Microbial Diversity in the Phyllosphere and Rhizosphere of an Apple Orchard Managed under Prolonged “Natural Farming” Practices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apple Orchards
2.2. Disease Incidence
2.3. Macroarray Analysis of Microbial Diversity in the Phyllosphere
2.4. Metagenome Analysis
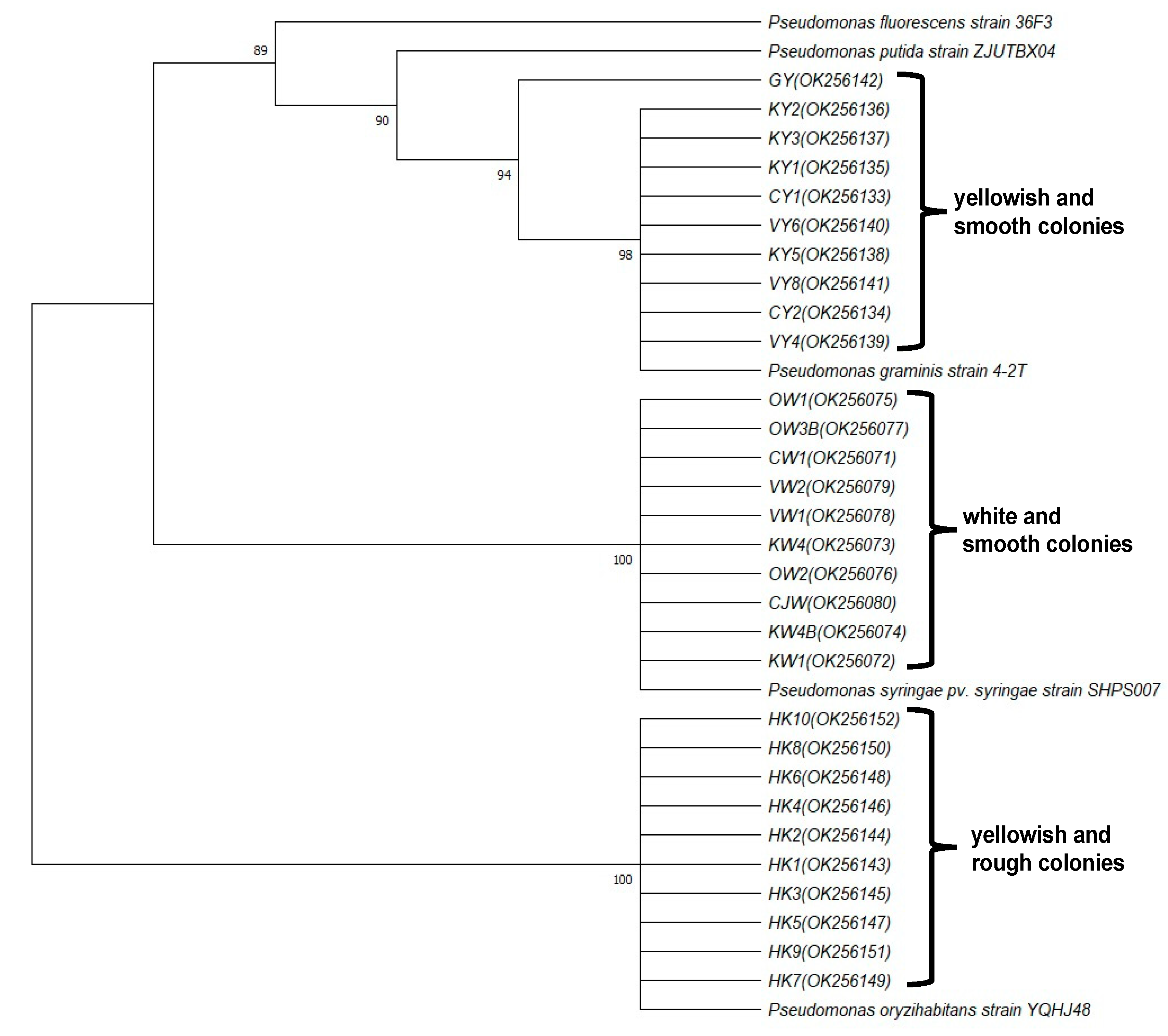
2.5. Isolation of Pseudomonas Species
2.6. Statistical Analyses
3. Results
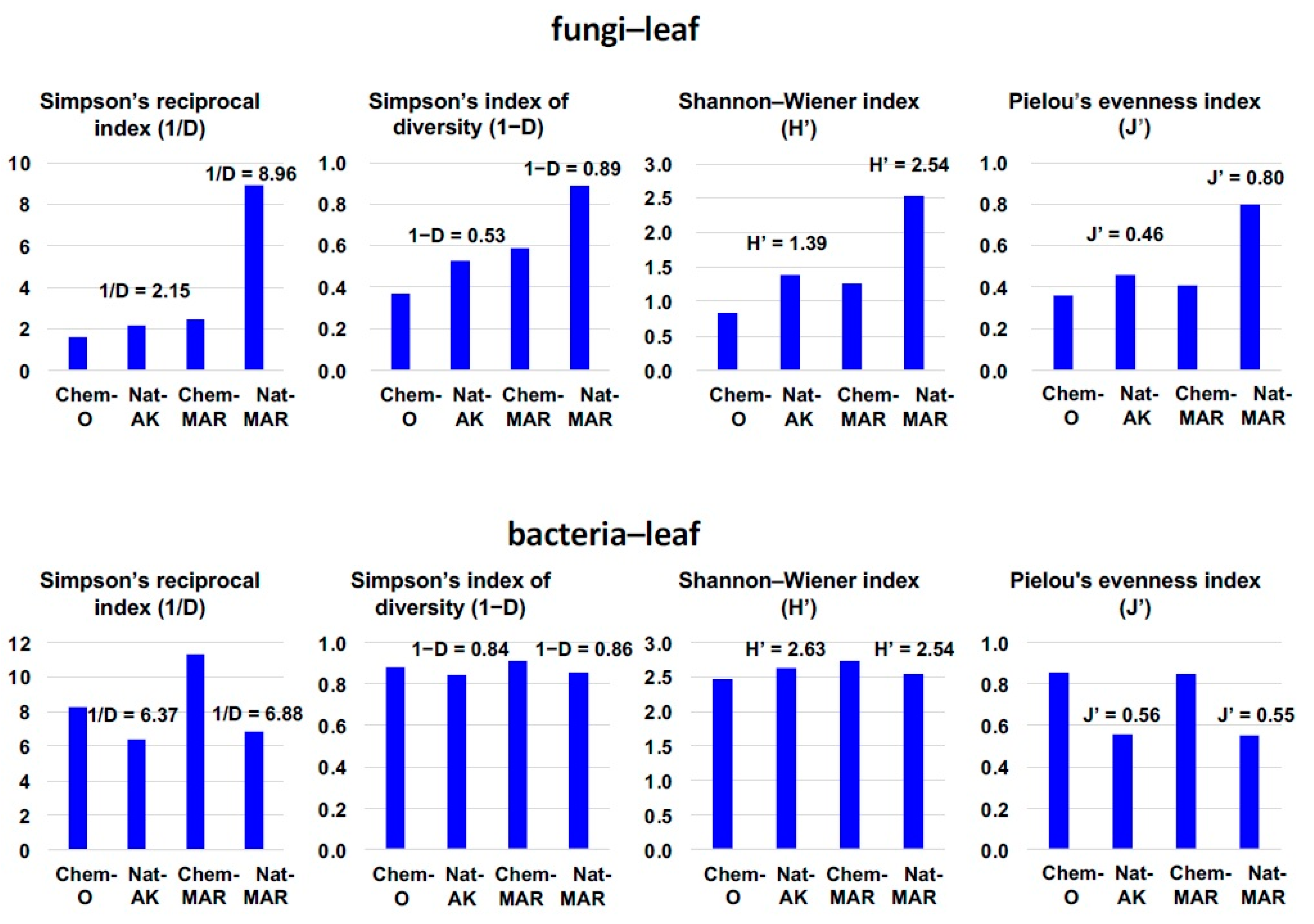
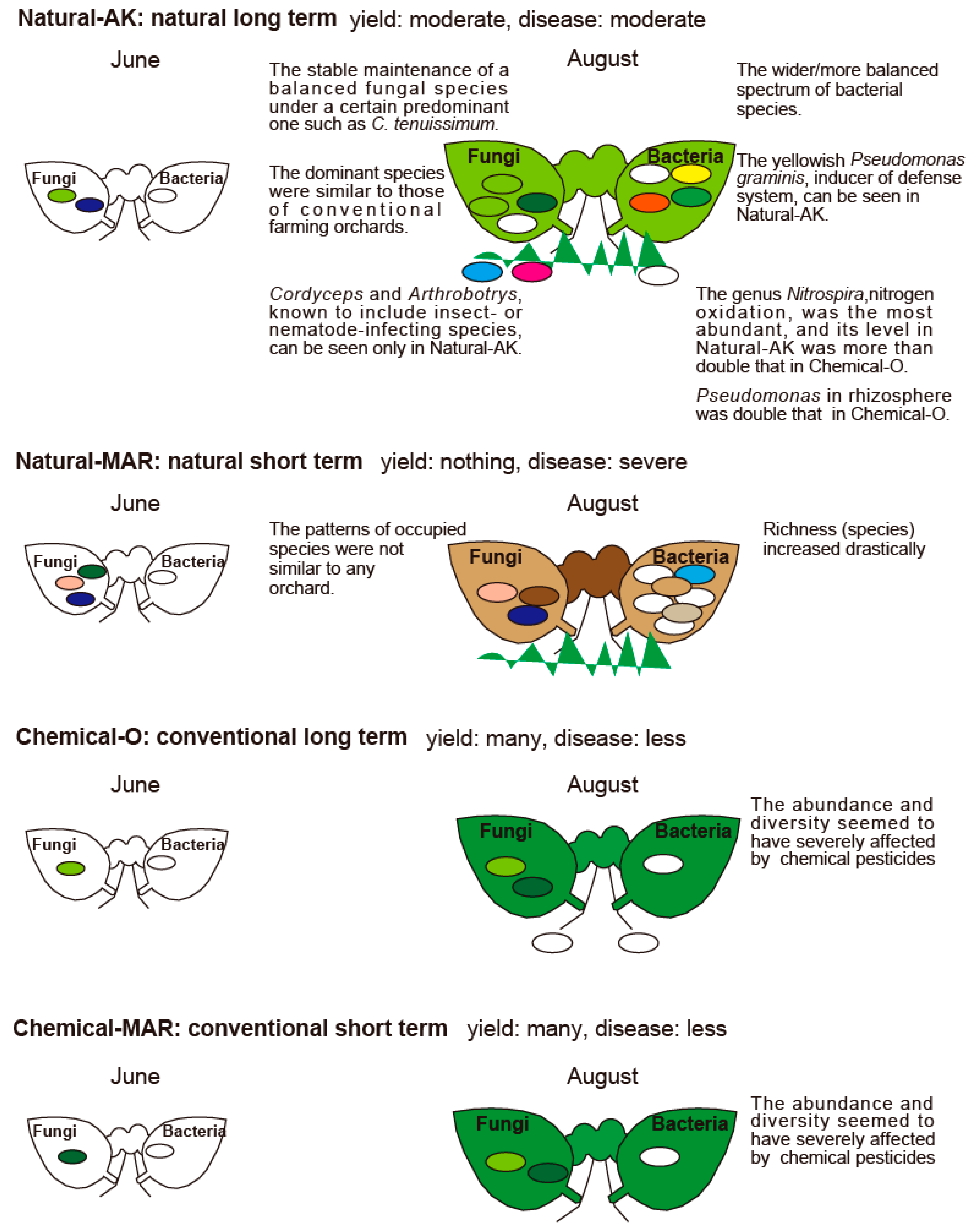
3.1. Disease Incidence and Microbial Diversity in the Phyllosphere of Natural and Conventional Apple Orchards
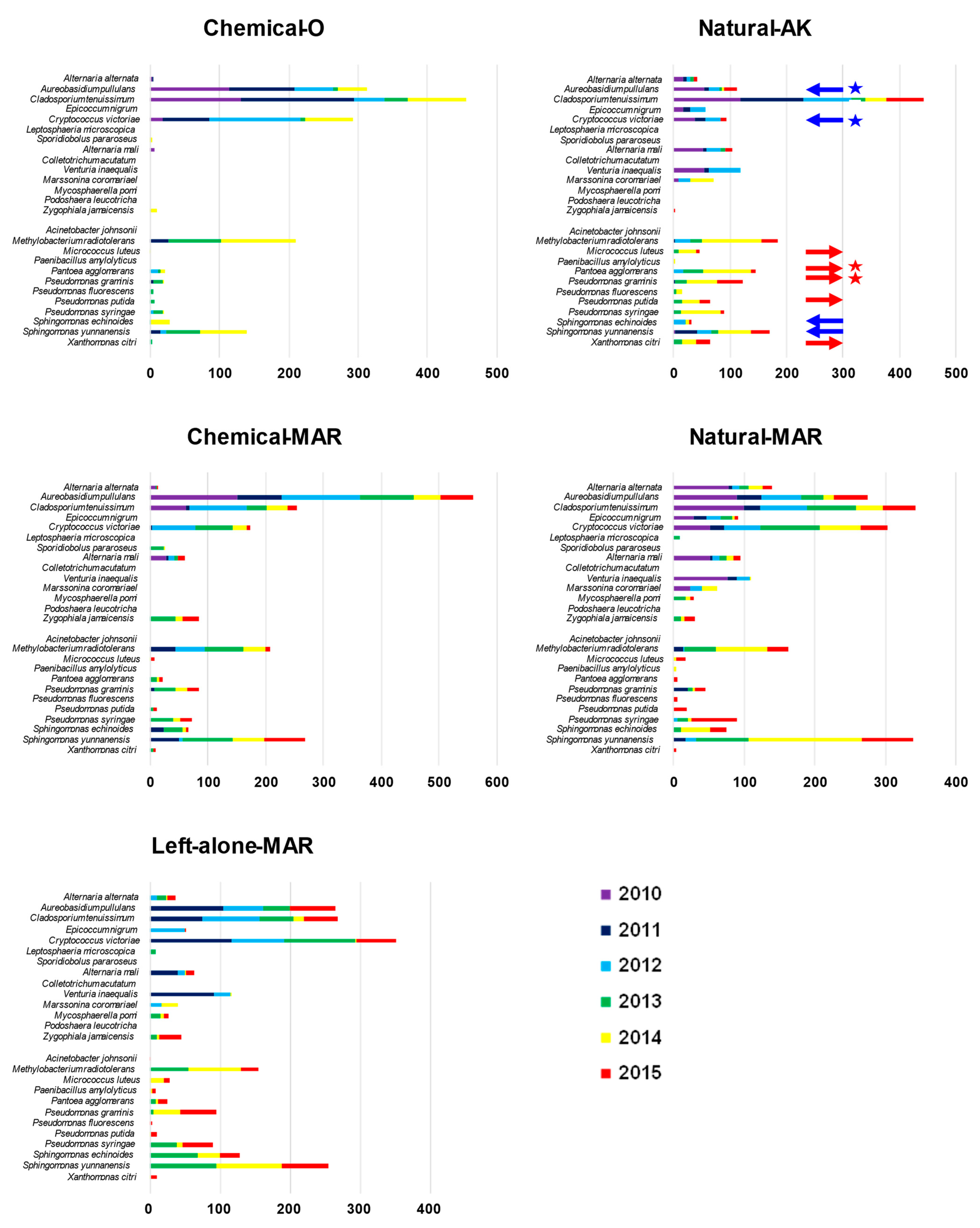
3.2. Microbial Diversity in the Apple Phyllosphere by Macroarray Analysis
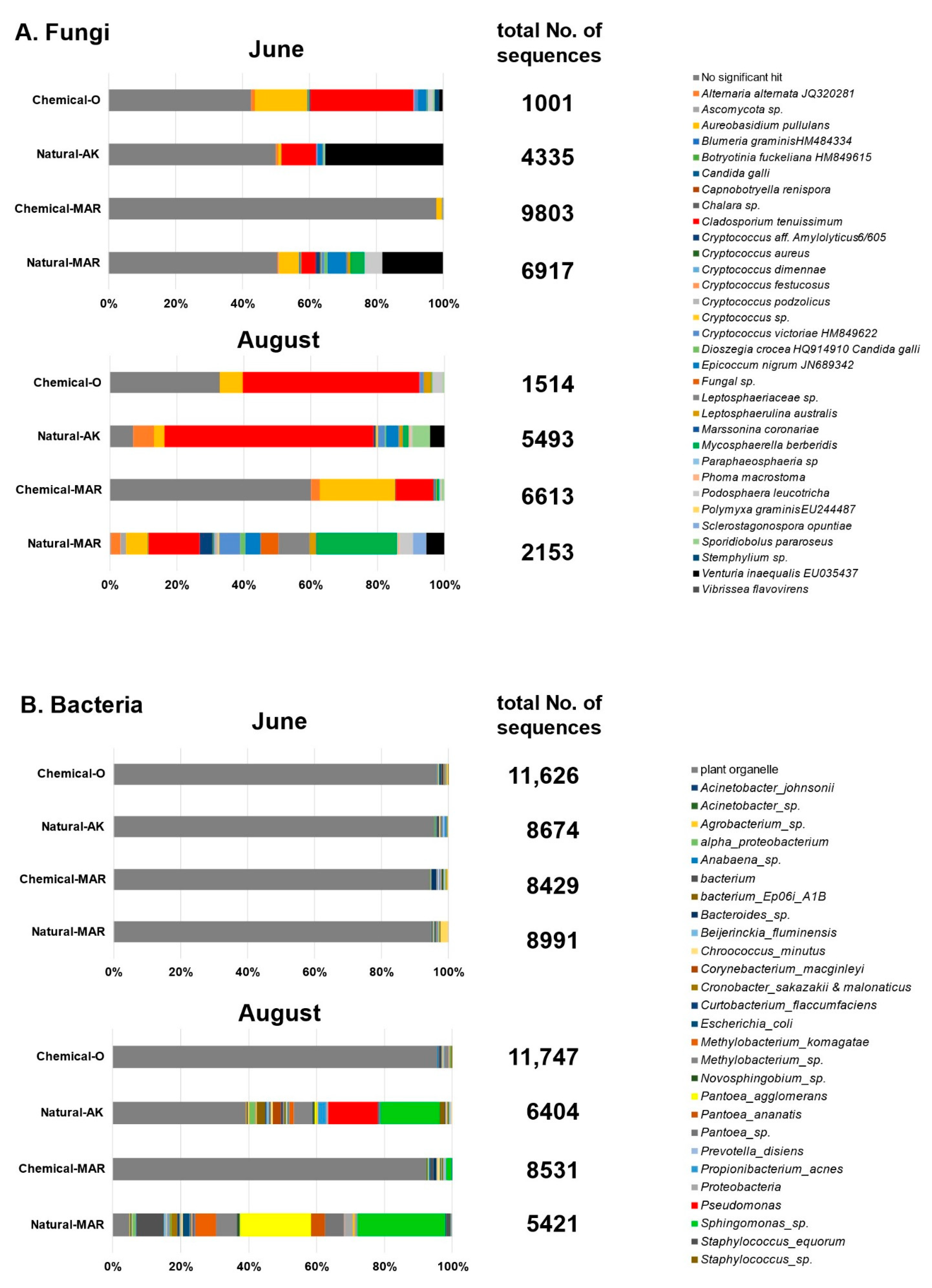
3.3. Metagenome Analysis by Next-Generation Sequencing
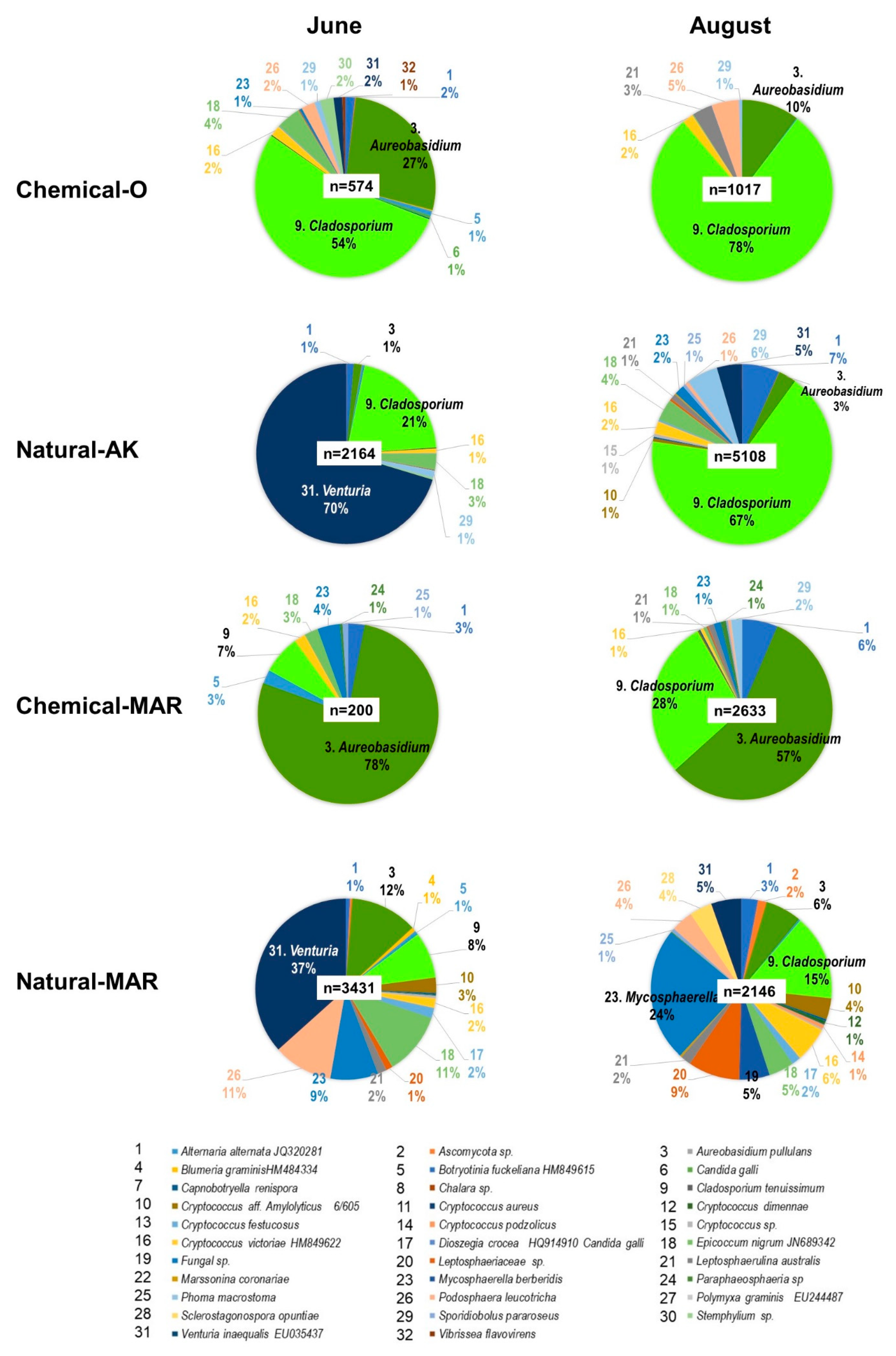
3.3.1. Large-Scale Sequencing Analysis of Microbial Inhabitants in the Apple Phyllosphere
