefly, 100 mg of the fecal sample was washed with TBS, and then genetic material (both chromosomal and non-chromosomal) were crosslinked in vivo while the bacterial cells were still intact using a formaldehyde solution, simultaneously digested using restriction enzymes Sau3AI and MlucI. The genetic materials were proximity ligated with biotinylated nucleotides to create chimeric molecules composed of fragments from different regions of genomes that were physically proximal in vivo according to the manufacturer’s instructions for the kit. The chance of inter-cellular interactions of genetic materials was negligible. Molecules were pulled down with streptavidin beads and processed into an Illumina-compatible sequencing library according to the protocol. Sequencing was performed on an Illumina HiSeq instrument (2 × 150 bp). The bioinformatic analyses were described in our recent publication [78].
- Quantitative real-time PCR (qPCR): To assess the dynamics of antimicrobial resistance genes following danofloxacin injection in the fecal samples, we ran qPCR using primers previously designed by Looft et al. [79] presented in Table 5. The target resistance genes were selected based on the metagenomic Hi-C results; accordingly, primers were ordered from the ISU DNA facility for tetW, tetO, tetX, ermB, and ermF. The DNA extracts (described above) from fecal samples collected on Day 18 for the pre-treatment and Day 28 for the post-treatment, were pooled together for each group separately. PCR assays were run using the SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA) and the CFX Maestro™ Real-Time PCR detection system (Bio-Rad). Dilutions of DNA template for both standards (16S and target genes) and all unknowns were run in triplicate with reaction volumes of 10 μL. Amplification of DNA occurred with 35 cycles of denaturation at 95 °C for 10 s and then annealing for each primer pair at 60 °C for 30 s. Both standard curves (16S and target genes) were experimentally validated to have high efficiency > 90% of amplification and precision R2 ~ 0.98 prior the analysis. Relative expression was normalized using 16S detection levels. The relative fold change of detection between the control and treatment groups (B and C) were calculated using the ISU Gallup Method Equation [80]. Statistical analysis was performed using Dunn pair-wise comparison test in Rstudio to determine significance changes in ARG levels between the pre-and post-treatment pooled samples. An adjusted p-value of < 0.05 was considered significant.
4.3. Bioinformatics and Data Analysis of 16S Data
Prediction Models Using Random Forest Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse Antibiotic Resistance Genes in Dairy Cow Manure. Mbio 2014, 5, e01017-13. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Call, D.R.; Davis, M.A.; Sawant, A.A. Antimicrobial resistance in beef and dairy cattle production. Anim. Health Res. Rev. 2008, 9, 159–167. [Google Scholar] [CrossRef]
- Catry, B.; Dewulf, J.; Maes, D.; Pardon, B.; Callens, B.; Vanrobaeys, M.; Opsomer, G.; de Kruif, A.; Haesebrouck, F. Effect of Antimicrobial Consumption and Production Type on Antibacterial Resistance in the Bovine Respiratory and Digestive Tract. PLoS ONE 2016, 11, e0146488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocejo, M.; Oporto, B.; Hurtado, A. 16S rRNA amplicon sequencing characterization of caecal microbiome composition of broilers and free-range slow-growing chickens throughout their productive lifespan. Sci. Rep. 2019, 9, 2506. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. Embo Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [Green Version]
- Udikovic-Kolic, N.; Wichmann, F.; Broderick, N.A.; Handelsman, J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. USA 2014, 111, 15202–15207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennani, H.; Mateus, A.; Mays, N.; Eastmure, E.; Staerk, K.D.C.; Hasler, B. Overview of Evidence of Antimicrobial Use and Antimicrobial Resistance in the Food Chain. Antibiotics 2020, 9, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, K.K.; Pendell, D.L. Market Impacts of Reducing the Prevalence of Bovine Respiratory Disease in United States Beef Cattle Feedlots. Front. Vet. Sci. 2017, 4, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.B.; Renter, D.G.; Cernicchiaro, N.; Shi, X.R.; Nickell, J.S.; Keil, D.J.; Nagaraja, T.G. A Randomized Trial to Assess the Effect of Fluoroquinolone Metaphylaxis on the Fecal Prevalence and Quinolone Susceptibilities of Salmonella and Campylobacter in Feedlot Cattle. Foodborne Pathog. Dis. 2017, 14, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Hilton, W.M. BRD in 2014: Where have we been, where are we now, and where do we want to go? Anim. Health Res. Rev. 2014, 15, 120–122. [Google Scholar] [CrossRef] [PubMed]
- USDA. Feedlot 2011 Part I: Management Practices on U.S. Feedlots with a Capacity of 1,000 or More Head; USDA-APHIS-VS-CEAH-NAHMS, Fort Collins, CO.; USDA: Washington, DC, USA, 2013.
