Carbapenem Resistance among Marine Bacteria—An Emerging Threat to the Global Health Sector
Abstract
:1. Introduction
2. Mechanisms of Carbapenem Resistance
3. Epidemiology and Distribution of Carbapenem Resistance
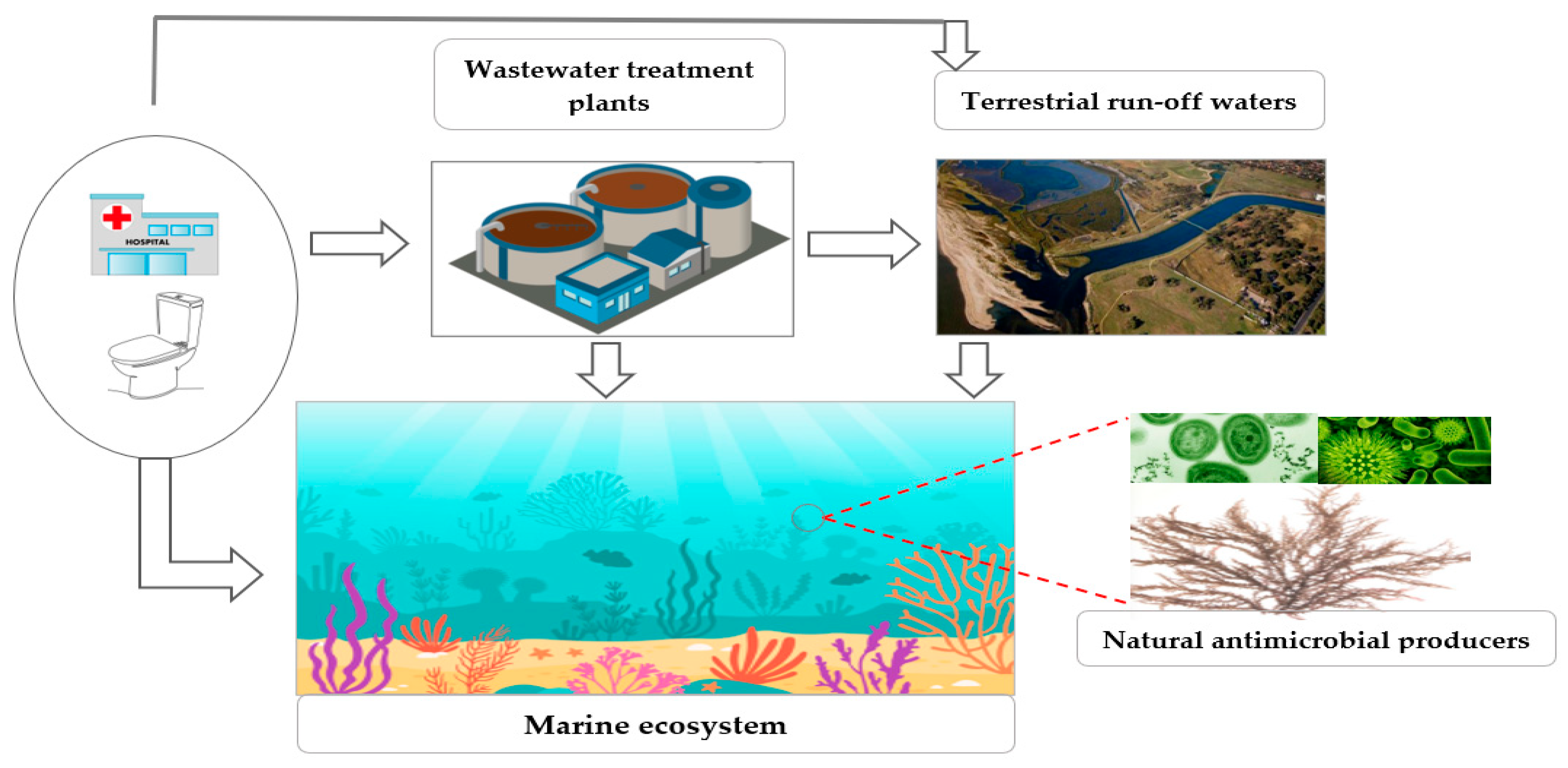
4. Distribution of Carbapenem Resistance in Marine Systems
5. Potential for CR Transfer and Reservoir in the Marine Environment
6. The Effect of Marine CRGs/CRB on Human Health
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; pp. 1–28. [Google Scholar]
- Centres for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2013. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 20 January 2021).