3.3.2. Large-Scale Sequencing Analysis of Microbial Inhabitants in the Apple Rhizosphere
3.4. Pseudomonas in Natural-AK
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kinkel, L.L. Microbial population dynamics on leaves. Annu. Rev. Phytopathol. 1997, 35, 327–347. [Google Scholar] [CrossRef]
- Lindow, S.E.; Brandl, M.T. Microbiology of the Phyllosphere. Appl. Environ. Microbiol. 2003, 64, 1875–1883. [Google Scholar] [CrossRef] [Green Version]
- Whipps, J.M.; Hand, P.; Pink, D.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langenheder, S.; Bulling, M.T.; Solan, M.; Prosser, J.I. Bacterial biodiversity-ecosystem functioning relations are modified by environmental complexity. PLoS ONE 2010, 5, e10834. [Google Scholar] [CrossRef] [Green Version]
- Pennycook, S.R.; Newhook, F.J. Seasonal changes in the apple phylloplane microflora. N. Z. J. Bot. 1981, 19, 273–283. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.; Frampton, C.M.; Boyd-Wilson, K.S.H.; Harris-Virgin, P.; Waipara, N.W. Agrichemical impact on growth and survival of non-target apple phyllosphere microorganisms. Can. J. Microbiol. 2007, 53, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Sessitsch, A.; Hardoim, P.; Doering, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant-Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil. Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebeis, S.L. The potential for give and take in plant–microbiome relationships. Front. Plant Sci. 2014, 5, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, S. Sugoi Hatake No Sugoi Tochi; Gentosha Inc.: Tokyo, Japan, 2013; ISBN 978-4-344-98306-9 C0295. (In Japanese) [Google Scholar]
- He, Y.-H.; Isono, S.; Shibuya, M.; Tsuji, M.; Purushothama, C.-R.A.; Tanaka, K.; Sano, T. Oligo-DNA Custom Macroarray for Monitoring Major Pathogenic and Non-Pathogenic Fungi and Bacteria in the Phyllosphere of Apple Trees. PLoS ONE 2012, 7, e34249. [Google Scholar] [CrossRef]
