Abstract
Consistent with their reported abundance in soils, several Burkholderia sensu lato strains were isolated from the rhizosphere of maize plants cultivated at different sites in central México. Comparative analysis of their 16S rRNA gene sequences permitted their separation into three distinctive clades, which were further subdivided into six other clusters by their close resemblance to (1) Trinickia dinghuensis; (2) Paraburkholderia kirstenboschensis, P. graminis, P. dilworthii and P. rhynchosiae; (3) B. gladioli; (4) B. arboris; (5) B. contaminans, or (6) B. metallica representative species. Direct confrontation assays revealed that these strains inhibited the growth of pathogenic Fusarium oxysporum f. sp. radicis-lycopersici, and F. verticillioides within a roughly 3–55% inhibition range. The use of a DIESI-based non-targeted mass spectroscopy experimental strategy further indicated that this method is an option for rapid determination of the pathogen inhibitory capacity of Burkholderia sensu lato strains based solely on the analysis of their exometabolome. Furthermore, it showed that the highest anti-fungal activity observed in B. contaminans and B. arboris was associated with a distinctive abundance of certain m/z ions, some of which were identified as components of the ornbactin and pyochelin siderophores. These results highlight the chemical diversity of Burkholderia sensu lato bacteria and suggest that their capacity to inhibit the Fusarium-related infection of maize in suppressive soils is associated with siderophore synthesis.
1. Introduction
Maize is extensively cultivated in the world, with a production rate of about 990.6 Mt/year, ranking as the third most important cereal for human consumption. During its vegetative cycle, the production of maize is conditioned by various biotic and abiotic factors. Fusarium spp. is a ubiquitous soil-borne plant pathogenic fungus that can cause highly damaging infections characterized by severe decay of seeds, roots, stems, ears or kernels [1,2]. Fusarium verticillioides (Gibberella fujikuroi mating population A; teleomorph, Gibberella moniliformis Wineland) is the prevalent species of Fusarium in distinct landscapes [3,4], and the losses caused by this fungus can vary from 7% to 38% [5].
A significant diversity of both deleterious and beneficial microbial species lives in association with the rhizosphere of most agricultural crops. Certain bacterial components of this root microbial community, known as plant growth promoting rhizobacteria (PGPR), are known to enhance plant growth and fitness [6]. Plant growth promotion can take place indirectly when PGPR prevent or diminish the damage caused by pathogenic microorganisms, an effect frequently produced through the production of secondary metabolites with strong inhibitory activity [7,8]. Examples of bacteria having this capability are found among species of the genera Serratia, Bacillus, Brevibacillus, Lysobacter, Pseudomonas and Burkholderia [9,10,11].
The genus Burkholderia belongs to a subphylum of the β-proteobacteria. It currently involves around 100 species with widespread distribution in frequently contrasting ecological niches [12]. The versatile lifestyles and the capability to develop in numerous habitats that characterize Burkholderia species are mainly due to their genomic and metabolic plasticity [13]. Several species of this genus have plant growth promoting activity, including several from the so-called Burkholderia cepacia complex (Bcc). Beneficial interactions with plants reported for Burkholderia species include nitrogen fixation, 1-aminocyclopropane-1-carboxylate (ACC) deaminase enzyme activity, phosphate solubilization and biological control of pathogens through the synthesis of siderophores, organic volatile compounds, antibiotics or lipopeptides [14,15,16,17,18,19].
Bcc members and other Burkholderia species, such as B. tropica, B. silvatlantica, B. sartisoli, B. graminis, B. phytofirmans, B. terrestris, B. telluris and B. unamae, represent a significant percentage of the total cultivable bacteria living within the rhizosphere of plants [20]. In this respect, various studies have reported that several species of the Bcc constitute an important segment of the cultivable root microbial communities found in the rhizosphere of plants belonging to the Gramineae family. For example, in the maize rhizosphere, B. cenocepacia, B. cepacia, B. multivorans, B. ambifaria and B. pyrrocinia represent between 0.5% and 6% of the total bacterial population [7,20,21].
Since its initial proposal in 1992, the taxonomic classification of Burkholderia has been constantly changing. This continuous flux has led to the suggestion of splitting this genus into two phylogenetic clusters [12,22]. Following this concept, the first clade should include only bacteria reported as plant or animal pathogens, such as B. rhizoxinica, B. gladioli, B. mallei and B. pseudomallei, along with the 20 species of the Bcc associated with serious respiratory infections in humans [20,23,24]. Other bacteria isolated from fungi or environmental samples (e.g., soil and water), as well as endophytic and free-living rhizosphere strains able to establish beneficial interactions with plants, should be incorporated into the second taxonomic clade [13,25,26]. The last clade has been accepted by the International Committee on Systematics of Prokaryotes as a new genus denominated Paraburkholderia, which includes several species having a potential biotechnological use in agriculture. This designation, however, remains controversial [12,22,27,28].
Current molecular classification methods allow an accurate classification of different environmental Burkholderia strains. Nevertheless, novel approaches in microbial characterization may help explore the largely unexplored exometabolome of these important rhizosphere microorganisms, which are known to secrete a wide array of secondary metabolites (e.g., cepalycin, pyrrolnitrin, components of the xylocandin complex, occidiofungin and ornibactin) that are active against soil-borne fungal plant pathogens [29,30,31]. In this respect, metabolic foot-printing was developed as a convenient, reproducible and high-throughput technique for the physiological-level characterization of microorganisms [32]. Recently, the use of untargeted mass spectrometry methods, particularly direct liquid injection electrospray ionization (DIESI-MS), for the study of plant–microbe interactions has greatly increased in the past few years [33]. In this context, the present study describes a metabolic foot-printing approach to explore the exometabolome secreted by different Burkholderia sensu lato strains isolated from the rhizosphere of maize plants. Its main objectives were to develop a rapid, economical and accurate methodology to determine the chemical basis of the differential anti-fungal activity observed in Burkholderia environmental strains isolated from maize plant rhizospheres.
2. Materials and Methods
2.1. Soil Samples
A collection of bacteria of the genus Burkholderia was generated from the rhizosphere of maize plants. They were isolated from 60 different soil samples obtained from 4 maize cultivation sites located in the state of Guanajuato, Mexico (coordinates: 20°38′19.2″ N 101°28′06.6″ W; 20°45′05.6″ N 101°23′30.6″ W; 20°39′21.6″ N 101°24′ 13.2″ W and 20°32′08.8″ N 101°32′53.2″ W). Thus, in each site, representative samples of the rhizosphere were taken from three neighboring plants situated at five randomly distributed points in the cultivation area. The samples were transported in refrigerated boxes lined with cooling gel packs and were immediately stored at 4 °C upon arrival to the laboratory.
2.2. Bacteria Isolation
The isolation of bacteria was performed using BAz medium, which is widely used for Burkholderia spp. [14]. However, some modifications in the formulation were applied to expand the diversity of the isolated species. Briefly, the media contained the following components (in g/L): carbon source, 2.0; K2HPO4, 0.4; KH2PO4, 0.4; MgSO4·7H2O, 0.2; CaCl2, 0.02; Na2MoO4·H2O, 0.002; FeCl3, 0.01; bromothymol blue, 0.075; yeast extract, 0.5; and agar, 15.0. It was adjusted to a pH of 5.7 with KOH (50%). In this study, azelaic acid or mannitol were used as carbon sources. After autoclaving (121 °C during 20 min), 70 mg/L of cycloheximide sterilized by microfiltration (0.22 µm) was added to inhibit fungal growth.
Ten grams of soil from each sample were placed in 90 mL of a 0.85% NaCl solution and were homogenized with constant stirring at 150 rpm for 60 min. Next, the denser soil aggregates were allowed to settle for 30 min at room temperature. Serial dilutions (10−2; 10−3 and 10−4) of the clarified soil suspensions were prepared, and 1 mL of each dilution was spread on the BAz solid media. Petri dishes were incubated for 48 h at 28 °C. Three independent replicates of each dilution and carbon source were used. Single, morphologically distinct colonies were selected for purification. These were streak-plated repeatedly onto fresh BAz media until axenic cultures were obtained.
2.3. Phylogenetic Identification and Classification
To obtain genomic DNA, the selected strains were grown in a nutrient broth for 24 h at 100 rpm and 28 °C. Then, 1 mL aliquots of the cultures were centrifuged at 5000 rpm for 10 min. Genomic DNA extraction from the bacterial cell pellets was performed using the PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA). The amplification of 16S rRNA gene fragments was performed by PCR amplification using the bacterial universal primers 27F and 1492R. The reaction conditions were the following: an initial cycle of denaturation (95 °C/3 min), followed by 35 amplification cycles (95 °C/30 s; 48 °C/30 s and 72 °C/1.5 min) and a final elongation step at 72 °C for 2 min. The PCR products containing the amplified 16S rRNA gene fragments were purified using the PureLink Quick PCR commercial package (Invitrogen) and sent for sequencing to the National Laboratory of Genomics for Biodiversity (Langebio; Cinvestav; Advanced Genomics Unit at Irapuato, Mexico).
The sequences generated were manually edited and analyzed using the CLC Sequence Viewer 7 program (CLC Bio). To identify the bacteria, each sequence was aligned using the EzBioCloud 16S database [34]. Phylogenetic analyses were performed using the maximum likelihood method along with the MEGA-X program [35]. The “find best model” tool was used to evaluate the substitution models. The approximation analysis (or “bootstrap”) was performed with 1000 replicates.
2.4. Fungal Strain Culture Conditions
Two fungal strains of the genus Fusarium were used: (1) Fusarium oxysporum f. sp. radicis-lycopersici (ATCC 60095 collection), kindly provided by Dr. Dora Linda Guzman (Mycotoxins laboratory, Cinvestav at Irapuato, Mexico), and (2) Fusarium verticillioides (Saccardo) Nirenberg MF-257 (ATCC 36792 collection). A conidia suspension of each strain was prepared from mature potato dextrose agar (PDA) in 10-day cultures. Briefly, the PDA plates were covered with 5 mL of sterile 0.01% Tween 20, and the fungal colonies were gently scraped using a sterile slide. After 5 min, the suspension was transferred to a tube and centrifuged at 5000 rpm for 10 min. The supernatant was discarded, and the remaining conidia were resuspended in 10 mL of 0.01% Tween 20. Aliquots containing 1 × 106 conidia/mL were prepared and stored at 4 °C.