- Humphreys, G.; Fleck, F. United Nations Meeting on Antimicrobial Resistance. World Health Organ. Bull. World Health Organ. 2016, 94, 638. [Google Scholar]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 13 February 2021).
- Chavda, K.D.; Chen, L.; Fouts, D.E.; Sutton, G.; Brinkac, L.; Jenkins, S.G.; Bonomo, R.A.; Adams, M.D.; Kreiswirth, B. Comprehensive Genome Analysis of Carbapenemase-Producing Enterobacter spp.: New Insights into Phylogeny, Population Structure, and Resistance Mechanisms. mBio 2016, 7, e02093-16. [Google Scholar] [CrossRef] [Green Version]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic Resistance Genes and Bacterial Community Composition in a River Influenced by a Wastewater Treatment Plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef] [PubMed]
- Mokracka, J.; Koczura, R.; Kaznowski, A. Multiresistant Enterobacteriaceae with class 1 and class 2 integrons in a municipal wastewater treatment plant. Water Res. 2012, 46, 3353–3363. [Google Scholar] [CrossRef] [PubMed]
- Ocampo-Sosa, A.A.; Guzmán-Gómez, L.P.; Fernández-Martínez, M.; Román, E.; Rodríguez, C.; Marco, F.; Vila, J.; Martínez-Martínez, L. Isolation of VIM-2-Producing Pseudomonas monteilii Clinical Strains Disseminated in a Tertiary Hospital in Northern Spain. Antimicrob. Agents Chemother. 2015, 59, 1334–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Baek, J.Y.; Cho, S.Y.; Huh, H.J.; Lee, N.Y.; Song, J.-H.; Chung, D.R.; Ko, K.S. bla NDM-5 -Bearing IncFII-Type Plasmids of Klebsiella pneumoniae Sequence Type 147 Transmitted by Cross-Border Transfer of a Patient. Antimicrob. Agents Chemother. 2016, 60, 1932–1934. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Lourida, P.; Poulikakos, P.; Rafailidis, P.I.; Tansarli, G.S. Antibiotic Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae: Systematic Evaluation of the Available Evidence. Antimicrob. Agents Chemother. 2014, 58, 654–663. [Google Scholar] [CrossRef] [Green Version]
- Fritzenwanker, M.; Imirzalioglu, C.; Herold, S.; Wagenlehner, F.M.; Zimmer, K.P.; Chakraborty, T. Treatment Options for Carbapenem-Resistant Gram-Negative Infections. Dtsch. Ärzteblatt Int. 2018, 115, 345–352. [Google Scholar] [CrossRef]
- Trecarichi, E.M.; Tumbarello, M. Therapeutic options for carbapenem-resistant Enterobacteriaceae infections. Virulence 2017, 8, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirel, L.; Pitout, J.D.; Nordmann, P. Carbapenemases: Molecular diversity and clinical consequences. Future Microbiol. 2007, 2, 501–512. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile beta-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuste, E.; López-Jiménez, L.; Segura, C.; Gainza, E.; Vinuesa, T.; Vinas, M. Carbapenem-resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa. J. Med. Microbiol. 2013, 62, 1317–1325. [Google Scholar] [CrossRef]
- Partridge, S.R.; Ginn, A.N.; Wiklendt, A.M.; Ellem, J.; Wong, J.S.J.; Ingram, P.; Guy, S.; Garner, S.; Iredell, J.R. Emergence of blaKPC carbapenemase genes in Australia. Int. J. Antimicrob. Agents 2015, 45, 130–136. [Google Scholar] [CrossRef]
- Tacão, M.; Correia, A.; Henriques, I.S. Low Prevalence of Carbapenem-Resistant Bacteria in River Water: Resistance Is Mostly Related to Intrinsic Mechanisms. Microb. Drug Resist. 2015, 21, 497–506. [Google Scholar] [CrossRef]
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital Wastewater Releases of Carbapenem-Resistance Pathogens and Genes in Urban India. Environ. Sci. Technol. 2017, 51, 13906–13912. [Google Scholar] [CrossRef] [Green Version]
- Scotta, C.; Juan, C.; Cabot, G.; Oliver, A.; Lalucat, J.; Bennasar, A.; Albertí, S. Environmental Microbiota Represents a Natural Reservoir for Dissemination of Clinically Relevant Metallo-beta-Lactamases. Antimicrob. Agents Chemother. 2011, 55, 5376–5379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Søraas, A.; Sundsfjord, A.; Sandven, I.; Brunborg, C.; Jenum, P.A. Risk Factors for Community-Acquired Urinary Tract Infections Caused by ESBL-Producing Enterobacteriaceae –A Case–Control Study in a Low Prevalence Country. PLoS ONE 2013, 8, e69581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bier, N.; Schwartz, K.; Guerra, B.; Strauch, E. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front. Microbiol. 2015, 6, 1179. [Google Scholar] [CrossRef]
- Dewi, D.A.P.R.; Götz, B.; Thomas, T. Diversity and Genetic Basis for Carbapenem Resistance in a Coastal Marine Environment. Appl. Environ. Microbiol. 2020, 86, e02939-19. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Singer, A.; Ukoumunne, O.C.; Gaze, W.H.; Garside, R. Is it safe to go back into the water? A systematic review and meta-analysis of the risk of acquiring infections from recreational exposure to seawater. Int. J. Epidemiol. 2018, 47, 572–586. [Google Scholar] [CrossRef]
- Bassetti, M.; Nicolini, L.; Esposito, S.; Righi, E.; Viscoli, C. Current Status of Newer Carbapenems. Curr. Med. Chem. 2009, 16, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Kattan, J.N.; Villegas, M.V.; Quinn, J.P. New developments in carbapenems. Clin. Microbiol. Infect. 2008, 14, 1102–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, M.L.; Qin, X.; Zerr, D.M.; Weissman, S.J. Molecular diversity in mechanisms of carbapenem resistance in paediatric Enterobacteriaceae. Int. J. Antimicrob. Agents 2012, 39, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Sho, T.; Muratani, T.; Hamasuna, R.; Yakushiji, H.; Fujimoto, N.; Matsumoto, T. The Mechanism of High-Level Carbapenem Resistance in Klebsiella pneumoniae: Underlying Ompk36-Deficient Strains Represent a Threat of Emerging High-Level Carbapenem-ResistantK. pneumoniaewith IMP-1 β-Lactamase Production in Japan. Microb. Drug Resist. 2013, 19, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.M.; Yang, Q.; Duval, V.; Chen, M.; Xu, Y.; Levy, S.B. Involvement of MarR and YedS in Carbapenem Resistance in a Clinical Isolate of Escherichia coli from China. Antimicrob. Agents Chemother. 2013, 57, 1935–1937. [Google Scholar] [CrossRef] [Green Version]
- Aubron, C.; Poirel, L.; Ash, R.J.; Nordmann, P. Carbapenemase-producing Enterobacteriaceae, U.S. Rivers. Emerg. Infect. Dis. 2005, 11, 260–264. [Google Scholar] [CrossRef]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- Naas, T.; Vandel, L.; Sougakoff, W.; Livermore, D.M.; Nordmann, P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 1994, 38, 1262–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queenan, A.M.; Torres-Viera, C.; Gold, H.S.; Carmeli, Y.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Quinn, J.P.; Hindler, J.; Medeiros, A.A.; Bush, K. SME-Type Carbapenem-Hydrolyzing Class A β-Lactamases from Geographically Diverse Serratia marcescens Strains. Antimicrob. Agents Chemother. 2000, 44, 3035–3039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, B.A.; Bush, K.; Keeney, D.; Yang, Y.; Hare, R.; O’Gara, C.; Medeiros, A.A. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 1996, 40, 2080–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giakkoupi, P.; Xanthaki, A.; Kanelopoulou, M.; Vlahaki, A.; Miriagou, V.; Kontou, S.; Papafraggas, E.; Malamou-Lada, H.; Tzouvelekis, L.S.; Legakis, N.J.; et al. VIM-1 Metallo-β-Lactamase-Producing Klebsiella pneumoniae Strains in Greek Hospitals. J. Clin. Microbiol. 2003, 41, 3893–3896. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tórtola, M.T.; Lavilla, S.; Miró, E.; González, J.J.; Larrosa, N.; Sabaté, M.; Navarro, F.; Prats, G. First Detection of a Carbapenem-Hydrolyzing Metalloenzyme in Two Enterobacteriaceae Isolates in Spain. Antimicrob. Agents Chemother. 2005, 49, 3492–3494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De La Cadena, E.; Correa, A.; Muñoz, J.S.; Rojas, L.J.; Hernández-Gómez, C.; Pallares, C.; Perez, F.; Bonomo, R.A.; Villegas, M.V. Molecular characterisation of carbapenem-resistant Enterobacter cloacae complex in Colombia: blaKPC and the ‘changing landscape. J. Glob. Antimicrob. Resist. 2018, 13, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, Y.; Shen, M.; Du, J.; Zhang, L.; Wang, D. Draft genome sequence of Enterobacter cloacae HBY, a ST128 clinical strain co-producing KPC-2 and NDM-1 carbapenemases. J. Glob. Antimicrob. Resist. 2018, 12, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Nasri, E.; Subirats, J.; Sànchez-Melsió, A.; Ben Mansour, H.; Borrego, C.M.; Balcázar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374. [Google Scholar] [CrossRef]
- Poirel, L.; Le Thomas, I.; Naas, T.; Karim, A.; Nordmann, P. Biochemical Sequence Analyses of GES-1, a Novel Class A Extended-Spectrum β-Lactamase, and the Class 1 Integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2000, 44, 622–632. [Google Scholar] [CrossRef] [Green Version]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995, 39, 1211–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordmann, P.; Poirel, L.; Walsh, T.R.; Livermore, D.M. The emerging NDM carbapenemases. Trends Microbiol. 2011, 19, 588–595. [Google Scholar] [CrossRef]
- Moellering, R.C., Jr. NDM-1—A Cause for Worldwide Concern. N. Engl. J. Med. 2010, 363, 2377–2379. [Google Scholar] [CrossRef] [PubMed]
- Giske, C.G.; Martinez, L.M.; Cantón, R.; Stefani, S.; Skov, R.; Glupczynski, Y.; Nordmann, P.; Wootton, M.; Miriagou, V.; Simonsen, G.S.; et al. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. 2017. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 23 May 2021).
- Guh, A.Y.; Limbago, B.M.; Kallen, A.J. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev. Anti Infect. Ther. 2014, 12, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Wailan, A.M.; Paterson, D.L.; Caffery, M.; Sowden, D.; Sidjabat, H.E. Draft Genome Sequence of NDM-5-Producing Escherichia coli Sequence Type 648 and Genetic Context of bla NDM-5 in Australia. Genome Announc. 2015, 3, e00194-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peleg, A.Y.; Franklin, C.; Bell, J.M.; Spelman, D.W. Dissemination of the Metallo-beta-Lactamase Gene blaIMP-4 among Gram-Negative Pathogens in a Clinical Setting in Australia. Clin. Infect. Dis. 2005, 41, 1549–1556. [Google Scholar] [CrossRef] [Green Version]
- Peleg, A.Y.; Franklin, C.; Walters, L.J.; Bell, J.M.; Spelman, D.W. OXA-58 and IMP-4 Carbapenem-Hydrolyzing beta-Lactamases in an Acinetobacter junii Blood Culture Isolate from Australia. Antimicrob. Agents Chemother. 2006, 50, 399–400. [Google Scholar] [CrossRef] [Green Version]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect. 2018, 24, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.-T.; Song, F.-J.; Zou, M.; Hao, Z.-H.; Shan, H. Emergence of Colistin Resistance Gene mcr-1 in Cronobacter sakazakii Producing NDM-9 and in Escherichia coli from the Same Animal. Antimicrob. Agents Chemother. 2017, 61, e01444-16. [Google Scholar] [CrossRef] [Green Version]
- Yousfi, M.; Touati, A.; Muggeo, A.; Mira, B.; Asma, B.; Brasme, L.; Guillard, T.; De Champs, C. Clonal dissemination of OXA-48-producing Enterobacter cloacae isolates from companion animals in Algeria. J. Glob. Antimicrob. Resist. 2018, 12, 187–191. [Google Scholar] [CrossRef]
- Abraham, S.; O’Dea, M.; Trott, D.J.; Abraham, R.J.; Hughes, D.; Pang, S.; McKew, G.; Cheong, E.Y.L.; Merlino, J.; Saputra, S. Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats. Sci. Rep. 2016, 6, 35527. [Google Scholar] [CrossRef]
- Lamba, M.; Gupta, S.; Shukla, R.; Graham, D.W.; Sreekrishnan, T.R.; Ahammad, S.Z. Carbapenem resistance exposures via wastewaters across New Delhi. Environ. Int. 2018, 119, 302–308. [Google Scholar] [CrossRef]
- Proia, L.; Anzil, A.; Borrego, C.; Farrè, M.; Llorca, M.; Sanchis, J.; Bogaerts, P.; Balcázar, J.L.; Servais, P. Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J. Hazard. Mater. 2018, 358, 33–43. [Google Scholar] [CrossRef]
- Yang, F.; Mao, D.; Zhou, H.; Wang, X.; Luo, Y. Propagation of New Delhi Metallo-β-lactamase Genes (blaNDM-1) from a Wastewater Treatment Plant to Its Receiving River. Environ. Sci. Technol. Lett. 2016, 3, 138–143. [Google Scholar] [CrossRef]
- Zong, Z.; Zhang, X. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J. Antimicrob. Chemother. 2013, 68, 1007–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Zhang, X.; Zhou, Y.; Zhang, Z.; Zheng, X. Whole-genome sequencing of an NDM-1- and OXA-58-producing Acinetobacter towneri isolate from hospital sewage in Sichuan Province, China. J. Glob. Antimicrob. Resist. 2019, 16, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Poirel, L.; Nordmann, P. Novel Ambler Class A Carbapenem-Hydrolyzing β-Lactamase from a Pseudomonas fluorescens Isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 2010, 54, 328–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picão, R.C.; Cardoso, J.P.; Campana, E.H.; Nicoletti, A.G.; Petrolini, F.V.B.; Assis, D.M.; Juliano, L.; Gales, A.C. The route of antimicrobial resistance from the hospital effluent to the environment: Focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 2013, 76, 80–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Ma, X.; Luo, L.; Hu, N.; Duan, J.; Tang, Z.; Zhong, R.; Li, Y. The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. Int. J. Environ. Res. Public Health 2020, 17, 1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, A.L.; Carter, C.; McLeod, H.; Wright, M.; Sritharan, P.; Tamber, S.; Wong, A.; Carrillo, C.D.; Blais, B.W. Detection of carbapenem-resistance genes in bacteria isolated from wastewater in Ontario. FACETS 2021, 6, 569–591. [Google Scholar] [CrossRef]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Tanner, W.D.; VanDerslice, J.A.; Goel, R.K.; Leecaster, M.K.; Fisher, M.A.; Olstadt, J.; Gurley, C.M.; Morris, A.G.; Seely, K.A.; Chapman, L. Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in U.S. drinking water. Sci. Rep. 2019, 9, 3938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Buergmann, H. Increased Levels of Multiresistant Bacteria and Resistance Genes after Wastewater Treatment and Their Dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriques, I.; Jucá Ramos, R.T.; Baraúna, R.A.; de Sá, P.H.; Marinho Almeida, D.; Carneiro, A.R.; Barbosa, S.; Pereira, A.; Alves, A.; Saavedra, M.J. Draft Genome Sequence of Serratia fonticola UTAD54, a Carbapenem-Resistant Strain Isolated from Drinking Water. Genome Announc. 2013, 1, e00970-13. [Google Scholar] [CrossRef] [Green Version]