- Ikeda, S.; Kaneko, T.; Okubo, T.; Rallos, L.E.E.; Eda, S.; Mitsui, H.; Sato, S.; Nakamura, Y.; Tabata, S.; Minamisawa, K. Development of a bacterial cell enrichment method and its application to the community analysis in soybean stems. Microbiol. Ecol. 2009, 58, 703–714. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Granado, J.; Thürig, B.; Kieffer, E.; Petrini, L.; Flieβbach, A.; Tamm, L.; Weibel, F.P.; Wyss, G.S. Culturable fungi of stored ‘golden delicious’ apple fruits: A one-season comparison study of organic and integrated production systems in Switzerland. Microb Ecol 2008, 56, 720–732. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Wees, S.; Van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; Van Loon, L.C. A novel signaling pathway controlling ISR in Ara bidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Sticher, L.; Mauch-Mani, B.; Métraux, J.P. Systemic acquired resistance. Annu. Rev. Phytopathol. 1997, 35, 235–270. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; Bakker, P.A.; Pieterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collazoa, C.; Giné-Bordonaba, J.; Aguiló-Aguayo, I.; Povedano, I.; Bademunt, A.; Viñas, I. Pseudomonas graminis strain CPA-7 differ entially modulates the oxidative response in fresh-cut ‘Golden delicious’ apple depending on the storage conditions. Postharvest Biol. Technol. 2018, 138, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iffland, D.W.; Allison, P. Nematode-trapping fungi: Evaluation of axenic healthy and galled roots as trap inducers. Science 1964, 146, 547–548. [Google Scholar] [CrossRef] [PubMed]
- Lücker, S.; Wagner, M.; Maixner, F.; Pelletier, E.; Koch, H.; Vacherie, B.; Rattei, T.; Damste, J.S.; Spieck, E.; Le Paslier, D.; et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 13479–13484. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Fungi | Bacteria | |||||||
|---|---|---|---|---|---|---|---|---|
| No. Genus | No. Species | No. Fungal Sequence | Total No. of Read * | No. Genus | No. Species | No. Bacterial Sequence | Total No. of Reads * | |
| June | ||||||||
| Chemical-O | 18 | 20 | 574 | 1001 | 23 | 29 | 402 | 11,626 |
| Natural-AK | 18 | 20 | 2164 | 4335 | 18 | 19 | 369 | 8674 |
| Chemical-MAR | 9 | 10 | 200 | 9803 | 16 | 17 | 465 | 8429 |
| Natural-MAR | 15 | 21 | 3431 | 6917 | 25 | 28 | 473 | 8991 |
| August | ||||||||
| Chemical-O | 10 | 10 | 1017 | 1514 | 17 | 18 | 537 | 11,747 |
| Natural-AK | 18 | 22 | 5108 | 5493 | 107 | 112 | 3887 | 6404 |
| Chemical-MAR | 18 | 21 | 2633 | 6613 | 23 | 25 | 661 | 8531 |
| Natural-MAR | 18 | 25 | 2146 | 2153 | 94 | 102 | 5158 | 5421 |
| Genus | Natural-AK | Chemi-O | |
|---|---|---|---|
| % | % | ||
| Major fungi | Emericella | 2.17 | 0.19 |
| Fusarium | 0.94 | 4.95 | |
| Mortierella | 0.51 | 0.23 | |
| Leohumicola | 0.34 | 0.36 | |
| Cordyceps | 0.34 | 0 | |
| Leptosphaeria | 0.26 | 0.02 | |
| Glomus | 0.26 | 0 | |
| Cryptococcus | 0.21 | 1.5 | |
| Zalerion | 0.17 | 0.13 | |
| Ophiosphaerella | 0.17 | 0 | |
| Cryptococcus | 0.21 | 1.50 | |
| Verticillium | 0.09 | 0.62 | |
| Dipodascus | 0 | 0.43 | |
| Sporidiobolus | 0 | 0.23 | |
| Pichia | 0 | 0.19 | |
| Cladosporium | 0 | 0.13 | |
| Major bacteria | Nitrospira | 6.12 | 2.12 |
| Bradyrhizobium | 1.85 | 1.19 | |
| Cupriavidus | 1.13 | 0.32 | |
| Burkholderia | 1.00 | 0.76 | |
| Pseudomonas | 0.96 | 0.47 | |
| Stigmatella | 0.84 | 0.16 | |
| Methylibium | 0.80 | 0.08 | |
| Flavobacterium | 0.77 | 0.26 | |
| Methylosinus | 0.68 | 0.84 | |
| Anaeromyxobacter | 0.65 | 0.26 | |
| Ktedonobacter | 0.45 | 0.99 | |
| Bacillus | 0.12 | 0.96 | |
| Thermosporothrix | 0.16 | 0.82 | |
| Methylocystis | 0.32 | 0.77 | |
| Legionella | 0.27 | 0.76 | |
| Candidatus Solibacter | 0.38 | 0.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.-H.; Adkar-Purushothama, C.R.; Ito, T.; Shirakawa, A.; Yamamoto, H.; Kashiwagi, A.; Tatewaki, A.; Fujibayashi, M.; Sugiyama, S.; Yaginuma, K.; et al. Microbial Diversity in the Phyllosphere and Rhizosphere of an Apple Orchard Managed under Prolonged “Natural Farming” Practices. Microorganisms 2021, 9, 2056. https://doi.org/10.3390/microorganisms9102056
He Y-H, Adkar-Purushothama CR, Ito T, Shirakawa A, Yamamoto H, Kashiwagi A, Tatewaki A, Fujibayashi M, Sugiyama S, Yaginuma K, et al. Microbial Diversity in the Phyllosphere and Rhizosphere of an Apple Orchard Managed under Prolonged “Natural Farming” Practices. Microorganisms. 2021; 9(10):2056. https://doi.org/10.3390/microorganisms9102056
Chicago/Turabian StyleHe, Ying-Hong, Charith Raj Adkar-Purushothama, Tsutae Ito, Asuka Shirakawa, Hideki Yamamoto, Akiko Kashiwagi, Ayumu Tatewaki, Misato Fujibayashi, Shuichi Sugiyama, Katsuhiko Yaginuma, and et al. 2021. "Microbial Diversity in the Phyllosphere and Rhizosphere of an Apple Orchard Managed under Prolonged “Natural Farming” Practices" Microorganisms 9, no. 10: 2056. https://doi.org/10.3390/microorganisms9102056