2.5. In Vitro Evaluation of Antagonistic Activity
The in vitro antagonistic activity was assessed by the dual culture technique [36]. Thus, each soil Burkholderia strain isolate was confronted against the two Fusarium strains mentioned above. A pre-inoculum of each Burkholderia strain was prepared in 10 mL of nutrient broth. After 24 h of incubation (150 rpm at 28 °C), 1 mL aliquots were centrifuged at 5000 rpm for 5 min. The pellets were suspended in a sterile 0.85% NaCl solution to obtain an OD595 of 0.2. The bacterial inocula were always prepared fresh just before the confrontation assay. Dual culture assays were performed in 8 cm Petri dishes containing PDA. Twenty µL aliquots of bacterial inocula were streaked on the agar, and then 10 µL of the conidia suspension was inoculated on sterile filter paper discs (0.7 cm in diameter) that were placed 5 cm from the bacteria streak on the opposite side of each plate. The control treatments consisted of plates inoculated with the respective fungal pathogens only. Fungal growth inhibition was recorded as the percentage reduction of the fungal colony’s radial growth 10 days after incubation at 28 °C. Five (F. verticillioides) and three (F. oxysporum) biological replicates of fungal inhibition growth assays per bacterial isolate were performed. The antagonistic activity data was analyzed by a standard analysis of variance (ANOVA) and Tukey’s post hoc tests with a significance level of p ≤ 0.05. These were performed using R Studio software [37].
2.6. Evaluation of Plant Growth Promotion Traits
Indoleacetic acid (IAA) production. IAA production was evaluated in Erlenmeyer flasks containing 50 mL of nutrient broth supplemented with 100 µg/L of L-tryptophan (Sigma, St. Louis, MO, USA). Aliquots (100 µL) of bacterial inocula were incubated for 72 h (150 rpm at 28 °C). Culture supernatants were obtained by centrifugation at 10,000 rpm for 5 min. IAA quantification was performed according to the procedure of Glickmann and Dessaux [38]. Briefly, 1 mL of supernatant was mixed with 2 mL of a Salkowski reagent (FeCl3 4.5 g/L in 10.8 M H2SO4). The mixture was left in the dark for 30 min at room temperature, followed by measurement of the absorbance at 450 nm using a microplate spectrophotometer (TECAN GENios; Groedig, Austria). Three biological replicates per bacterial isolate were performed.
Phosphate solubilization. The quantitative estimation of phosphate solubilization (PS) was conducted using Erlenmeyer flasks containing 100 mL of PVK medium [39]. These were performed in triplicate with inocula of each bacterial strain. The cultures were incubated in a rotatory shaker (150 rpm) for 4 days at 28 °C. The cell biomass was separated by centrifugation (10,000 rpm for 15 min), and the amount of soluble phosphate in the supernatants was determined by the ascorbic acid colorimetric method at 880 nm [40]. These assays were performed in a microplate spectrophotometer (Bio-Rad, Hercules, CA, USA).
Nitrogen fixation activity. To determine the nitrogen fixing capability, an acetylene reduction assay (ARA) was performed essentially as described by Cavalcante and Döbereiner [41]. Briefly, 10 mL serum vials were filled with 5 mL of a semi-solid LGI medium prepared with some modifications. Its composition in g/L was as follows: K2HPO4, 0.2; KH2PO4, 0.6; MgSO4, 0.2; CaCl2, 0.02; Na2MoO4, 0.002; FeCl3, 0.01; bromothymol blue, 0.075; agar, 3.0; and sucrose, 5.0. The medium was adjusted to a pH of 6.0 with KOH. It was inoculated with 25 µL of bacterial inocula and incubated for 48 h at 28 °C. The acetylene reduction was estimated by changing the cotton plugs of the vials with rubber covers immediately after the injection of 13% v/v acetylene (C2H2) gas. After 24 h, the ethylene (C2H4) concentration was estimated in a gas chromatograph (YL6500-GC; Anyang, Korea) equipped with a 30 m long HP-Plot/Q column (0.53 mm internal diameter) and a flame-ionization detector. The gas chromatograph was operated at 120 °C using helium as the carrier gas. Three biological replicates of each bacterium were analyzed.
Exopolysaccharide (EPS) production. The Congo red agar method was used as a qualitative test to detect the capacity to produce exopolysaccharides [42,43]. A single colony of each strain was streaked on an ATCC no. 14 medium supplemented with 0.8 g/L of Congo red stain. The cultures were incubated for 48 h at 28 °C. An EPS positive result was indicated by the development of black colonies, while the colonies of non-slime producers remained pink.
2.7. Analysis of Extracellular Metabolite Production
Sample preparation. The secreted metabolites produced by the Burkholderia strains analyzed were obtained from bacteria cultured in M9 minimal medium. Thus, 100 µL aliquots of bacterial inocula were cultured in 150 mL of M9 medium containing the following in g/L: Na2HPO4, 6.0; KH2PO4, 3.0; NaCl, 0.5; NH4Cl, 1.0; MgSO4, 0.2; CaCl2, 0.01; and glucose, 2.0. The medium was adjusted to a pH of 7.4 with 50% KOH. After 48 h, with stirring at 150 rpm and at 28 °C, the cultures were centrifuged at 10,000 rpm for 30 min. The cell-free supernatants were filter-sterilized twice to remove bacterial biomass, first with a Nalgene 0.45-µm nylon syringe filter and then using a 0.22-µm polyvinylidene fluoride membranes (Millex GV; Carrigwohill, Cork, Ireland). The controls consisted of uninoculated M9 medium.
Solid phase separation (SPE) using 300 mg Maxi-Clean Sep-Pak C18 sorbent cartridges (Waters Corporation; Milford, MA, USA) was used as the sample preparation method. The cartridges were conditioned at a flow rate of 1 mL/min with 5 mL of methanol and 5 mL of ultra-pure water. The samples (100 mL) were loaded into the cartridges using a vacuum to achieve a flow rate of 10 mL/min. The sample concentration was achieved by eluting the material held by the SPE cartridges with 1 mL of 50% chromatography-grade methanol (LiChrosolv; Merck, Darmstadt, Germany). Prior to DIESI-MS analysis, the metabolites were filtered through 0.22 µm pore size Captiva Econofilter polytetrafluoroethylene membranes (Agilent Technologies Inc.; Santa Clara, CA, USA), and the samples were stored at −60 °C until used. Biological replicas were obtained from three independent bacterial cultures and extractions.
Untargeted DIESI-MS analysis. DIESI-MS was used as previously described [44,45]. The samples were injected directly with a constant 10 μL/min flow rate with a syringe pump loaded with 100 μL of the sample extract. The settings for the SQ2 equipment (Waters) were the following: 3 kV voltage for the capillary and 30 V for the cone and temperatures set to 100 °C at the source and 250 °C for the solvation gas. The desolvation gas flow was 250 L/h and 50 L/h in the cone. The mass software implemented for data acquisition was MassLynx™ 4.1 software (Waters). Continuous spectra were collected in a range of 0–800 m/z in positive ion mode only. Each reading lasted 5 min and always included at least six technical replicates of three biological replicates per bacterial cell-free extract. Due to ion suppression effects, no collection of data in the negative ionization mode was possible.
MS data treatment and evaluation. MS raw data preprocessing for peak alignment and binning was performed with R statistical software [46] using ChemometricsWithR [47] and previously reported scripts [44,48]. MetaboAnalyst 5.0 [49] was implemented for data transformation and multivariate statistical analyses. The MS data were scaled using the autoscaling function (mean-centered and divided by the standard deviation of each variable). One-way ANOVA was executed with a Tukey’s post hoc test and a p-value threshold of 0.001 to assign statistical significance to the difference in ion intensity signals detected in the profiles produced by each of the strains studied. Multivariate unsupervised and supervised methods such as principal component analysis (PCA), random forest (RF) and hierarchical cluster analysis were employed to group the Burkholderia strains according to their exometabolome footprints.
MS/MS method. XEVO TQD equipment (Waters) was used for the MS/MS analysis, which was set to the following parameters: a 3 kV capillary voltage and 30 V for the cone, 150 °C source temperature and 250 °C for the desolvation gas. The desolvation gas flow rate was 250 L/h and reduced to 50 L/h at the cone. The samples were injected directly with a constant 10 μL/min flow rate. MS fragmentation allowed the identification of metabolites by employing the Daughters method of Masslynx 4.1 as described by Peniche-Pavía and Tiessen [50]. Six energy levels were applied: 10, 20, 40, 50, 60 and 70 V. Parent ion selection was performed based on ions with a significant difference in ANOVA MS analysis and included differential (highest mean decrease in accuracy value) ions obtained in the RF tests. A commercial standard of pyochelin (Toronto Research Chemicals, Toronto, ON, Canada), a known Burkholderia siderophore, was used for identification purposes.
3. Results
3.1. Identification and Classification of the Isolated Rhizobacteria
Twelve Burkholderia and Paraburkholderia strains were isolated from the soil samples using the semi-selective BAz medium. High sequence similarities (>97%) with the 16S rRNA genes compiled in the EzBioCloud 16S database permitted the identification of the Burkholderia sensu lato strains listed in Table S1. Ten different species were identified: Burkholderia contaminans MSR2; B. arboris 1Ac4; B. metallica 1Ac2; B. ubonensis PEI4; B. gladioli 1Ac1; B. gladioli 2 Ma15; B. gladioli 1Ma4; Paraburkholderia kirstenboschensis SCV25; P. graminis SCV16; P. dilworthii FCV2; P. rhynchosiae SCV21; and Trinickia dinghuensis 2Ma17 [51] basonym P. dinghuensis [52].
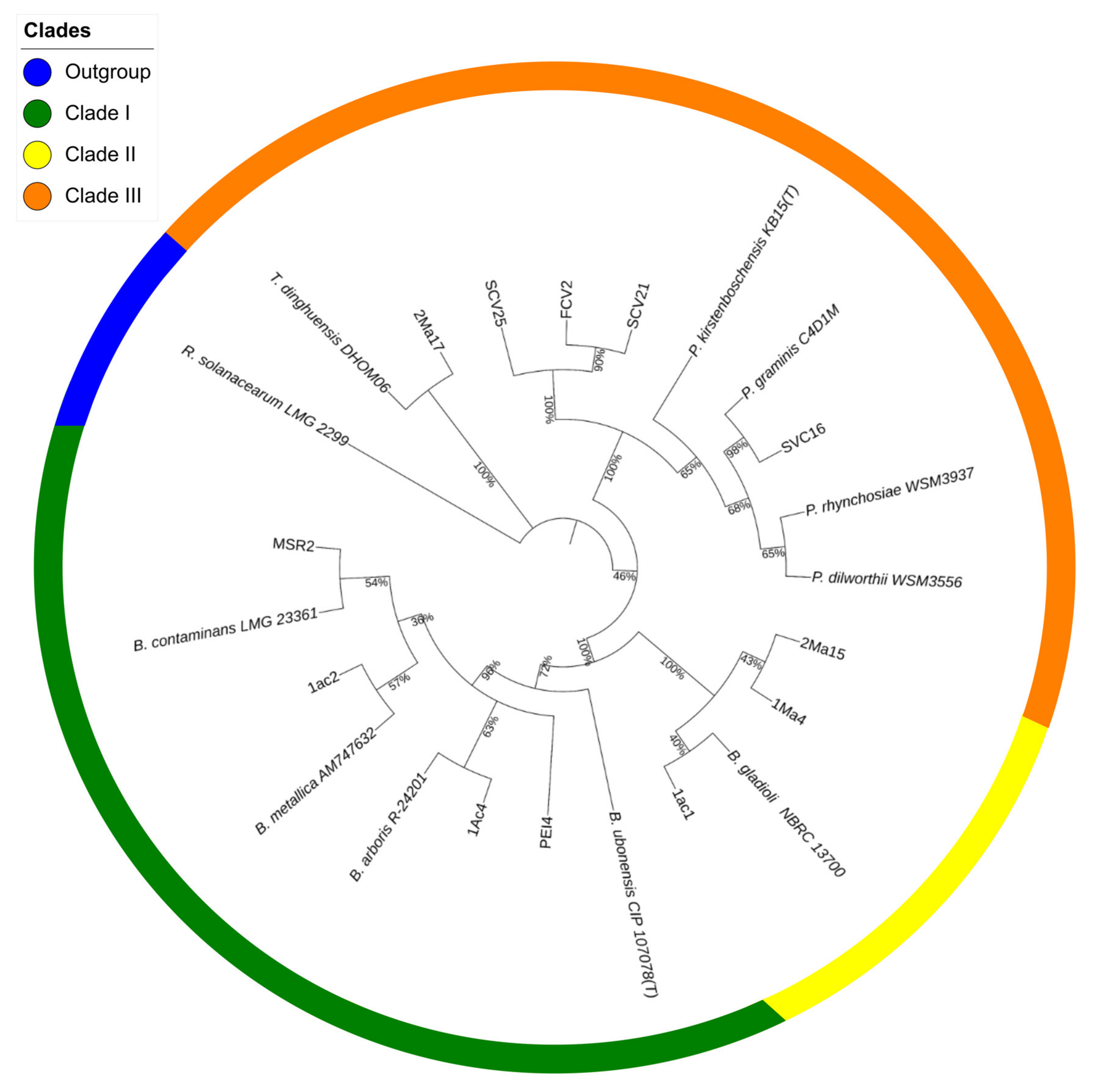
The sequencing data were used to construct a phylogenetic tree showing the position of the rhizosphere strains in relation to the type strains of species of the Burkholderia, Paraburkholderia, and Trinickia genera that conformed to the tree’s major clusters (Figure 1).
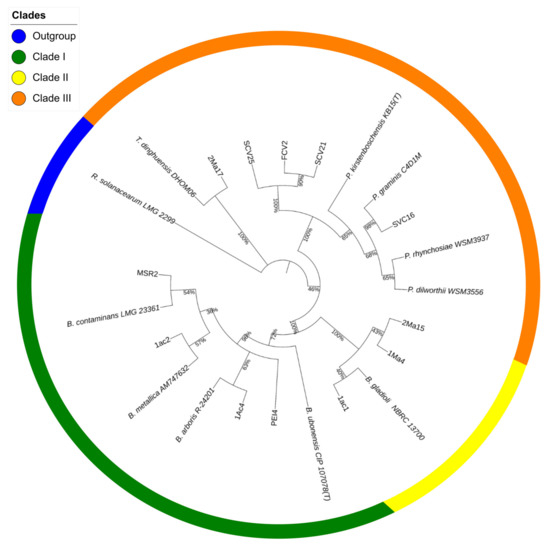
Figure 1.
Maximum likelihood phylogenetic tree of rhizosphere Burkholderia sensu lato strains. The tree was constructed based on a comparative analysis of 16S rRNA gene sequences. Shown is the phylogenetic position of isolated Burkholderia sensu lato strains in relation to Burkholderia-type strains. The numbers at the nodes represent percentage levels of bootstrap support from 1000 replications. The sequence of Ralstonia solanarum LMG 2299, used as an outgroup, roots the tree.
Cluster I showed two subdivisions: one grouped the PEI4 and 1Ac4 strains together with the B. arboris R-24201 type strain, while the other placed the 1Ac2 and MSR2 strains along with the B. contaminans LMG 23361- and B. metallica AM747632-type strains. Cluster II, closely related to the B. gladioli NBRC 13700-type strain, was formed with the 1Ma4, 2Ma15 and 1Ac1 strains. Strain 2Ma17 was grouped separately in cluster III with the T. dinghuensis DHOM06-type strain. The SCV25, SCV21, FCV2 and SVC16 strains were grouped in a subdivision of this cluster together with the type strains of the Paraburkholderia genus. The percentage of similarity of each sequence with respect to the type strains is shown Table S1.
3.2. In Vitro Antagonistic Activity against Fusarium spp.
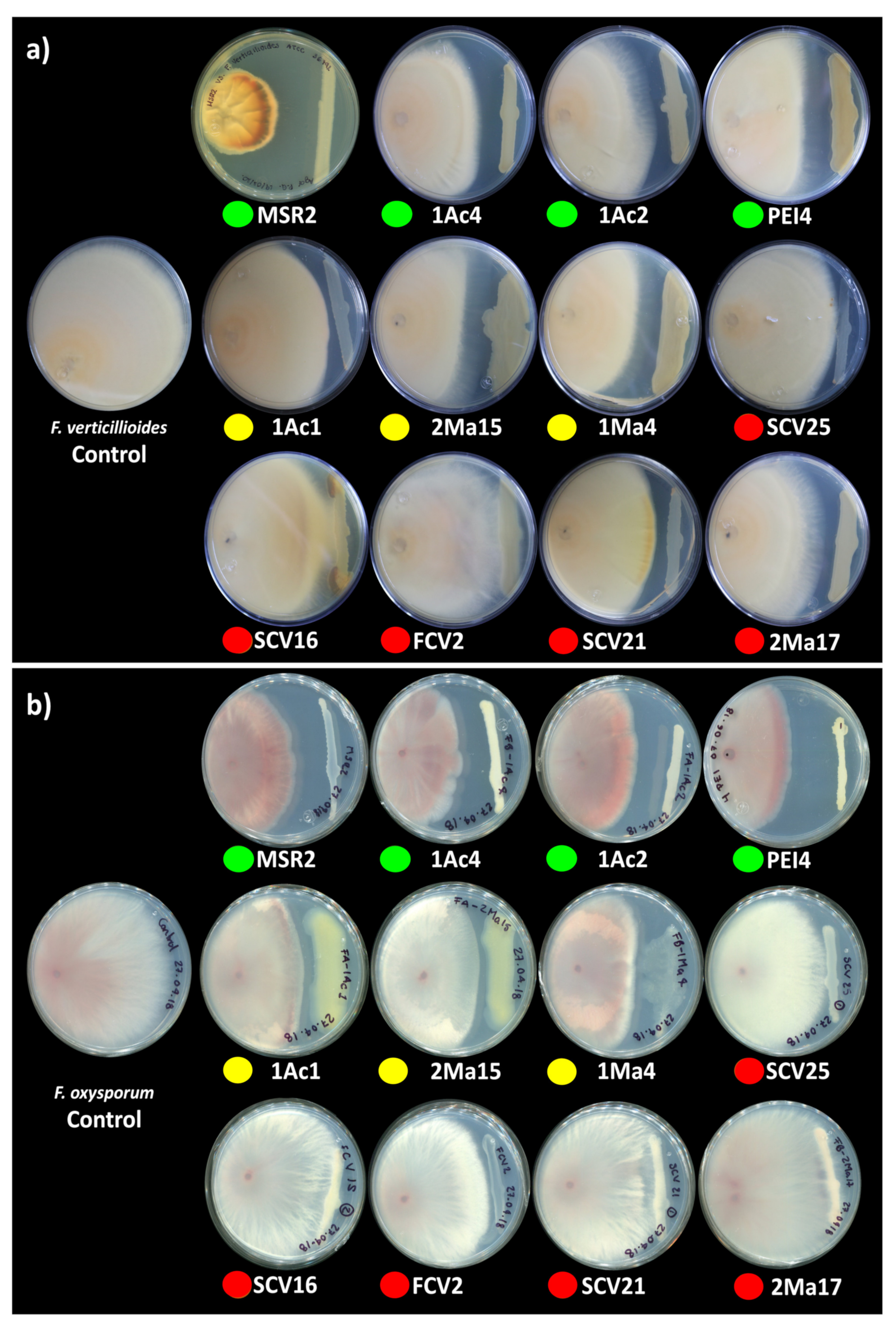
The potential biological control capacity of each Burkholderia sensu lato strain was tested by measuring their capability to inhibit the hyphal growth of F. verticillioides MF-257 and F. oxysporum f. sp. radicis-lycopersici. The antagonism of Burkholderia sensu lato strains against these phytopathogens was scored by measuring the inhibition zones produced between the fungal mycelium and the bacterial streaks (Figure 2).
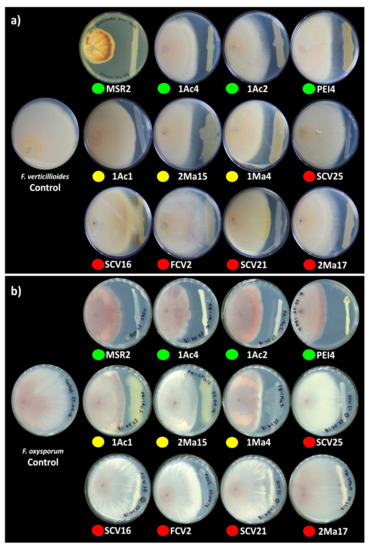
Figure 2.
Antagonism of rhizosphere Burkholderia sensu lato strains against phytopathogenic Fusarium. Antagonism vs. (a) F. verticillioides MF-257 and (b) F. oxysporum f. sp. radicis-lycopersici was determined by the capacity of rhizosphere Burkholderia strains to inhibit their radial hyphal growth when placed in direct confrontation. The single plates in the left end of the image correspond to the negative controls. The colored circles below the plates represent the different Burkholderia sensu lato groups: green = Clade I; yellow = Clade II; and orange-red = Clade III. The results shown are representative of experiments that were performed with five (F. verticillioides) and three (F. oxysporum) replicates per bacterial isolate.