- Novo, A.; André, S.; Viana, P.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Res. 2013, 47, 1875–1887. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Yang, L.; Cai, J.C.; Zhou, H.W.; Chen, G.-X. High-level carbapenem resistance in a Citrobacter freundii clinical isolate is due to a combination of KPC-2 production and decreased porin expression. J. Med. Microbiol. 2008, 57, 332–337. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Zhang, Q.; Lobos, A.; Senkbeil, J.; Sadowsky, M.J.; Harwood, V.J.; Saeidi, N.; Marinoni, O.; Ishii, S. Precipitation influences pathogenic bacteria and antibiotic resistance gene abundance in storm drain outfalls in coastal sub-tropical waters. Environ. Int. 2018, 116, 308–318. [Google Scholar] [CrossRef]
- Montezzi, L.F.; Campana, E.H.; Corrêa, L.L.; Justo, L.H.; Paschoal, R.P.; da Silva, I.L.V.D.; Souza, M.D.C.M.; Drolshagen, M.; Picão, R.C. Occurrence of carbapenemase-producing bacteria in coastal recreational waters. Int. J. Antimicrob. Agents 2015, 45, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.M.; Gast, R.J.; Bogomolni, A.; Ellis, J.C.; Lentell, B.J.; Touhey, K.; Moore, M. Occurrence and patterns of antibiotic resistance in vertebrates off the Northeastern United States coast. FEMS Microbiol. Ecol. 2009, 67, 421–431. [Google Scholar] [CrossRef]
- Sidhu, J.P.S.; Hodgers, L.; Ahmed, W.; Chong, M.N.; Toze, S. Prevalence of human pathogens and indicators in stormwater runoff in Brisbane, Australia. Water Res. 2012, 46, 6652–6660. [Google Scholar] [CrossRef] [PubMed]
- Paschoal, R.P.; Campana, E.H.; Corrêa, L.L.; Montezzi, L.F.; Barrueto, L.R.L.; da Silva, I.R.; Bonelli, R.R.; Castro, L.D.S.; Picão, R.C. Concentration and Variety of Carbapenemase Producers in Recreational Coastal Waters Showing Distinct Levels of Pollution. Antimicrob. Agents Chemother. 2017, 61, e01963-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, R.; Paikin, S.; Rokney, A.; Rubin-Blum, M.; Astrahan, P. Multidrug-resistant enterobacteriaceae in coastal water: An emerging threat. Antimicrob. Resist. Infect. Control 2020, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.W.; Cheng, H.; Huo, Y.Y.; Xu, L.; Wu, Y.H.; Liu, W.H.; Tao, F.F.; Cui, X.J.; Zheng, B.W. Biochemical and genetic characterization of a novel metallo-β-lactamase from marine bacterium Erythrobacter litoralis HTCC 2594. Sci. Rep. 2018, 8, 803. [Google Scholar] [CrossRef] [Green Version]
- Sellera, F.P.; Fernandes, M.R.; Moura, Q.; Souza, T.A.; Cerdeira, L.; Lincopan, N. Draft genome sequence of Enterobacter cloacae ST520 harbouring bla KPC-2, bla CTX-M-15 and bla OXA-17 isolated from coastal waters of the South Atlantic Ocean. J. Glob. Antimicrob. Resist. 2017, 10, 279–280. [Google Scholar] [CrossRef]
- Zheng, B.; Jiang, X.; Xu, Z.; Fang, Y.; Li, L. Characterization of a novel metallo-β-lactamases fold hydrolase from Pelagibacterium halotolerans, a marine halotolerant bacterium isolated from East China Sea. Extremophiles 2016, 20, 37–44. [Google Scholar] [CrossRef]
- Harmon, D.E.; Miranda, O.A.; McCarley, A.; Eshaghian, M.; Carlson, N.; Ruiz, C. Prevalence and characterization of carbapenem-resistant bacteria in water bodies in the Los Angeles–Southern California area. Microbiologyopen 2019, 8, e00692. [Google Scholar] [CrossRef] [Green Version]
- Marqué, S.; Poirel, L.; Héritier, C.; Brisse, S.; