Bacterial Membrane-Derived Vesicles Attenuate Vancomycin Activity against Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Materials & Methods
2.1. Bacterial Strains, Media and Antibiotics
2.2. Isolation and Purification of MVs
2.3. Minimum Inhibitory Concentration and Checkerboard Assays
2.4. Ethical Approval
2.5. Whole Blood Killing Assay
2.6. Neutrophil Killing Assay
2.7. Serum Killing Assay
2.8. Growth Curve
2.9. Proteomic and Bioinformatic Analysis of MVs Containing Protein
2.10. Statistical Analysis
3. Results
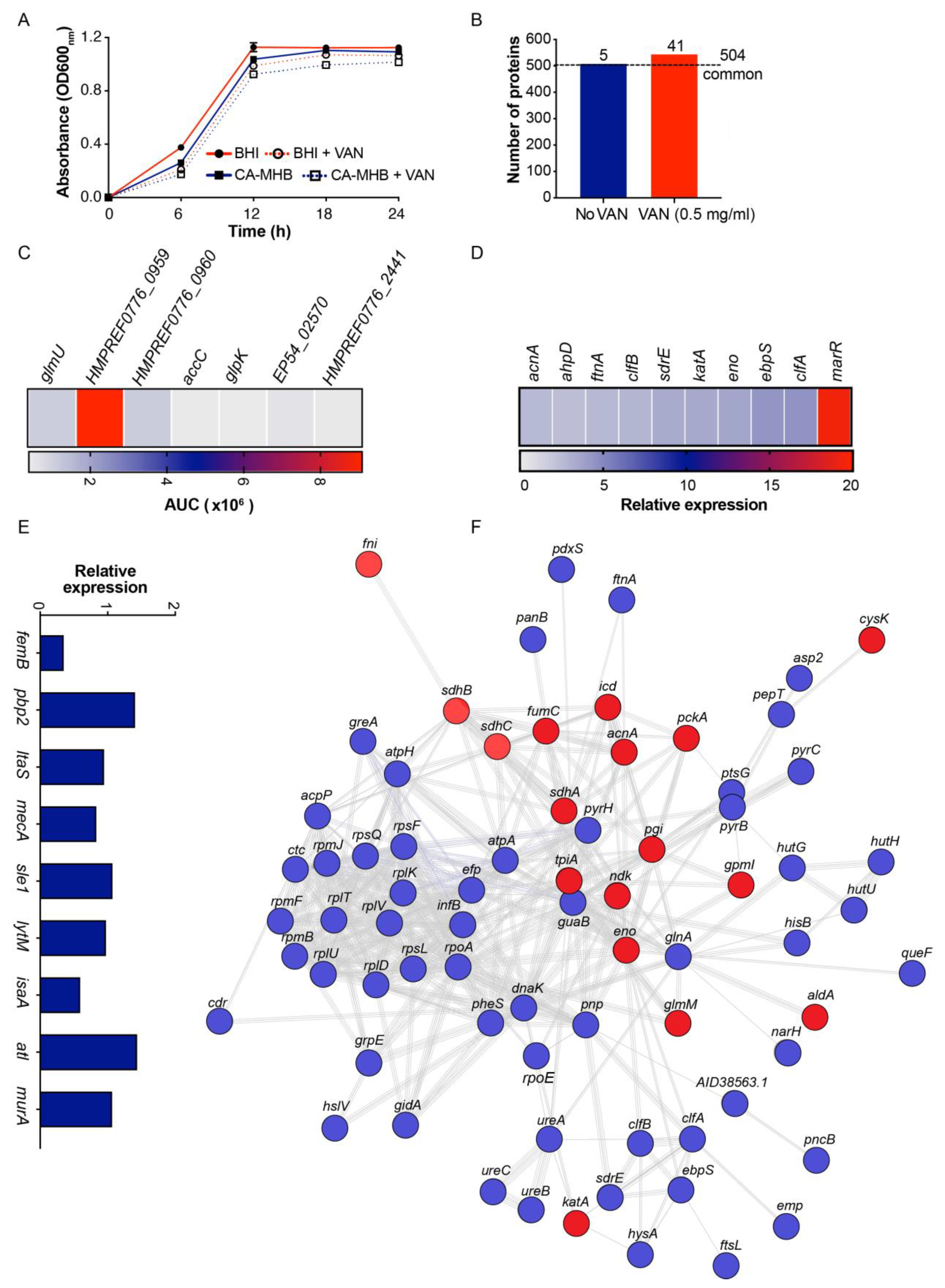
3.1. Exogenous MVs Promote MRSA Survival in the Presence of VAN
3.2. MVs, VAN and Innate Immune Mediated Killing of MRSA
3.3. VAN Exposure Influences MRSA MV Proteome Content
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mertz, D.; Frei, R.; Periat, N.; Zimmerli, M.; Battegay, M.; Fluckiger, U.; Widmer, A.F. Exclusive Staphylococcus aureus throat carriage: At-risk populations. Arch. Intern. Med. 2009, 169, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.F.; Leech, J.M.; Rogers, T.R.; McLoughlin, R.M. Staphylococcus aureus Colonization: Modulation of Host Immune Response and Impact on Human Vaccine Design. Front. Immunol. 2014, 4, 507. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef]
- Askarian, F.; Lapek, J.D., Jr.; Dongre, M.; Tsai, C.M.; Kumaraswamy, M.; Kousha, A.; Valderrama, J.A.; Ludviksen, J.A.; Cavanagh, J.P.; Uchiyama, S.; et al. Staphylococcus aureus Membrane-Derived Vesicles Promote Bacterial Virulence and Confer Protective Immunity in Murine Infection Models. Front. Microbiol. 2018, 9, 262. [Google Scholar] [CrossRef]
- Toyofuku, M.; Carcamo-Oyarce, G.; Yamamoto, T.; Eisenstein, F.; Hsiao, C.C.; Kurosawa, M.; Gademann, K.; Pilhofer, M.; Nomura, N.; Eberl, L. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 2017, 8, 481. [Google Scholar] [CrossRef] [Green Version]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.J.; Wang, X.H.; Fan, G.C. Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases. Acta Pharmacol. Sin. 2018, 39, 514–533. [Google Scholar] [CrossRef] [PubMed]
- Caruana, J.C.; Walper, S.A. Bacterial Membrane Vesicles as Mediators of Microbe—Microbe and Microbe—Host Community Interactions. Front. Microbiol. 2020, 11, 432. [Google Scholar] [CrossRef] [Green Version]
- Andreoni, F.; Toyofuku, M.; Menzi, C.; Kalawong, R.; Mairpady Shambat, S.; Francois, P.; Zinkernagel, A.S.; Eberl, L. Antibiotics Stimulate Formation of Vesicles in Staphylococcus aureus in both Phage-Dependent and -Independent Fashions and via Different Routes. Antimicrob. Agents Chemother. 2019, 63, e01439-18. [Google Scholar] [CrossRef] [Green Version]
- Pader, V.; Hakim, S.; Painter, K.L.; Wigneshweraraj, S.; Clarke, T.B.; Edwards, A.M. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat. Microbiol. 2016, 2, 16194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.W.; Seo, J.S.; Park, S.B.; Lee, A.R.; Lee, J.S.; Jung, J.W.; Chun, J.H.; Lazarte, J.M.S.; Kim, J.; Kim, J.H.; et al. Significant increase in the secretion of extracellular vesicles and antibiotics resistance from methicillin-resistant Staphylococcus aureus induced by ampicillin stress. Sci. Rep. 2020, 10, 21066. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Barna, J.C.; Williams, D.H. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 1984, 38, 339–357. [Google Scholar] [CrossRef]
- Walraven, C.J.; North, M.S.; Marr-Lyon, L.; Deming, P.; Sakoulas, G.; Mercier, R.C. Site of infection rather than vancomycin MIC predicts vancomycin treatment failure in methicillin-resistant Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 2011, 66, 2386–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumaraswamy, M.; Lin, L.; Olson, J.; Sun, C.F.; Nonejuie, P.; Corriden, R.; Dohrmann, S.; Ali, S.R.; Amaro, D.; Rohde, M.; et al. Standard susceptibility testing overlooks potent azithromycin activity and cationic peptide synergy against MDR Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2016, 71, 1264–1269. [Google Scholar] [CrossRef] [Green Version]
- Askarian, F.; Uchiyama, S.; Valderrama, J.A.; Ajayi, C.; Sollid, J.U.; van Sorge, N.M.; Nizet, V.; van Strijp, J.A.; Johannessen, M. Serine-aspartate-repeat protein D Increases Staphylococcus aureus Virulence and Survival in Blood. Infect. Immun. 2017, 85, e00559-16. [Google Scholar] [CrossRef] [Green Version]
- Kumaraswamy, M.; Do, C.; Sakoulas, G.; Nonejuie, P.; Tseng, G.W.; King, H.; Fierer, J.; Pogliano, J.; Nizet, V. Listeria monocytogenes endocarditis: Case report, review of the literature, and laboratory evaluation of potential novel antibiotic synergies. Int. J. Antimicrob. Agents 2018, 51, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Joshi, B.; Janice, J.; Askarian, F.; Škalko-Basnet, N.; Hagestad, O.; Mekhlif, A.; Wai, S.; Hegstad, K.; Johannessen, M. Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J. Proteom. 2018, 187, 28–38. [Google Scholar] [CrossRef]
- Cavanagh, J.P.; Pain, M.; Askarian, F.; Bruun, J.-A.; Urbarova, I.; Wai, S.N.; Schmidt, F.; Johannessen, M. Comparative exoproteome profiling of an invasive and a commensal Staphylococcus haemolyticus isolate. J. Proteom. 2018, 197, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. String V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibek, G.C.; Sahukhal, G.S.; Elasri, M.O. Role of the msaABCR Operon in Cell Wall Biosynthesis, Autolysis, Integrity, and Antibiotic Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Utaida, S.; Dunman, P.M.; Macapagal, D.; Murphy, E.; Projan, S.J.; Singh, V.K.; Jayaswal, R.K.; Wilkinson, B.J. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 2003, 149, 2719–2732. [Google Scholar] [CrossRef] [Green Version]
- Labischinski, H.; Ehlert, K.; Berger-Bachi, B. The targeting of factors necessary for expression of methicillin resistance in staphylococci. J. Antimicrob. Chemother. 1998, 41, 581–584. [Google Scholar] [CrossRef]
- Severin, A.; Wu, S.W.; Tabei, K.; Tomasz, A. Penicillin-binding protein 2 is essential for expression of high-level vancomycin resistance and cell wall synthesis in vancomycin-resistant Staphylococcus aureus carrying the enterococcal vanA gene complex. Antimicrob. Agents Chemother. 2004, 48, 4566–4573. [Google Scholar] [CrossRef] [Green Version]
- Leski, T.A.; Tomasz, A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 2005, 187, 1815–1824. [Google Scholar] [CrossRef] [Green Version]
- Choo, E.J.; Chambers, H.F. Treatment of Methicillin-Resistant Staphylococcus aureus Bacteremia. Infect. Chemother. 2016, 48, 267–273. [Google Scholar] [CrossRef] [Green Version]
- Sakoulas, G.; Moise-Broder, P.A.; Schentag, J.; Forrest, A.; Moellering, R.C., Jr.; Eliopoulos, G.M. Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 2004, 42, 2398–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.H.; Kim, M.; Kim, C.J.; Cho, J.E.; Choi, Y.J.; Park, J.S.; Ahn, S.; Jang, H.C.; Park, K.H.; Jung, S.I.; et al. Impact of Vancomycin MIC on Treatment Outcomes in Invasive Staphylococcus aureus Infections. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodise, T.P.; Graves, J.; Evans, A.; Graffunder, E.; Helmecke, M.; Lomaestro, B.M.; Stellrecht, K. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 2008, 52, 3315–3320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.C.; Sy, C.L.; Huang, Y.C.; Shie, S.S.; Shu, J.C.; Hsieh, P.H.; Hsiao, C.H.; Chen, C.J. Risk factors of treatment failure and 30-day mortality in patients with bacteremia due to MRSA with reduced vancomycin susceptibility. Sci. Rep. 2018, 8, 7868. [Google Scholar] [CrossRef] [Green Version]
- Choe, D.; Szubin, R.; Dahesh, S.; Cho, S.; Nizet, V.; Palsson, B.; Cho, B.K. Genome-scale analysis of Methicillin-resistant Staphylococcus aureus USA300 reveals a tradeoff between pathogenesis and drug resistance. Sci. Rep. 2018, 8, 2215. [Google Scholar] [CrossRef] [Green Version]
- Hebrard, M.; Viala, J.P.; Meresse, S.; Barras, F.; Aussel, L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J. Bacteriol. 2009, 191, 4605–4614. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; De Angelis, G.; Cataldo, M.A.; Pozzi, E.; Cauda, R. Does antibiotic exposure increase the risk of methicillin-resistant Staphylococcus aureus (MRSA) isolation? A systematic review and meta-analysis. J. Antimicrob. Chemother. 2008, 61, 26–38. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.R.; Bae, T.; Williams, W.A.; Duguid, E.M.; Rice, P.A.; Schneewind, O.; He, C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2006, 2, 591–595. [Google Scholar] [CrossRef] [PubMed]


| MRSA (TCH 1516) | −MVs (0 mg/L) | +MVs (1.56–100 mg/L) | ||||||
|---|---|---|---|---|---|---|---|---|
| VAN | DAP | CFZ | NAF | VAN a | DAP | CFZ | NAF | |
| CA-MHB | 1 | 2 | ≥64 | ≥64 | 2 | 2 | ≥64 | ≥64 |
| RPMI + 5%BHI | 1 | 0.5 | 4 | 4 | 4 | 0.5 | 4 | 4 |
| Gene Symbol/ORF | Description |
|---|---|
| dut | dUTP diphosphatase |
| purE | N5-carboxyaminoimidazole ribonucleotide mutase |
| valS | Valine—tRNA ligase |
| rocF | Arginase |
| SAUSA300_2144 | Uncharacterized protein |
| glmU | Bifunctional protein GlmU |
| HMPREF0776_2553 | Dehydrogenase E1 component |
| alaS | Alanine—tRNA ligase |
| HMPREF0776_0959 | Acetyl-CoA C-acetyltransferase |
| SAUSA300_2328 | Uncharacterized protein |
| dmpI | Tautomerase |
| mtlD | Mannitol-1-phosphate 5-dehydrogenase |
| SAUSA300_0003 | Uncharacterized protein |
| SAUSA300_1690 | Putative thioredoxin |
| HMPREF0776_2533 | Alpha-amylase |
| SAUSA300_0289 | Uncharacterized protein |
| lysS | Lysine—tRNA ligase |
| pyrG | CTP synthase |
| SAUSA300_0706 | Putative osmoprotectant ABC transporter, ATP-binding protein |
| SA0224 | 3-hydroxyacyl-CoA dehydrogenase, NAD binding domain protein |
| recA | Protein RecA |
| HMPREF0776_0345 | Accessory regulator family |
| SAUSA300_2132 | UPF0457 protein SAUSA300_2132 |
| accC | Acetyl-CoA carboxylase, biotin carboxylase |
| HMPREF0776_2830 | Excalibur domain protein |
| SAUSA300_0460 | Uncharacterized protein |
| hflB ftsH | ATP-dependent zinc metalloprotease FtsH |
| SAUSA300_2289 | Uncharacterized protein |
| isdB | Iron-regulated surface determinant protein B |
| srrA | Staphylococcal respiratory response protein, SrrA |
| codY | GTP-sensing transcriptional pleiotropic repressor CodY |
| SAUSA300_0816 | UPF0337 protein SAUSA300_0816 |
| rpsD | 30S ribosomal protein S4 |
| HMPREF0776_2441 | Peptidase, S41 family |
| rpsR | 30S ribosomal protein S18 |
| HMPREF0776_1767 | HD domain protein |
| SAUSA300_1674 | Putative serine protease HtrA |
| HMPREF0776_0647 | Uncharacterized protein |
| EP54_02570 | Lipoprotein |
| glpK | Glycerol kinase |
| HMPREF0776_2430 | Glyoxalase family protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumaraswamy, M.; Wiull, K.; Joshi, B.; Sakoulas, G.; Kousha, A.; Vaaje-Kolstad, G.; Johannessen, M.; Hegstad, K.; Nizet, V.; Askarian, F. Bacterial Membrane-Derived Vesicles Attenuate Vancomycin Activity against Methicillin-Resistant Staphylococcus aureus. Microorganisms 2021, 9, 2055. https://doi.org/10.3390/microorganisms9102055
Kumaraswamy M, Wiull K, Joshi B, Sakoulas G, Kousha A, Vaaje-Kolstad G, Johannessen M, Hegstad K, Nizet V, Askarian F. Bacterial Membrane-Derived Vesicles Attenuate Vancomycin Activity against Methicillin-Resistant Staphylococcus aureus. Microorganisms. 2021; 9(10):2055. https://doi.org/10.3390/microorganisms9102055
Chicago/Turabian StyleKumaraswamy, Monika, Kamilla Wiull, Bishnu Joshi, George Sakoulas, Armin Kousha, Gustav Vaaje-Kolstad, Mona Johannessen, Kristin Hegstad, Victor Nizet, and Fatemeh Askarian. 2021. "Bacterial Membrane-Derived Vesicles Attenuate Vancomycin Activity against Methicillin-Resistant Staphylococcus aureus" Microorganisms 9, no. 10: 2055. https://doi.org/10.3390/microorganisms9102055
APA StyleKumaraswamy, M., Wiull, K., Joshi, B., Sakoulas, G., Kousha, A., Vaaje-Kolstad, G., Johannessen, M., Hegstad, K., Nizet, V., & Askarian, F. (2021). Bacterial Membrane-Derived Vesicles Attenuate Vancomycin Activity against Methicillin-Resistant Staphylococcus aureus. Microorganisms, 9(10), 2055. https://doi.org/10.3390/microorganisms9102055