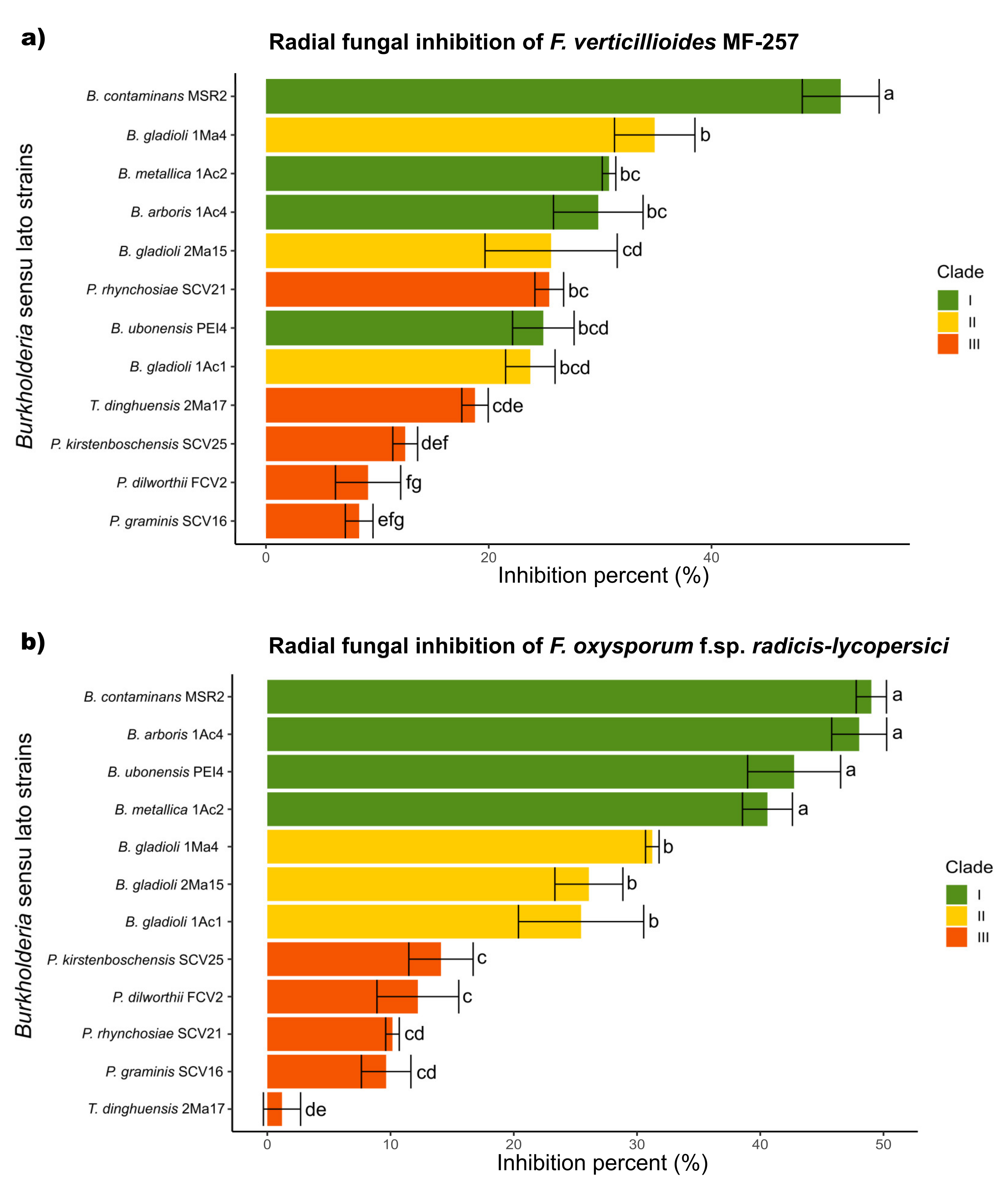
In general, the mycelial radial growth of both pathogenic fungi was reduced by all the Burkholderia sensu lato strains evaluated. The inhibition percent ranged from 3 to 52%, as summarized in Figure 3. Burkholderia contaminans MSR2 produced the significantly highest inhibition of the radial growth of F. verticillioides (52.0 ± 1.7) and F. oxysporum (48.9 ± 1.4%). Furthermore, visible changes in mycelial coloration were observed when B. contaminans MSR2 was confronted with each phytopathogen, as shown in Figure 2. The Burkholderia arboris 1Ac4, B. metallica 1Ac2 and B. ubonensis PEI4 strains also had elevated antagonistic activity (i.e., 41–47%) against F. oxysporum, while their capacity to suppress the mycelial growth of F. verticillioides was lower (i.e., 25–30%). The least inhibitory activity was observed in the clade grouping the Paraburkholderia species (Figure 2 and Figure 3).
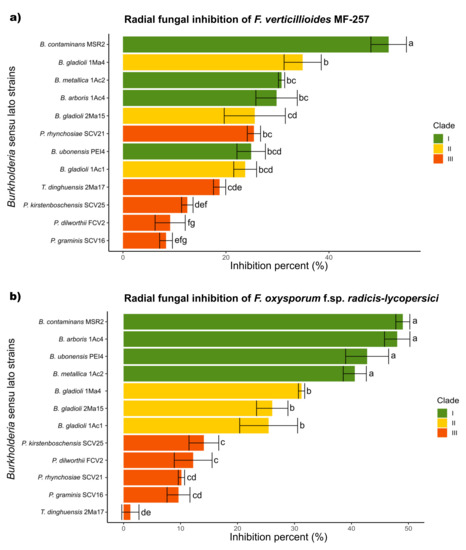
Figure 3.
Antifungal activity of Burkholderia sensu lato strains against Fusarium pathogens. The colored bars represent the percentage of inhibition produced against (a) F. verticillioides MF-257 and (b) F. oxysporum f. sp. radicis-lycopersici by Burkholderia sensu lato bacteria isolated from the rhizosphere of maize plants. The bar colors represent their different phylogenetic classifications: green = Clade I; yellow = Clade II; and orange-red = Clade III. Data are the mean values (±std. error) representative of experiments that were performed with five (F. verticillioides) and three (F. oxysporum) biological replicates per bacterial isolate. Different letters alongside the inhibition percent values represent statistically significant differences (one-way ANOVA, Tukey test, p < 0.05).
3.3. Plant Growth-Promoting Capability
The in vitro plant growth promotion attributes of the Burkholderia sensu lato strains are presented in Table 1. Significant IAA production was detected in all clade II strains. Burkholderia gladioli 1Ma4 at 114.6 ± 3 μg/mL was, by far, the highest IAA producer among all the rhizosphere Burkholderia sensu lato strains analyzed, whose IAA levels fluctuated between 0.5 and 10.2 µg/mL. Seven Burkholderia sensu lato strains were able to solubilize inorganic phosphate. Although phosphate-solubilizing strains were distributed in all Burkholderia groups, a significantly major solubilization capability was observed in clade III, specifically in P. rhynchosiae SCV2, P. graminis SCV16 and P. kirstenboschensis SCV25. The ARA yielded nitrogenase activity that ranked between 8.2 and 12.5 nmol/mL·h. The highest activity was detected in the B. gladioli 1Ac1 strain, whereas the EPS qualitative screening indicated that eight Burkholderia sensu lato strains were positive for the test, as verified by changes in the colonial morphology and in the culture media (Figure S1).

Table 1.
Plant growth-promoting activities of rhizosphere Burkholderia sensu lato strains.
3.4. DIESI-MS Metabolic Footprinting
3.4.1. Principal Component Analysis
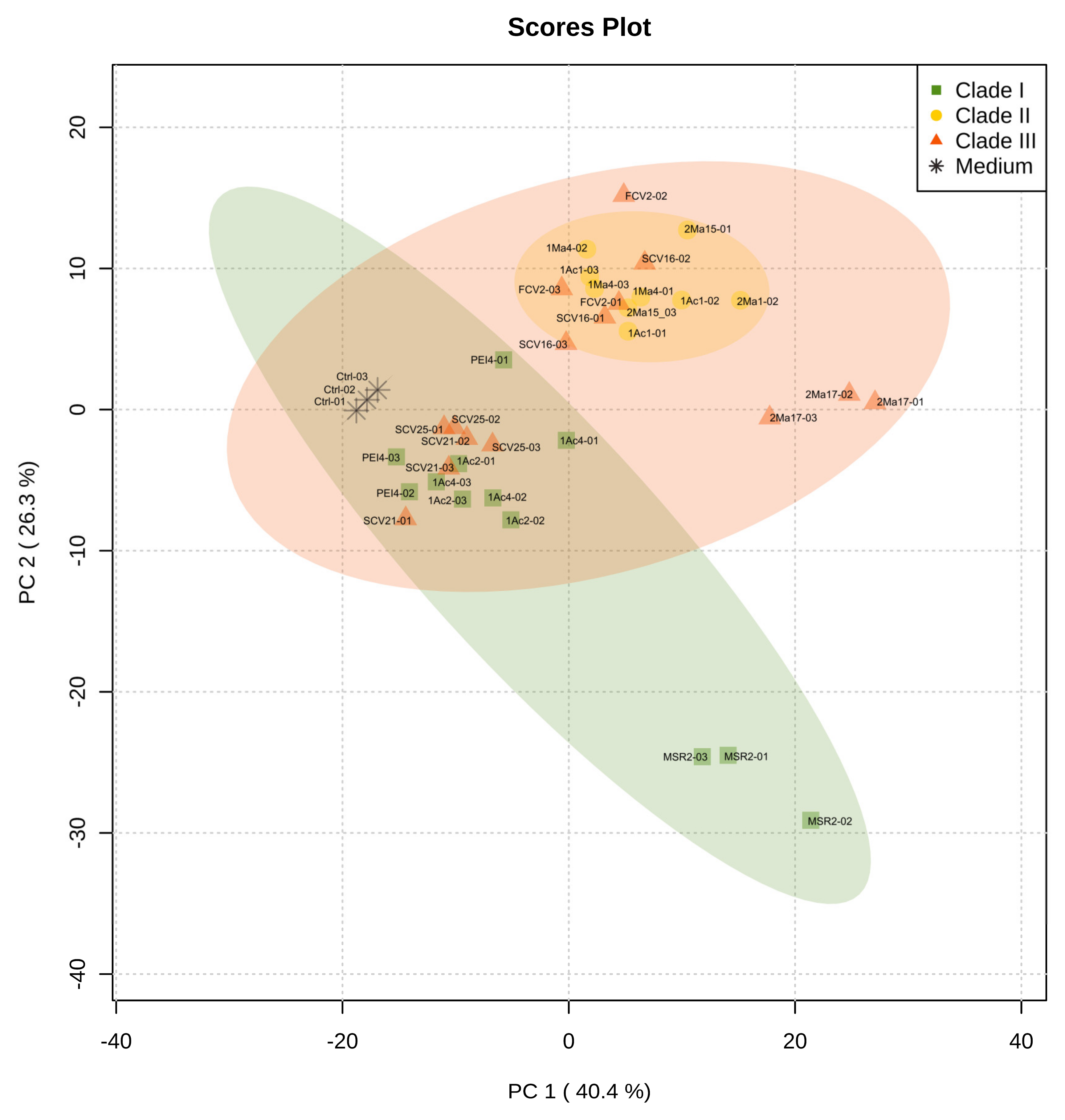
Metabolic footprinting of Burkholderia sensu lato strains by DIESI-MS resulted in mass spectra that highlighted the similarities and differences between the 12 strains analyzed (Figure S2). After data binning and filtering, 364 ions within an m/z range of 200–800 were selected for further analysis. Exo-metabolome comparisons among Burkholderia strains were performed by unsupervised multivariate PCA to assess the similarities between the bacteria. As illustrated in Figure 4, the most important sources of data variability (66.7%) were explained by components PC1 (40.4%) and PC2 (26.3%). Clade I and II strains clearly separated into two well-defined groups. In contrast, the bacteria of clade III grouped with the isolates belonging to either clade I or clade II. Thus, P. kirstenboschensis SCV25 and P. rhynchosiae SCV21 were grouped together with members of the clade I strains, whereas P. dilworthii FCV2 and P. graminis SCV16 were closer to the strains of clade II. The T. dinghuensis 2Ma17 and B. contaminans MSR2 strains clearly separated from the rest of the Burkholderia strains (Figure 4).
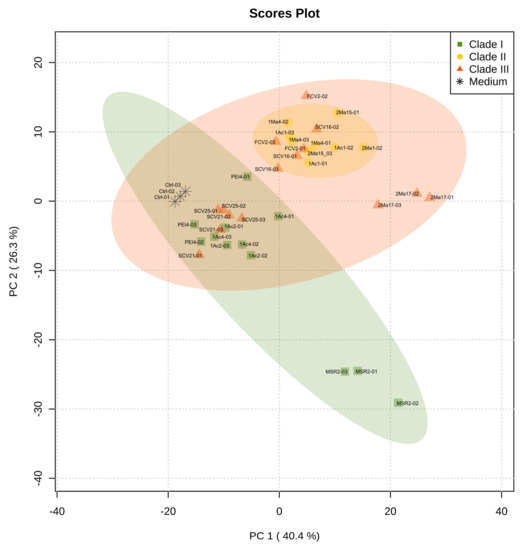
Figure 4.
Principal component analysis (PCA) obtained from DIESI-MS analysis of culture extracts of the rhizosphere Burkholderia sensu lato strains. Different colored symbols represent the phylogenetic clustering resulting from previous 16S rRNA analysis: green squares (clade I), yellow circles (clade II) and orange-red triangles (clade III). Black asterisks correspond to uninoculated M9 medium controls.
3.4.2. Hierarchical Clustering Analysis
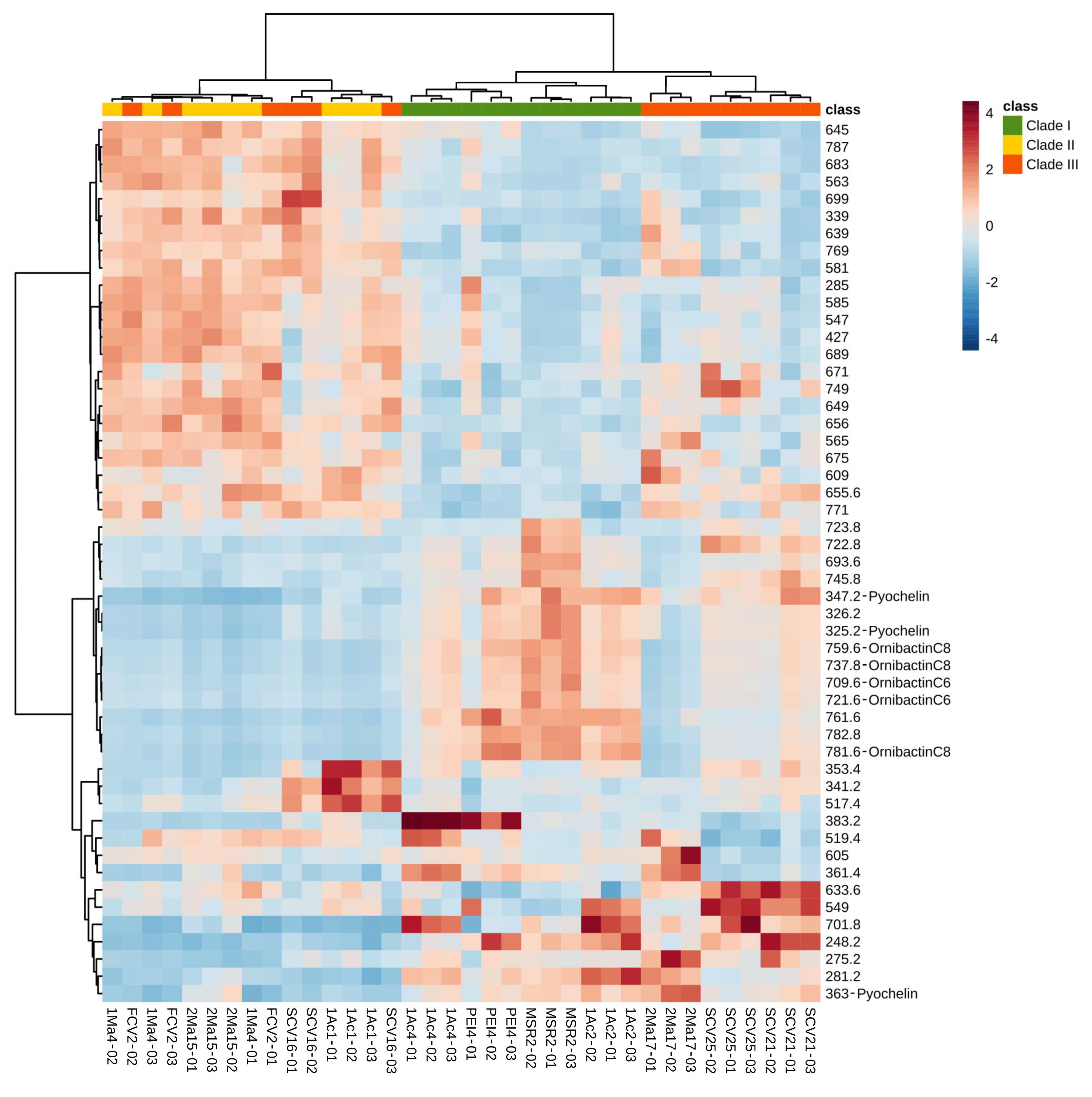
To categorize and select the important metabolic features of the Burkholderia sensu lato strains, 50 discriminant ions were chosen according to their mean decrease accuracy valued, as determined by the RF model. This strategy’s objective was to reduce the data complexity and detect significant differences among the DIESI-MS profiles. The Euclidian distance and Ward’s algorithm were applied to the ions selected in order to obtain the chemical profiles of the Burkholderia sensu lato strains. The results are presented as a heatmap depicting the abundance of these metabolite ions in the strain’s cultures (Figure 5). Hierarchical clustering of the Burkholderia sensu lato strains (top of Figure 5) showed that all clade I strains (B. contaminans MSR2, B. arboris 1Ac4, B. metallica 1Ac2 and B. ubonensis PEI4) grouped together. Conversely, some clade II strains (e.g., B. gladioli 1Ac1, B. gladioli 2Ma15 and B. gladioli 1Ma4) clustered together with two clade III strains (P. dilworthii FCV2 and P. graminis SCV16), while the rest of the clade III strains (i.e., P. rhynchosiae SCV21, P. kirstenboschensis SCV25 and T. dinghuensis 2Ma17) formed a distinct and chemically related group. As observed, the grouping of these Burkholderia sensu lato strains based on metabolic attributes showed variations from the phylogenetic classification based on the analysis of their respective 16S rRNA sequences. This difference most likely reflected the metabolic diversity that characterizes the Trinickia, Burkholderia and Paraburkholderia genera [53].
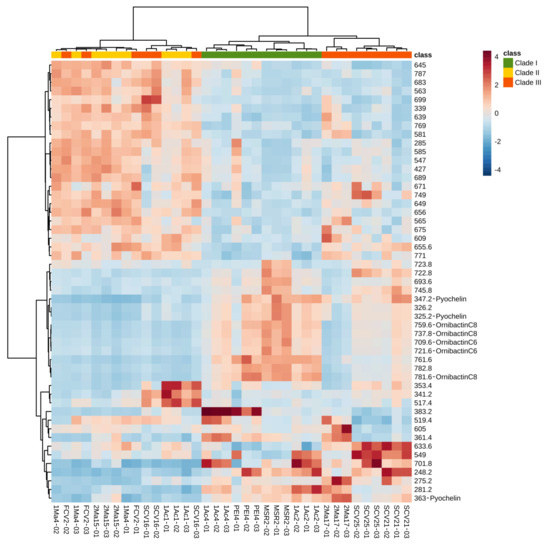
Figure 5.
Metabolic heat map based on DIESI-MS data of 50 discriminant ions selected by using the random forest model. The Euclidean distance and Ward’s algorithm were used for the hierarchical classification of rhizosphere Burkholderia sensu lato strains, listed on the bottom end of the chart, based on the respective abundance of the discriminant ions selected. Some of these were assigned an identity, as shown for ions corresponding to pyochelin and ornibactin C8 and C6. The colors at the top of the chart are representative of the grouping of the different Burkholderia sensu lato strains: clade I (green), clade II (yellow) and clade III (orange-red). The colored bars (in the blue-red range) indicate the abundance of each metabolite ion, where red indicates higher signal intensity.
3.4.3. DIESI-MSQD Analysis and MS/MS Siderophore Identification
Representative DIESI-MS spectra of the 200–800 m/z region showed the most intense ions corresponding to the exo-metabolites secreted to the culture medium by each strain (Figure S2). The identity of the most intense ions, as determined by the RF model (Figure 5), was obtained by their subsequent fragmentation (see Table 2 and Table S2 [54,55,56]). Remarkably, B. contaminans MSR2 generated ion signals with m/z values of 737.7, 759.7 and 781.6, whose maximal intensity was confirmed by an ANOVA test. Interestingly, the first ion was found to correspond to the [M+H]+ adduct of the ornibactin C8 siderophore, while the 759 and 781 m/z signals corresponded to the sodium (+23 Da) and potassium (+39 Da) adducts of ornibactin C8, respectively (Figure 5 and Table 2).

Table 2.
Metabolites secreted by Burkholderia sensu lato maize rhizosphere strains as identified by MS/MS analysis.
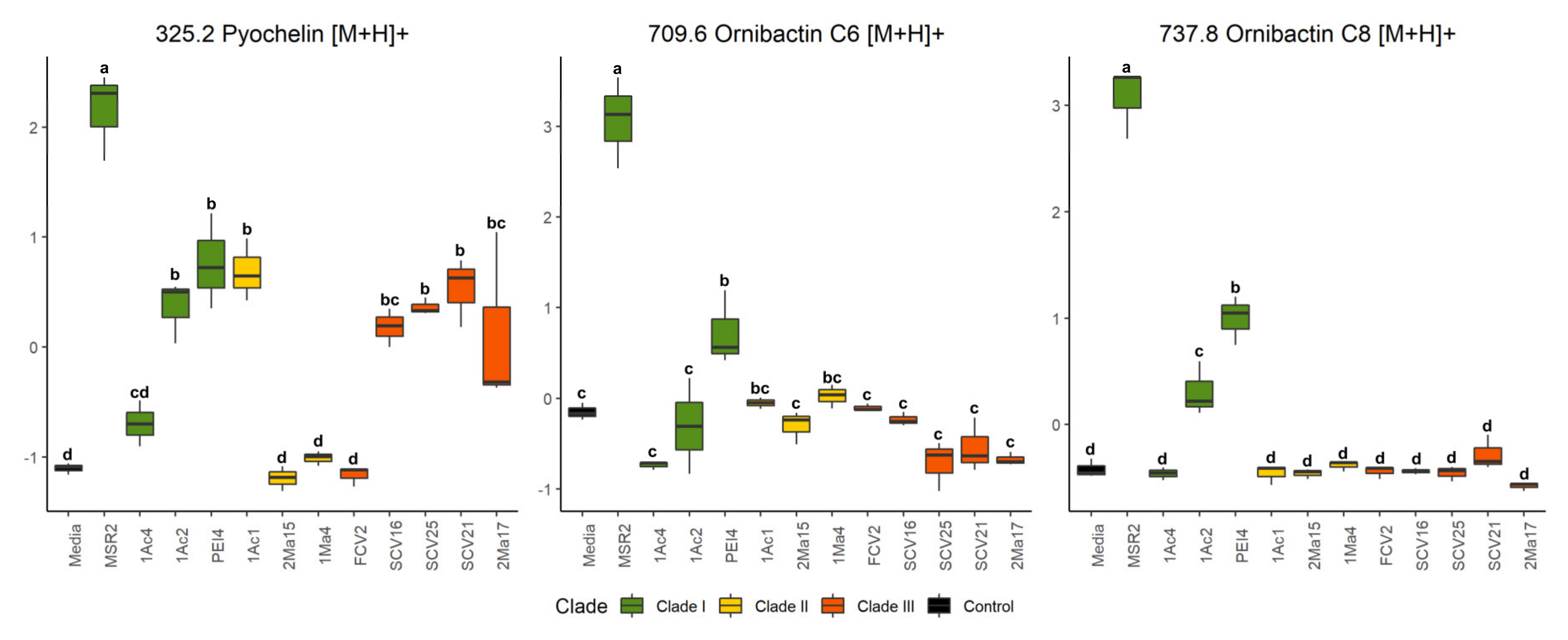
In addition to B. contaminans MSR2, the signature ions of this metabolite were also intense in clade I B. ubonensis PEI4 and B. arboris 1Ac2 (Figure 6). The secretion of ornibactin C6 (709 m/z, [M+H]+ and 721 m/z, [M+Na]+; Table 2) was also detected in the cultures of the B. contaminans MSR2 strain, while no significant accumulation of these siderophore signals was detected in the rest of clade I strains (Figure 6). The intensity of these ions was also very weak in the samples of strains conforming clades II and III. Conversely, the pyochelin siderophore (i.e., 325 [M+H]+ and 347 [M+Na]+ m/z ions; Table 2) was produced by most of the strains analyzed.
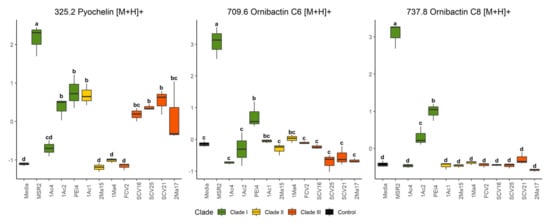
Figure 6.
Quantification of siderophore accumulation in the culture media of rhizosphere Burkholderia sensu lato strains. The box plots represent the normalized intensities of the ion signals identified as part of the pyochelin (m/z 325.2), ornibactin C6 (709.6) and ornibactin C8 (737.8) siderophores. These were detected in the culture media used to grow different rhizosphere Burkholderia sensu lato strains. Different letters over the box plots represent statistically significant differences (one-way ANOVA, Tukey test, p < 0.05). The box plot colors are representative of the grouping of the different Burkholderia sensu lato strains: clade I (green), clade II (yellow) and clade III (orange-red).
The ANOVA test showed that, like ornibactin C6 and C8, the MSR2 strain secreted the highest concentration of pyochelin compared with all other Burkholderia strains analyzed (Figure 6). Among these, the clade III Paraburkholderia strains secreted intermediate amounts of the latter siderophore. Included in this group were P. graminis SCV16, P. rhynchosiae SCV21, P. kirstenboschensis SCV25 and T. dinghuensis 2Ma17 but not P. dilworthii FCV2. A characteristic of clade II strains was their inability to secrete at least one of these three siderophores.
4. Discussion
This study identified rhizobacteria associated with suppressive soils able to reduce the incidence of highly damaging fungal infections of cultivated maize in central Mexico. It concentrated on those caused by F. verticillioides and F. oxysporum, which are known to severely impact worldwide maize production in tropical and subtropical latitudes [3,57]. Fusarium verticillioides is frequently reported as the causative agent of severe maize plant diseases characterized by extensive stalk, ear or root decay that may also lead to reduced grain quality [58,59]. Consistent with the high diversity of Burkholderia bacteria that is found in soils [60], the analysis of several maize rhizospheres obtained from plants sampled from diverse agricultural soils of the Bajío region of central Mexico led to the isolation and identification of several Burkholderia sensu lato strains. Traditional sequence analysis of the 16S rRNA gene was used for the identification of these strains, which were further subdivided into three clades on the basis of their sequence similarity with the strain types. This classification grouped strain 2Ma17 and the T. dinghuensis DHOM06-type strain within clade III, together with the Paraburkholderia strains. However, the deep branching observed suggests that the alternative of placing the 2Ma17 strain in a different clade is a plausible possibility. This proposal is supported by a recent study in which a maximum likelihood analysis of conserved genes from more than 100 Burkholderia sensu lato species strongly supported the grouping of the novel Mycetohabitans and Trinickia genera described into two distinct and unique clades [53]. Similar to the present study, the Trinickia cluster was positioned between those grouping the Burkholderia and Paraburholderia strains.
This and other molecular biology methods have also been used to identify Burkholderia bacteria [61,62]. However, they appear to be insufficient for correctly classifying those belonging to the Bcc, whose taxonomic status remains problematic [63]. In this respect, the analysis of whole genome sequences was proposed as the method of choice for the identification of Bcc bacteria and others that are difficult to identify by conventional techniques [64]. On the other hand, other more practical approaches based on the use of mass spectrometry methods have been developed to obtain rapid and reliable diagnostic results in clinical microbiology. These are based on the generation and interpretation of mass spectra for the fast identification of indicative pathogenic microorganisms, including bacteria relevant to this study, such as B. contaminans [65,66,67,68,69,70]. In this context, the results obtained in the present study revealed that, in contrast to the MALDI-MS-based methods mentioned above, the utilization of untargeted DIESI-MS analysis offered a rapid, accurate and cost-effective alternative for the detection and quantification of siderophores in Burkholderia cell-free cultures. Thus, MALDI-TOF analysis of the acidified ethyl acetate extracts of B. contaminans MSR2 cell-free extracts incubated in minimum M9 media supplemented with 100 µM of FeCl3 to inhibit siderophore synthesis was unable to appropriately identify ornibactin C6, ornibactin C8 and pyochelin in the relatively complex bacterial extracts, even in the absence of Fe3+. Moreover, DIESI-MS proved itself to be equivalent to the laborious and time-consuming UPLC-MS analysis for the identification of these siderophores in cell-free bacterial cultures (data not shown).
In accordance with the previously published data, the maize rhizosphere Burkholderia strains had properties known to be associated with plant growth promotion [26,71,72,73,74,75]. These were unevenly distributed among the different groups, as shown by (1) the detection of clade II B. gladioli 1Ma4 as a lone IAA hyper-accumulator; (2) the dispersal of PS and nitrogen-fixing strains in clades I and III and (3) the dominance of EPS production in the clade I strains. The detection of IAA and N-fixing activity in Bcc strains belonging to clade I and in the two others was in accordance with the studies by Suárez-Moreno et al. [26], Caballero-Mellado et al. [72], Angus et al. [73], Martínez-Aguilar et al. [76] and Estrada de los Santos et al. [77], while PS and EPS production by Bcc and Paraburkholderia bacteria similar to those grouped in clades I and III, respectively, was associated with the growth promotion of C4 plants [77,78,79,80,81]. These results strongly suggest that these Burkholderia strains could be important maize plant growth-promoting bacteria. Some of them also revealed a potential capacity to protect these plants against aggressive soil-borne fungi, such as specific Fusarium pathogens.
Thus, the in vitro confrontation assays indicated that some isolated maize rhizosphere Burkholderia strains, mainly B. contaminans and B. arboris, strongly inhibited the growth of the two pathogenic Fusarium genotypes tested. The suppression effect was associated with the accumulation of ornibactin and pyochelin siderophores, in accordance with reports linking the antifungal behavior of some Burkholderia strains via iron sequestration through the synthesis of ornibactin [82,83], malleobactin and pyochelin [19,84]. Supporting evidence for this proposal was provided by a recent study that attributed the antimicrobial activity versus antibiotic resistant pathogens detected in Burkholderia sensu stricto bacteria to several metabolites, including the ornibactin and pyochelin siderophores [85]. Additional evidence showing that these metabolites were rarely present in strains classified within the Paraburkholderia, Trinickia and other genera was also in agreement with their lack of biocontrol capacity against phytopathogenic fungi, as reported in this and other studies. In this respect, the finding that the pyochelin-producing Paraburkholderia and Trinickia strains had the lowest inhibition activity versus Fusarium suggests that this metabolite, despite its siderophore activity, has a low biocontrol potential, at least against these fungal pathogens. In addition, data from recent reports support the argument that other unidentified metabolites could have been contributing factors to the different antifungal effect observed in the present study between B. contaminans and B. arboris on one hand and the Paraburkholderia and Trinickia isolates on the other. In this respect, the superior Rhizoctonia-suppressive activity produced by P. graminis PHS1 compared with several other isolates of the Paraburkholderia and Burkholderia species was found to not depend on diffusible antimicrobial compounds but on the emission of sulfurous volatile compounds [86]. This study also suggested that differences found in the gene clusters encoding polyketide synthases and non-ribosomal peptide synthetases of unknown functions between these bacteria could lead to the synthesis of still unknown metabolites able to increase the suppressive potential of soils against fungal pathogens.
The results of the present study complement related studies that identified certain traits in Burkholderia and other rhizosphere bacteria that were linked to the control of Fusarium-related infections in maize and other cereals. Thus, activity against Aspergillus flavus and F. verticillioides was associated to the production of extracellular chitinolytic enzymes or the accumulation of antifungal metabolites, mainly by Burkholderia and Pseudomonas bacteria [87]. Siderophores and auxins were also identified as components responsible for reducing the disease severity caused by F. verticillioides in maize plants [57]. Likewise, two growth-promoting Burkholderia strains isolated from maize rhizospheres and identified as B. cenocepacia and B. contaminans showed broad spectrum antifungal activity against several pathogenic Fusarium species [88], whereas the capacity to degrade fusaric acid by a B. ambifaria strain isolated from the rhizosphere of barley contributed to the biocontrol of Fusarium wilt in barley seedlings [75]. Finally, the moderate capacity to inhibit F. verticillioides and the plant growth-promoting potential of the Paraburkholderia strains reported in this study, in addition to previously reported data [86], indicate that this marginally studied genus represents an interesting subject for future research that could employ untargeted metabolomic approaches as experimental tools. This effort could compensate for the scarce information regarding the metabolites synthesized by the numerous species that conform to the Paraburkholderia genus.
From a technical perspective, it may be concluded that the DIESI MS methodology offers a rapid and cost-effective approach for the identification of key metabolite-associated pathogen inhibitory effects and, potentially, the growth promotion capability of novel isolates, such as the rhizosphere Burkholderia sensu lato strains described in this study. This was demonstrated by the identification of biocontrol-related metabolites, such as siderophores, which were associated with the capacity of B. contaminans, B. ubonensis and B. metallica to suppress the growth of Fusarium phytopathogens.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9102061/s1. Figure S1: Congo red agar assay for the qualitative detection of exopolysaccharide (EPS) production. Figure S2: DIESI-MSQD spectra of exometabolomes from rhizosphere Burkholderia sensu lato strains. Table S1: Phylogenetic similarity of bacteria isolated from maize rhizospheric soil. Table S2: Fragmentation ion pattern of selected signals obtained from DIESI-MS analysis of rhizospheric Burkholderia sensu lato culture supernatants.
Author Contributions
Conceptualization, G.C.B.-G., J.P.D.-F. and J.J.P.-C.; methodology, G.C.B.-G., J.P.D.-F. and H.A.P.-P.; software, G.C.B.-G., S.A.C. and H.A.P.-P.; validation, G.C.B.-G., S.A.C. and J.P.D.-F.; formal analysis, G.C.B.-G., S.A.C. and H.A.P.-P.; investigation, G.C.B.-G.; resources, J.J.P.-C., J.A.V.-N. and J.P.D.-F.; data curation, G.C.B.-G., S.A.C. and H.A.P.-P.; writing—original draft preparation, G.C.B.-G. and J.P.D.-F.; writing—review and editing, J.P.D.-F.; visualization, G.C.B.-G.; supervision, J.J.P.-C.; project administration, J.J.P.-C. and J.P.D.-F.; funding acquisition, J.J.P.-C. and J.P.D.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Council for Science and Technology (CONACYT, México, SEP-CONACYT 2012-179319). G.C.B.-G. and H.A.P.-P. were supported by postgraduate scholarships (Code Nos. 449551 and 492183, respectively).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The bacterial 16S rRNA gene sequencing data reported in this paper have been deposited in the GenBank® genetic sequence database under the accession numbers MZ275276 to MZ275286.
Acknowledgments
The authors thank Luis E. Hernández Rodríguez from Cinvestav and Oscar Ivan Rangel Díaz from the Autonomous University of San Luis Potosi (UASLP) for their technical assistance in the laboratory and programming work, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Coninck, E.; Scauflaire, J.; Gollier, M.; Liénard, C.; Foucart, G.; Manssens, G.; Munaut, F.; Legrève, A. Trichoderma atroviride as a promising biocontrol agent in seed coating for reducing Fusarium damping-off on maize. J. Appl. Microbiol. 2020, 129, 637–651. [Google Scholar] [CrossRef]
- Oldenburg, E.; Höppner, F.; Ellner, F.; Weinert, J. Fusarium diseases of maize associated with mycotoxin contamination of agricultural products intended to be used for food and feed. Mycotoxin. Res. 2017, 33, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Madrigal, K.Y.; Larralde-Corona, C.P.; Apodaca-Sánchez, M.A.; Quiroz-Figueroa, F.R.; Mexia-Bolaños, P.A.; Portillo-Valenzuela, S.; Ordaz-Ochoa, J.; Maldonado-Mendoza, I.E. Fusarium species from the Fusarium fujikuroi species complex involved in mixed infections of maize in Northern Sinaloa, Mexico. J. Phytopathol. 2015, 163, 486–497. [Google Scholar] [CrossRef]
- Reyes-Velázquez, W.P.; Figueroa-Gómez, R.M.; Barberis, M.; Reynoso, M.M.; Rojo, F.G.A.; Chulze, S.N.; Torres, A.M. Fusarium species (section Liseola) occurrence and natural incidence of beauvericin, fusaproliferin and fumonisins in maize hybrids harvested in Mexico. Mycotoxin. Res. 2011, 27, 187–194. [Google Scholar] [CrossRef] [PubMed]
- González-Huerta, A.; Vázquez-García, L.M.; Sahagún-Castellanos, J.; Rodríguez-Pérez, J.E.; Pérez-López, D.J. Rendimiento del maíz de temporal y su relación con la pudrición de la mazorca. Agric. Tec. Mex. 2007, 33, 33–42. [Google Scholar]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils. 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Arruda, L.; Beneduzi, A.; Martins, A.; Lisboa, B.; Lopes, C.; Bertolo, F.; Passaglia, L.M.P.; Vargas, L.K. Screening of rhizobacteria isolated from maize (Zea mays L.) in Rio Grande do Sul State (South Brazil) and analysis of their potential to improve plant growth. Appl. Soil. Ecol. 2013, 63, 15–22. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Alías-Villegas, C.; Bellogín, R.A.; del Cerro, P.; Espuny, M.R.; Jiménez-Guerrero, I.; López-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef] [Green Version]
- Colombo, E.M.; Kunova, A.; Cortesi, P.; Saracchi, M.; Pasquali, M. Critical assessment of Streptomyces spp. able to control toxigenic fusaria in cereals: A literature and patent review. Int. J. Mol. Sci. 2019, 20, 6119. [Google Scholar] [CrossRef] [Green Version]
- Raaijmakers, J.M.; Vlami, M.; de Souza, J.T. Antibiotic production by bacterial biocontrol agents. Antonie Leeuwenhoek 2002, 81, 537–547. [Google Scholar] [CrossRef]
- Soenens, A.; Imperial, J. Biocontrol capabilities of the genus Serratia. Phytochem. Rev. 2019, 19, 577–587. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Rojas-Rojas, F.U.; Tapia-García, E.Y.; Vásquez-Murrieta, M.S.; Hirsch, A.M. To split or not to split: An opinion on dividing the genus Burkholderia. Ann. Microbiol. 2016, 66, 1303–1314. [Google Scholar] [CrossRef] [Green Version]
- Mannaa, M.; Park, I.; Seo, Y.S. Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated Burkholderia species. Int. J. Mol. Sci. 2018, 20, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-de los Santos, P.; Bustillos-Cristales, R.; Caballero-Mellado, J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 2001, 67, 2790–2798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, V.M.; Estrada-de los Santos, P.; Tenorio-Salgado, S.; Vogel, J.; Stoffels, M.; Guyon, S.; Mavingui, P.; Baldani, V.L.D.; Schmid, M.; Baldani, J.I.; et al. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 2155–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero-Mellado, J.; Onofre-Lemus, J.; Estrada-de los Santos, P.; Martínez-Aguilar, L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 2007, 73, 5308–5319. [Google Scholar] [CrossRef] [Green Version]
- De los Santos-Villalobos, S.; Barrera-Galicia, G.C.; Miranda-Salcedo, M.A.; Peña-Cabriales, J.J. Burkholderia cepacia XXVI siderophore with biocontrol capacity against Colletotrichum gloeosporioides. World J. Microbiol. Biotechnol. 2012, 28, 2615–2623. [Google Scholar] [CrossRef]
- Parra-Cota, F.I.; Peña-Cabriales, J.J.; de los Santos-Villalobos, S.; Martínez-Gallardo, N.A.; Délano-Frier, J.P. Burkholderia ambifaria and B. caribensis promote growth and increase yield in grain amaranth (Amaranthus cruentus and A. hypochondriacus) by improving plant nitrogen uptake. PLoS ONE 2014, 9, e88094. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Pupin, M.; Kieu, N.P.; Chataigné, G.; Béchet, M.; Deravel, J.; Krier, F.; Höfte, M.; Jacques, P.; Leclère, V. Burkholderia genome mining for nonribosomal peptide synthetases reveals a great potential for novel siderophores and lipopeptides synthesis. Microbiol. Open 2016, 5, 512–526. [Google Scholar] [CrossRef] [Green Version]
- Vial, L.; Chapalain, A.; Groleau, M.C.; Déziel, E. The various lifestyles of the Burkholderia cepacia complex species: A tribute to adaptation. Environ. Microbiol. 2011, 13, 1–12. [Google Scholar] [CrossRef]
- Vandamme, P.; Eberl, L. Burkholderia. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Hedlund, B., Dedysh, S., Eds.; John Wiley & Sons Ltd.: Chichester, UK, 2018; pp. 1–45. [Google Scholar] [CrossRef]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, L.; Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Visca, P. Burkholderia cepacia complex species: Health hazards and biotechnological potential. Trends Microbiol. 2006, 14, 277–286. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Partida-Martinez, L.P.; Groth, I.; Schmitt, I.; Richter, W.; Roth, M.; Hertweck, C. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int. J. Syst. Evol. Microbiol. 2007, 57, 2583–2590. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Moreno, Z.R.; Caballero-Mellado, J.; Coutinho, B.G.; Mendonça-Previato, L.; James, E.K.; Venturi, V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberl, L.; Vandamme, P. Members of the genus Burkholderia: Good and bad guys. F1000Research 2016, 5, 1007. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Liu, A.X.; Guerrero, A.; Liu, J.; Yu, X.Q.; Deng, P.; Ma, L.; Baird, S.M.; Smith, L.; Li, X.D.; et al. Occidiofungin is an important component responsible for the antifungal activity of Burkholderia pyrrocinia strain Lyc2. J. Appl. Microbiol. 2015, 120, 607–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Pupin, M.; Jacques, P.; Leclère, V. Nonribosomal peptide and polyketides of Burkholderia: New compounds potentially implicated in biocontrol and pharmaceuticals. Environ. Sci. Pollut. Res. 2017, 25, 29794–29807. [Google Scholar] [CrossRef]
- Kell, D.B.; Brown, M.; Davey, H.M.; Dunn, W.B.; Spasic, I.; Oliver, S.G. Metabolic footprinting and systems biology: The medium is the message. Nat. Rev. Microbiol. 2005, 3, 557–565. [Google Scholar] [CrossRef]
- Ramírez-Ordorica, A.; Valencia-Cantero, E.; Flores-Cortez, I.; Carrillo-Rayas, M.T.; Elizarraraz-Anaya, M.I.C.; Montero-Vargas, J.; Winkler, R.; Macías-Rodríguez, L. Metabolomic effects of the colonization of Medicago truncatula by the facultative endophyte Arthrobacter agilis UMCV2 in a foliar inoculation system. Sci. Rep. 2020, 10, 8426. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- De Costa, D.M.; Erabadupitiya, H.R.U.T. An integrated method to control postharvest diseases of banana using a member of the Burkholderia cepacia complex. Postharvest Biol. Technol. 2005, 36, 31–39. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development Environment for R. RStudio, PBC. Available online: http://www.rstudio.com (accessed on 20 May 2019).
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Watanabe, F.S.; Olsen, S.R. Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from soil. Soil Sci. Soc. Ame. J. 1965, 29, 677–678. [Google Scholar] [CrossRef]
- Cavalcante, V.A.; Dobereiner, J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 1988, 108, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef] [Green Version]
- Muzammil, S.; Khurshid, M.; Nawaz, I.; Siddique, M.H.; Zubair, M.; Nisar, M.A.; Imran, M.; Hayat, S. Aluminium oxide nanoparticles inhibit EPS production, adhesion and biofilm formation by multidrug resistant Acinetobacter baumannii. Biofouling 2020, 36, 492–504. [Google Scholar] [CrossRef]
- García-Flores, M.; Juárez-Colunga, S.; García-Casarrubias, A.; Trachsel, S.; Winkler, R.; Tiessen, A. Metabolic profiling of plant extracts using direct-injection electrospray ionization mass spectrometry allows for high-throughput phenotypic characterization according to genetic and environmental effects. J. Agric. Food. Chem. 2015, 63, 1042–1052. [Google Scholar] [CrossRef]
- García-Casarrubias, A.; Winkler, R.; Tiessen, A. Mass fingerprints of tomatoes fertilized with different nitrogen sources reveal potential biomarkers of organic farming. Plant Foods Hum. Nutr. 2019, 74, 247–254. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://cran.r-project.org (accessed on 20 May 2019).
- Wehrens, R. Chemometrics with R; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar] [CrossRef]
- García-Flores, M.; Juárez-Colunga, S.; Montero-Vargas, J.M.; López-Arciniega, J.A.I.; Chagolla, A.; Tiessen, A.; Winkler, R. Evaluating the physiological state of maize (Zea mays L.) plants by direct-injection electrospray mass spectrometry (DIESI-MS). Mol. BioSyst. 2012, 8, 1658–1660. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, 1–128. [Google Scholar] [CrossRef] [PubMed]
- Peniche-Pavía, H.; Tiessen, A. Anthocyanin profiling of maize grains using DIESI-MSQD reveals that cyanidin-based derivatives predominate in purple corn, whereas pelargonidin-based molecules occur in red-pink varieties from Mexico. J. Agric. Food. Chem. 2020, 68, 5980–5994. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Lv, Y.; You, J.; Gao, Z.; Wang, B.; Qiu, L. Paraburkholderia dinghuensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1613–1620. [Google Scholar] [CrossRef]
- Gao, Z.; Zhong, S.; Qin, Y.; Yang, Z.; Lv, Y.; Qiu, L. Trinickia dinghuensis sp. nov. and Trinickia fusca sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1390–1397. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef] [Green Version]
- Lybert, A.C.; Williams, J.; Raghuvanshi, R.; Jones, A.D.; Quinn, R.A. Mining public mass spectrometry data to characterize the diversity and ubiquity of P. aeruginosa specialized metabolites. Metabolites 2020, 10, 445. [Google Scholar] [CrossRef]
- Talbot, H.M.; Sidgwich, F.R.; Bischoff, J.; Osborne, K.A.; Rush, D.; Sherry, A.; Spencer-Jones, C.L. Analysis of non-derivatised bacteriohopanepolyols by ultra high performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 2087–2098. [Google Scholar] [CrossRef] [Green Version]
- Stephan, H.; Freund, S.; Beck, W.; Jung, G.; Meyer, J.M.; Winkelmann, G. Ornibactins—A new family of siderophores from Pseudomonas. BioMetals 1993, 6, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Lopez, A.M.; Cordero-Ramirez, J.D.; Martinez-Alvarez, J.C.; Lopez-Meyer, M.; Lizarraga-Sanchez, G.J.; Felix-Gastelum, R.; Castro-Martinez, C.; Maldonado-Mendoza, I.E. Rhizospheric bacteria of maize with potential for biocontrol of Fusarium verticillioides. SpringerPlus 2016, 5, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Rodríguez, A.; Heydrich-Pérez, M.; Acebo-Guerrero, Y.; Velazquez del Valle, M.G.; Hernández-Lauzardo, A.N. Antagonistic activity of Cuban native rhizobacteria against Fusarium verticillioides (Sacc.) Nirenb. in maize (Zea mays L.). Appl. Soil Ecol. 2008, 39, 180–186. [Google Scholar] [CrossRef]
- Czembor, E.; Stępień, Ł.; Waśkiewicz, A. Effect of environmental factors on Fusarium species and associated mycotoxins in maize grain grown in Poland. PLoS ONE 2015, 10, e0133644. [Google Scholar] [CrossRef]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Barka, E.A. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef] [Green Version]
- Karlin, S.; Weinstock, G.M.; Brendel, V. Bacterial classifications derived from recA protein sequence comparisons. J. Bacteriol. 1995, 177, 6881–6893. [Google Scholar] [CrossRef] [Green Version]
- Eisen, J.A. The RecA protein as a model molecule for molecular systematic studies of bacteria: Comparison of trees of RecAs and 16S rRNAs from the same species. J. Mol. Evol. 1995, 41, 1105–1123. [Google Scholar] [CrossRef] [Green Version]
- Furlan, J.P.R.; Pitondo-Silva, A.; Braz, V.S.; Gallo, I.F.L.; Stehling, E.G. Evaluation of different molecular and phenotypic methods for identification of environmental Burkholderia cepacia complex. World J. Microbiol. Biotechnol. 2019, 35, 39. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhou, J.; Zhou, J.; Hu, M.; Zhang, Q.; Kong, N.; Ren, H.; Liang, L.; Yue, J. Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status. Biol. Direct 2020, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Keys, C.J.; Dare, D.J.; Sutton, H.; Wells, G.; Lunt, M.; McKenna, T.; McDowall, M.; Shah, H.N. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterization of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 2004, 4, 221–242. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Bizzini, A.; Greub, G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 2010, 16, 1614–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cipolla, L.; Rocca, F.; Armitano, R.I.; Martinez, C.; Almuzara, M.; Faccone, D.; Vay, C.; Prieto, M. Desarrollo y evaluación de una base de datos in house para la identificación rápida de Burkholderia contaminans por EM MALDI-TOF. Rev. Argent. Microbiol. 2019, 51, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Rocca, M.F.; Barrios, R.; Zintgraff, J.; Martínez, C.; Irazu, L.; Vay, C.; Prieto, M. Utility of platforms Viteks MS and Microflex LT for the identification of complex clinical isolates that require molecular methods for their taxonomic classification. PLoS ONE 2019, 14, e0218077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiamanya, S.; Cipolla, L.; Prieto, M.; Stelling, J. Exploring the value of MALDI-TOF MS for the detection of clonal outbreaks of Burkholderia contaminans. J. Microbiol. Methods 2021, 181, 106130. [Google Scholar] [CrossRef]
- Gyaneshwar, P.; Kumar, G.N.; Parekh, L. Role of soil microorganisms in improving P nutrition of plants. Plant Soil 2002, 245, 83–93. [Google Scholar] [CrossRef]
- Caballero-Mellado, J.; Martínez-Aguilar, L.; Parades-Valdez, G.; Estrada-de los Santos, P. Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 2004, 54, 1165–1172. [Google Scholar] [CrossRef]
- Angus, A.A.; Lee, A.; Lum, M.R.; Shehayeb, M.; Hessabi, R. Nodulation and effective nitrogen fixation of Macroptilium atropurpureum (siratro) by Burkholderia tuberum, a nodulating and plant growth promoting beta-proteobacterium, are influenced by environmental factors. Plant Soil 2013, 369, 543–562. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilising microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2013, 2, 587. [Google Scholar] [CrossRef] [Green Version]
- Simonetti, E.; Roberts, I.N.; Montecchia, M.S.; Gutierrez-Boem, F.H.; Gomez, F.M.; Ruiz, J.A. A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth. Microbiol. Res. 2018, 206, 50–59. [Google Scholar] [CrossRef]
- Martínez-Aguilar, L.; Salazar-Salazar, C.; Díaz-Méndez, R.; Caballero-Mellado, J.; Hirsch, A.M.; Vásquez-Murrieta, M.S.; Estrada-de los Santos, P. Burkholderia caballeronis sp. nov., a nitrogen fixing species isolated from tomato (Lycopersicon esculentum) with the ability to effectively nodulate Phaseolus vulgaris. Antonie Leeuwenhoek 2013, 104, 1063–1071. [Google Scholar] [CrossRef] [Green Version]
- Estrada-de los Santos, P.; Martínez-Aguilar, L.; Vinuesa, P.; Hirsch, A.M.; Caballero-Mellado, J. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr. Microbiol. 2013, 67, 51–60. [Google Scholar] [CrossRef]
- Da Silveira, A.P.D.; Iório, R.P.F.; Marcos, F.C.C.; Fernandes, A.O.; de Souza, S.A.C.; Kuramae, E.E.; Cipriano, M.A.P. Exploitation of new endophytic bacteria and their ability to promote sugarcane growth and nitrogen nutrition. Antonie Leeuwenhoek 2018, 112, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Schlemper, T.R.; Dimitrov, M.R.; Silva-Gutierrez, F.A.O.; van Veen, J.A.; Silveira, A.P.D.; Kuramae, E.E. Effect of Burkholderia tropica and Herbaspirillum frisingense strains on sorghum growth is plant genotype dependent. PeerJ 2018, 6, 5346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuramae, E.E.; Derksen, S.; Schlemper, T.R.; Dimitrov, M.R.; Costa, O.Y.A.; da Silveira, A.P.D. Sorghum growth promotion by Paraburkholderia tropica and Herbaspirillum frisingense: Putative mechanisms revealed by genomics and metagenomics. Microorganisms 2020, 8, 725. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Fang, S.; MacDonald, J.; Xu, J.; Yuan, Z. Isolation and characterization of Burkholderia cenocepacia CR318, a phosphate solubilizing bacterium promoting corn growth. Microbiol. Res. 2020, 233, 126395. [Google Scholar] [CrossRef] [PubMed]
- Lewenza, S.; Sokol, P.A. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 2001, 183, 2212–2218. [Google Scholar] [CrossRef] [Green Version]
- Deng, P.; Foxfire, A.; Xu, J.; Baird, S.M.; Jia, J.; Delgado, K.H.; Shin, R.; Smith, L.; Lu, S.E. The siderophore product ornibactin is required for the bactericidal activity of Burkholderia contaminans MS14. Appl. Environ. Microbiol. 2017, 83, e0005117. [Google Scholar] [CrossRef] [Green Version]
- Da Silva-Araújo, F.D.; Araújo, W.L.; Eberlin, M.N. Potential of Burkholderia seminalis TC3.4.2R3 as biocontrol agent against Fusarium oxysporum evaluated by mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 2017, 28, 901–907. [Google Scholar] [CrossRef]
- Depoorter, E.; De Canck, E.; Coenye, T.; Vandamme, P. Burkholderia bacteria produce multiple potentially novel molecules that inhibit carbapenem-resistant gram-negative bacterial pathogens. Antibiotics 2021, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Cordovez, V.; Tyc, O.; Etalo, D.W.; de Bruijn, I.; de Jager, V.C.L.; Medema, M.H.; Eberl, L.; Raaijmakers, J.M. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 2018, 12, 2307–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palumbo, J.D.; O’Keeffe, T.L.; Abbas, H.K. Isolation of maize soil and rhizosphere bacteria with antagonistic activity against Aspergillus flavus and Fusarium verticillioides. J. Food Prot. 2007, 70, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Tagele, S.B.; Kim, S.W.; Lee, H.G.; Lee, Y.S. Potential of novel sequence type of Burkholderia cenocepacia for biological control of root rot of maize (Zea mays L.) caused by Fusarium temperatum. Int. J. Mol. Sci. 2019, 20, 1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).