- Guterbock, W.M. The impact of BRD: The current dairy experience. Anim. Health Res. Rev. 2014, 15, 130–134. [Google Scholar] [CrossRef]
- USDA. Dairy 2007: Part V: Changes in Dairy Cattle Health and Management Practices in the United States, 1996–2007; USDA-APHIS National Animal Health Monitoring System. Info Sheet; USDA: Washington, DC, USA, 2009.
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can. Vet. J. -Rev. Vet. Can. 2010, 51, 1095–1102. [Google Scholar]
- FDA. 2018 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; FDA: Rockville, MD, USA, 2019.
- Hope, K.J.; Apley, M.D.; Schrag, N.F.D.; Lubbers, B.V.; Singer, R.S. Antimicrobial use in 22 US beef feedyards: 2016–2017. Zoonoses Public Health 2020, 67, 94–110. [Google Scholar] [CrossRef]
- Juntunen, P.; Olkkola, S.; Hanninen, M.-L. Longitudinal on-farm study of the development of antimicrobial resistance in Campylobacter coli from pigs before and after danofloxacin and tylosin treatments. Vet. Microbiol. 2011, 150, 322–330. [Google Scholar] [CrossRef]
- Dryden, M.S.; Gabb, R.J.E.; Wright, S.K. Empirical Treatment of Severe Acute Community-Acquired Gastroenteritis with Ciproftoxacin. Clin. Infect. Dis. 1996, 22, 1019–1025. [Google Scholar] [CrossRef] [Green Version]
- Nelson, J.M.; Chiller, T.M.; Powers, J.H.; Angulo, F.J. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: A public health success story. Clin. Infect. Dis. 2007, 44, 977–980. [Google Scholar] [CrossRef] [Green Version]
- Foditsch, C.; Pereira, R.V.V.; Siler, J.D.; Altier, C.; Warnick, L.D. Effects of treatment with enrofloxacin or tulathromycin on fecal microbiota composition and genetic function of dairy calves. PLoS ONE 2019, 14, e0219635. [Google Scholar] [CrossRef] [Green Version]
- Wieczorek, K.; Osek, J. Antimicrobial resistance mechanisms among Campylobacter. BioMed Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyselkova, M.; Kotrbova, L.; Bhumibhamon, G.; Chronakova, A.; Jirout, J.; Vrchotovaa, N.; Schmitt, H.; Elhottovaa, D. Tetracycline resistance genes persist in soil amended with cattle feces independently from chlortetracycline selection pressure. Soil Biol. Biochem. 2015, 81, 259–265. [Google Scholar] [CrossRef]
- Yin, J.B.; Zhang, X.X.; Wu, B.; Xian, Q.M. Metagenomic insights into tetracycline effects on microbial community and antibiotic resistance of mouse gut. Ecotoxicology 2015, 24, 2125–2132. [Google Scholar] [CrossRef]
- Cheng, P.; Yang, Y.; Li, F.; Li, X.; Liu, H.; Fazilani, S.A.; Guo, W.; Xu, G.; Zhang, X. The prevalence and mechanism of fluoroquinolone resistance in Escherichia coli isolated from swine farms in China. BMC Vet. Res. 2020, 16, 258. [Google Scholar] [CrossRef]
- Li, J.; Hao, H.; Cheng, G.; Liu, C.; Ahmed, S.; Shabbir, M.A.B.; Hussain, H.I.; Dai, M.; Yuan, Z. Microbial Shifts in the Intestinal Microbiota of Salmonella Infected Chickens in Response to Enrofloxacin. Front. Microbiol. 2017, 8, 1711. [Google Scholar] [CrossRef] [Green Version]
- Francino, M.P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 2016, 7, 01543. [Google Scholar] [CrossRef] [Green Version]
- Panda, S.; El Khader, I.; Casellas, F.; Vivancos, J.L.; Cors, M.G.; Santiago, A.; Cuenca, S.; Guarner, F.; Manichanh, C. Short-Term Effect of Antibiotics on Human Gut Microbiota. PLoS ONE 2014, 9, e95476. [Google Scholar] [CrossRef]
- Schokker, D.; Jansman, A.J.M.; Veninga, G.; de Bruin, N.; Vastenhouw, S.A.; de Bree, F.M.; Bossers, A.; Rebel, J.M.J.; Smits, M.A. Perturbation of microbiota in one-day old broiler chickens with antibiotic for 24 hours negatively affects intestinal immune development. BMC Genom. 2017, 18, 241. [Google Scholar] [CrossRef] [Green Version]
- Schokker, D.; Zhang, J.; Zhang, L.-l.; Vastenhouw, S.A.; Heilig, H.G.H.J.; Smidt, H.; Rebel, J.M.J.; Smits, M.A. Early-Life Environmental Variation Affects Intestinal Microbiota and Immune Development in New-Born Piglets. PLoS ONE 2014, 9, e100040. [Google Scholar] [CrossRef]
- Zaura, E.; Brandt, B.W.; de Mattos, M.J.T.; Buijs, M.J.; Caspers, M.P.M.; Rashid, M.-U.; Weintraub, A.; Nord, C.E.; Savell, A.; Hu, Y.; et al. Same Exposure but Two Radically Different Responses to Antibiotics: Resilience of the Salivary Microbiome versus Long-Term Microbial Shifts in Feces. Mbio 2015, 6, e01693-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronvold, A.M.R.; Mao, Y.J.; L’Abee-Lund, T.M.; Sorum, H.; Sivertsen, T.; Yannarell, A.C.; Mackie, R.I. Fecal microbiota of calves in the clinical setting: Effect of penicillin treatment. Vet. Microbiol. 2011, 153, 354–360. [Google Scholar] [CrossRef]
- Ferguson, K.M.; Jacob, M.E.; Theriot, C.M.; Callahan, B.J.; Prange, T.; Papich, M.G.; Foster, D.M. Dosing Regimen of Enrofloxacin Impacts Intestinal Pharmacokinetics and the Fecal Microbiota in Steers. Front. Microbiol. 2018, 9, 2190. [Google Scholar] [CrossRef]
- Weese, J.S.; Jelinski, M. Assessment of the Fecal Microbiota in Beef Calves. J. Vet. Intern. Med. 2017, 31, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Durso, L.M.; Harhay, G.P.; Smith, T.P.L.; Bono, J.L.; DeSantis, T.Z.; Harhay, D.M.; Andersen, G.L.; Keen, J.E.; Laegreid, W.W.; Clawson, M.L. Animal-to-Animal Variation in Fecal Microbial Diversity among Beef Cattle. Appl. Environ. Microbiol. 2010, 76, 4858–4862. [Google Scholar] [CrossRef] [Green Version]
- Stalder, T.; Press, M.O.; Sullivan, S.; Liachko, I.; Top, E.M. Linking the resistome and plasmidome to the microbiome. ISME J. 2019, 13, 2437–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mzyk, D.A.; Bublitz, C.M.; Martinez, M.N.; Davis, J.L.; Baynes, R.E.; Smith, G.W. Impact of bovine respiratory disease on the pharmacokinetics of danofloxacin and tulathromycin in different ages of calves. PLoS ONE 2019, 14, e0218864. [Google Scholar] [CrossRef] [PubMed]
- Mestorino, N.; Marchetti, M.L.; Turic, E.; Pesoa, J.; Errecalde, J. Concentrations of danofloxacin 18% solution in plasma, milk and tissues after subcutaneous injection in dairy cows. Anal. Chim. Acta 2009, 637, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Ferran, A.A.; Bibbal, D.; Pellet, T.; Laurentie, M.; Gicquel-Bruneau, M.; Sanders, P.; Schneider, M.; Toutain, P.L.; Bousquet-Melou, A. Pharmacokinetic/pharmacodynamic assessment of the effects of parenteral administration of a fluoroquinolone on the intestinal microbiota: Comparison of bactericidal activity at the gut versus the systemic level in a pig model. Int. J. Antimicrob. Agents 2013, 42, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Khoder, M.; Tsapis, N.; Domergue-Dupont, V.; Gueutin, C.; Fattal, E. Removal of residual colonic ciprofloxacin in the rat by activated charcoal entrapped within zinc-pectinate beads. Eur. J. Pharm. Sci. 2010, 41, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Edlund, C.; Lindqvist, L.; Nord, C.E. Binding of norfloxacin to human fecal material. Rev. Infect. Dis. 1989, 11, S1237–S1238. [Google Scholar]
- Wiuff, C.; Lykkesfeldt, J.; Svendsen, O.; Aarestrup, F.M. The effects of oral and intramuscular administration and dose escalation of enrofloxacin on the selection of quinolone resistance among Salmonella and coliforms in pigs. Res. Vet. Sci. 2003, 75, 185–193. [Google Scholar] [CrossRef]
- Zaheer, R.; Lakin, S.M.; Polo, R.O.; Cook, S.R.; Larney, F.J.; Morley, P.S.; Booker, C.W.; Hannon, S.J.; Van Domselaar, G.; Read, R.R.; et al. Comparative diversity of microbiomes and Resistomes in beef feedlots, downstream environments and urban sewage influent. BMC Microbiol. 2019, 19, 197. [Google Scholar] [CrossRef] [Green Version]
- Durso, L.; Wells, J.; Kim, M.S. Diversity of Microbiomes in Beef Cattle. In Encyclopedia of Metagenomics; Nelson, K., Ed.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Valerio de Oliveira, M.N.; Jewell, K.A.; Freitas, F.S.; Benjamin, L.A.; Totola, M.R.; Borges, A.C.; Moraes, C.A.; Suen, G. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet. Microbiol. 2013, 164, 307–314. [Google Scholar] [CrossRef]
- Kim, M.; Kim, J.; Kuehn, L.A.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J.E. Investigation of bacterial diversity in the feces of cattle fed different diets. J. Anim. Sci. 2014, 92, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Rudi, K.; Moen, B.; Sekelja, M.; Frisli, T.; Lee, M.R.F. An eight-year investigation of bovine livestock fecal microbiota. Vet. Microbiol. 2012, 160, 369–377. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhu, Y.; Qiu, X.; Gao, C.; Wang, J.; Wang, H.; He, Y.; Ur Rahman, M.A.; Cao, B.; Su, H. Dynamic Variations in Fecal Bacterial Community and Fermentation Profile of Holstein Steers in Response to Three Stepwise Density Diets. Animals 2019, 9, 560. [Google Scholar] [CrossRef] [Green Version]
- McKellar, Q.; Gibson, I.; Monteiro, A.; Bregante, M. Pharmacokinetics of enrofloxacin and danofloxacin in plasma, inflammatory exudate, and bronchial secretions of calves following subcutaneous administration. Antimicrob. Agents Chemother. 1999, 43, 1988–1992. [Google Scholar] [CrossRef] [Green Version]
- Brooks, P.T.; Mansfield, L.S. Effects of antibiotic resistance (AR) and microbiota shifts on Campylobacter jejuni-mediated diseases. Anim. Health Res. Rev. 2017, 18, 99–111. [Google Scholar] [CrossRef]
- O’Loughlin, J.L.; Samuelson, D.R.; Braundmeier-Fleming, A.G.; White, B.A.; Haldorson, G.J.; Stone, J.B.; Lessmann, J.J.; Eucker, T.P.; Konkel, M.E. The Intestinal Microbiota Influences Campylobacter jejuni Colonization and Extraintestinal Dissemination in Mice. Appl. Environ. Microbiol. 2015, 81, 4642–4650. [Google Scholar] [CrossRef] [Green Version]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Soldan, M.M.P.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.F.; et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef]
- Sakaridis, I.; Ellis, R.J.; Cawthraw, S.A.; van Vliet, A.H.M.; Stekel, D.J.; Penell, J.; Chambers, M.; La Ragione, R.M.; Cook, A.J. Investigating the Association Between the Caecal Microbiomes of Broilers and Campylobacter Burden. Front. Microbiol. 2018, 9, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampmann, C.; Dicksved, J.; Engstrand, L.; Rautelin, H. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin. Microbiol. Infect. 2016, 22, 61.e1–61.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dicksved, J.; Ellstrom, P.; Engstrand, L.; Rautelin, H. Susceptibility to Campylobacter Infection Is Associated with the Species Composition of the Human Fecal Microbiota. Mbio 2014, 5, e01212-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Kuehn, L.A.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J.E. The impact of the bovine faecal microbiome on Escherichia coli O157:H7 prevalence and enumeration in naturally infected cattle. J. Appl. Microbiol. 2017, 123, 1027–1042. [Google Scholar] [CrossRef]
- Garber, J.M.; Nothaft, H.; Pluvinage, B.; Stahl, M.; Bian, X.; Porfirio, S.; Enriquez, A.; Butcher, J.; Huang, H.; Glushka, J.; et al. The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus. Commun. Biol. 2020, 3, 1–11. [Google Scholar] [CrossRef]
- Morishita, T.Y.; Aye, P.P.; Harr, B.S.; Cobb, C.W.; Clifford, J.R. Evaluation of an avian-specific probiotic to reduce the colonization and shedding of Campylobacter jejuni in broilers. Avian Dis. 1997, 41, 850–855. [Google Scholar] [CrossRef]
- Ghareeb, K.; Awad, W.A.; Mohnl, M.; Porta, R.; Biarnes, M.; Bohm, J.; Schatzmayr, G. Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poult. Sci. 2012, 91, 1825–1832. [Google Scholar] [CrossRef]
- Santini, C.; Baffoni, L.; Gaggia, F.; Granata, M.; Gasbarri, R.; Di Gioia, D.; Biavati, B. Characterization of probiotic strains: An application as feed additives in poultry against Campylobacter jejuni. Int. J. Food Microbiol. 2010, 141, S98–S108. [Google Scholar] [CrossRef]
- Brooks, P.T.; Bell, J.A.; Bejcek, C.E.; Malik, A.; Mansfield, L.S. An antibiotic depleted microbiome drives severe Campylobacter jejuni-mediated Type 1/17 colitis, Type 2 autoimmunity and neurologic sequelae in a mouse model. J. Neuroimmunol. 2019, 337, 577048. [Google Scholar] [CrossRef]
- FDA. 2019 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; FDA: Rockville, MD, USA, 2020.
- Rovira, P.; McAllister, T.; Lakin, S.M.; Cook, S.R.; Doster, E.; Noyes, N.R.; Weinroth, M.D.; Yang, X.; Parker, J.K.; Boucher, C.; et al. Characterization of the Microbial Resistome in Conventional and “Raised Without Antibiotics” Beef and Dairy Production Systems. Front. Microbiol. 2019, 10, 1980. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005, 41, S120–S126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holman, D.B.; Yang, W.; Alexander, T.W. Antibiotic treatment in feedlot cattle: A longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 2019, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Leclercq, S.O.; Wang, C.; Sui, Z.; Wu, H.; Zhu, B.; Deng, Y.; Feng, J. A multiplayer game: Species of Clostridium, Acinetobacter, and Pseudomonas are responsible for the persistence of antibiotic resistance genes in manure-treated soils. Environ. Microbiol. 2016, 18, 3494–3508. [Google Scholar] [CrossRef] [PubMed]
- Slizovskiy, I.B.; Mukherjee, K.; Dean, C.J.; Boucher, C.; Noyes, N.R. Mobilization of Antibiotic Resistance: Are Current Approaches for Colocalizing Resistomes and Mobilomes Useful? Front. Microbiol. 2020, 11, 1376. [Google Scholar] [CrossRef] [PubMed]
- Brenciani, A.; Bacciaglia, A.; Vecchi, M.; Vitali, L.A.; Varaldo, P.E.; Giovanettil, E. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 2007, 51, 1209–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, T.A.; Stedtfeld, R.D.; Wang, Q.; Cole, J.R.; Hashsham, S.A.; Looft, T.; Zhu, Y.-G.; Tiedje, J.M. Clusters of Antibiotic Resistance Genes Enriched Together Stay Together in Swine Agriculture. Mbio 2016, 7, e02214-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.W.; Shin, M.K.; Jung, M.; Belaynehe, K.M.; Yoo, H.S. Prevalence of Antimicrobial Resistance and Transfer of Tetracycline Resistance Genes in Escherichia coli Isolates from Beef Cattle. Appl. Environ. Microbiol. 2015, 81, 5560–5566. [Google Scholar] [CrossRef] [Green Version]
- Dewanckele, L.; Vlaeminck, B.; Fievez, V. Sharpea azabuensis: A ruminal bacterium that produces trans-11 intermediates from linoleic and linolenic acid. Microbiology 2019, 165, 772–778. [Google Scholar] [CrossRef]
- Whittle, G.; Shoemaker, N.B.; Salyers, A.A. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 2002, 59, 2044–2054. [Google Scholar] [CrossRef]
- Quesada-Gomez, C. Bacteroides mobilizable and conjugative genetic elements: Antibiotic resistance among clinical isolates. Rev. Esp. De Quimioter. 2011, 24, 184–190. [Google Scholar]
- Hanthorn, C.J.; Dewell, R.D.; Cooper, V.L.; Frana, T.S.; Plummer, P.J.; Wang, C.; Dewell, G.A. Randomized clinical trial to evaluate the pathogenicity of Bibersteinia trehalosi in respiratory disease among calves. BMC Vet. Res. 2014, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AVMA. AVMA Guidelines for the Euthanasia of Animals: Version 2020.0.1, 2020 Edition; AVMA: Schaumburg, IL, USA, 2020. [Google Scholar]
- Beyi, A.F.; Hassall, A.; Phillips, G.J.; Plummer, P.J. Tracking Reservoirs of Antimicrobial Resistance Genes in a Complex Microbial Community Using Metagenomic Hi-C: The Case of Bovine Digital Dermatitis. Antibiotics 2021, 10, 221. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallup, J.M.; Ackermann, M.R. Addressing fluorogenic real-time qPCR inhibition using the novel custom Excel file system ‘FocusField2-6GallupqPCRSet-upTool-001’ to attain consistently high fidelity qPCR reactions. Biol. Proced. Online 2006, 8, 87–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Genus | Relative Abundance (%) | p | Genus | Relative Abundance (%) | p | ||
|---|---|---|---|---|---|---|---|
| Pre | Post | Pre | Post | ||||
| Succinivibrio | 0.38 | 0.05 | 0.000 | Erysipelotrichales_RFN20 | 0.04 | 0.03 | 0.000 |
| Streptophyta_5-7N15 | 4.12 | 8.31 | 0.000 | Desulfovibrio | 0.51 | 0.27 | 0.000 |
| Enterobacteriaceae_unclassified | 0.08 | 0.01 | 0.000 | Elusimicrobiaceae_unclassified | 0.12 | 0.04 | 0.000 |
| Peptococcaceae_unclassified | 0.13 | 0.25 | 0.000 | Bacteroidaceae_unclassified | 1.53 | 2.54 | 0.000 |
| Alphaproteobacteria_unclassified | 0.38 | 0.14 | 0.000 | Synergistes | 0.03 | 0.01 | 0.000 |
| Eubacterium | 0.13 | 0.03 | 0.000 | Methanosphaera | 0.35 | 0.22 | 0.001 |
| Coprobacillus | 0.01 | 0.00 | 0.000 | Sutterella | 0.61 | 0.24 | 0.001 |
| Erysipelotrichales_p-75-a5 | 0.10 | 0.00 | 0.000 | Pseudoramibacter | 0.05 | 0.02 | 0.001 |
| Prevotella | 2.29 | 0.93 | 0.000 | Rikenellaceae_unclassified | 5.57 | 3.44 | 0.003 |
| Blautia | 0.27 | 0.05 | 0.000 | Epulopiscium | 0.04 | 0.08 | 0.004 |
| [Paraprevotellaceae]_unclassified | 0.04 | 0.00 | 0.000 | Treponema | 1.98 | 1.45 | 0.005 |
| Faecalibacterium | 0.15 | 0.00 | 0.000 | Clostridiales_unclassified | 9.72 | 8.65 | 0.005 |
| Phascolarctobacterium | 0.51 | 0.36 | 0.000 | [Barnesiellaceae]_unclassified | 0.19 | 0.32 | 0.008 |
| Anaerovibrio | 0.10 | 0.03 | 0.000 | Bifidobacterium | 0.37 | 0.12 | 0.013 |
| Mogibacteriaceae_unclassified | 0.69 | 0.50 | 0.000 | Desulfovibrionaceae_unclassified | 0.20 | 0.28 | 0.014 |
| Butyrivibrio | 0.09 | 0.16 | 0.000 | Akkermansia | 2.94 | 4.72 | 0.018 |
| Cyanobacteria_unclassified | 1.16 | 0.75 | 0.000 | Elusimicrobium | 0.22 | 0.29 | 0.022 |
| Veillonellaceae_unclassified | 0.18 | 0.13 | 0.000 | Peptostreptococcaceae_unclassified | 1.61 | 2.33 | 0.023 |
| Ruminococcaceae_unclassified | 27.17 | 31.87 | 0.000 | Bacteroides | 1.83 | 0.23 | 0.042 |
| Pirellulaceae_unclassified | 0.05 | 0.14 | 0.000 | Ruminobacter | 0.01 | 0.02 | 0.046 |
| Antibiotic Class | Resistance Gene | Number of Hits | Number of Hosts | ||
|---|---|---|---|---|---|
| Pre-trt a (165 *, 38 **) | Post-trt b (200, 64) | Pre-trt (165, 38) | Post-trt (200, 64) | ||
| Aminoglycoside | aac6 | 0 | 3 | 0 | 3 |
| aph2 | 61 | 58 | 14 | 16 | |
| aph3 | 51 | 55 | 11 | 13 | |
| ant6 | 75 | 84 | 22 | 29 | |
| ant9 | 39 | 189 | 12 | 23 | |
| sat | 51 | 56 | 11 | 12 | |
| Beta-lactams | aci | 1 | 4 | 1 | 2 |
| cfX | 5 | 8 | 5 | 3 | |
| Macrolides | ermB | 8 | 0 | 3 | 0 |
| ermF | 0 | 1 | 0 | 1 | |
| ermG | 24 | 5 | 3 | 3 | |
| ermQ | 1 | 0 | 1 | 0 | |
| mefE | 3 | 1 | 2 | 1 | |
| Phenicol | cfR | 15 | 24 | 3 | 14 |
| Tetracyclines | tet32 | 20 | 16 | 9 | 7 |
| tet40 | 112 | 1484 | 44 | 91 | |
| tet44 | 15 | 6 | 10 | 5 | |
| tetA | 8 | 4 | 5 | 3 | |
| tetB | 1 | 0 | 1 | 0 | |
| tetL | 0 | 69 | 0 | 5 | |
| tetO | 78 | 75 | 27 | 21 | |
| tetQ | 25 | 483 | 10 | 19 | |
| tetW | 178 | 2836 | 79 | 105 | |
| tetX | 0 | 1 | 0 | 1 | |
| Phylum | Pre-Treatment | Post-Treatment |
|---|---|---|
| Actinobacteria | ermB, ermG, tetW | ant9, tet40, tetQ, tetW |
| Bacteroidetes | tetQ | aph2, aph3, ant6, cfX, ermF, ermG, mefE, cfR, tet40, tetQ, tetW |
| Euryarchaeota | ant9, tetA, tetO | No ARGs |
| Firmicutes | aph2, aph3, ant6, ant9, sat, cfX, ermB, cfR, tet32, tet40, tet44, tetA, tetB, tetO, tetQ, tetW | aac6, aph2, aph3, ant6, ant9, sat, ermG, cfR, tet32, tet40, tetA, tetL, tetO, tetQ, tetW |
| Proteobacteria | aph2, aph3, ant6, ant9, sat, tet32, tet40, tetO, tetW | aac6, aph2, aph3, ant6, ant9, cfR, tet40, tetL, tetQ, tetW |
| Spirochaetes | NA | ant6, sat, tet40, tet44, tetW |
| Tenericutes | tet40 | No ARGs |
| Verrucomicrobia | tet40 | ant6, sat, tet40, tetW |
| Change | Control | Group B | Group C | |
|---|---|---|---|---|
| tetW | Mean (log2 *) | −0.21 | 0.58 | 0.34 |
| SD | 0.017 | 0.065 | 0.015 | |
| p value ** | NA | 0.034 a | 0.272 | |
| tetO | Mean | −0.01 | −0.15 | −0.32 |
| SD | 0.024 | 0.088 | 0.173 | |
| p value | NA | 0.272 | 0.0338 c | |
| tetX | Mean | −0.31 | −0.92 | 0.02 |
| SD | 0.081 | 0.058 | 0.039 | |
| p value | NA | 0.267 | 0.264 | |
| ermB | Mean | 0.52 | −0.76 | −0.15 |
| SD | 0.081 | 0.064 | 0.174 | |
| p value | NA | 0.022 b | 0.359 | |
| ermF | Mean | −0.68 | −0.44 | 0.19 |
| SD | 0.054 | 0.095 | 0.173 | |
| p value | NA | 0.359 | 0.022 d |
| ARGs | Forward Primer | Reverse Primer | Reference |
|---|---|---|---|
| ermB | TGAAAGCCATGCGTCTGACA | CCCTAGTGTTCGGTGAATATCCA | [79] |
| ermF | TTTCAAAGTGGTGTCAAATATTCCTT | GGACAATGGAACCTCCCAGAA | |
| tetO | ATGTGGATACTACAACGCATGAGATT | TGCCTCCACATGATATTTTTCCT | |
| tetW | TCCTTCCAGTGGCACAGATGT | GCCCCATCTAAAACAGCCAAA | |
| tetX | AAATTTGTTACCGACACGGAAGTT | CATAGCTGAAAAAATCCAGGACAGTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beyi, A.F.; Brito-Goulart, D.; Hawbecker, T.; Slagel, C.; Ruddell, B.; Hassall, A.; Dewell, R.; Dewell, G.; Sahin, O.; Zhang, Q.; et al. Danofloxacin Treatment Alters the Diversity and Resistome Profile of Gut Microbiota in Calves. Microorganisms 2021, 9, 2023. https://doi.org/10.3390/microorganisms9102023
Beyi AF, Brito-Goulart D, Hawbecker T, Slagel C, Ruddell B, Hassall A, Dewell R, Dewell G, Sahin O, Zhang Q, et al. Danofloxacin Treatment Alters the Diversity and Resistome Profile of Gut Microbiota in Calves. Microorganisms. 2021; 9(10):2023. https://doi.org/10.3390/microorganisms9102023
Chicago/Turabian StyleBeyi, Ashenafi Feyisa, Debora Brito-Goulart, Tyler Hawbecker, Clare Slagel, Brandon Ruddell, Alan Hassall, Renee Dewell, Grant Dewell, Orhan Sahin, Qijing Zhang, and et al. 2021. "Danofloxacin Treatment Alters the Diversity and Resistome Profile of Gut Microbiota in Calves" Microorganisms 9, no. 10: 2023. https://doi.org/10.3390/microorganisms9102023
APA StyleBeyi, A. F., Brito-Goulart, D., Hawbecker, T., Slagel, C., Ruddell, B., Hassall, A., Dewell, R., Dewell, G., Sahin, O., Zhang, Q., & Plummer, P. J. (2021). Danofloxacin Treatment Alters the Diversity and Resistome Profile of Gut Microbiota in Calves. Microorganisms, 9(10), 2023. https://doi.org/10.3390/microorganisms9102023






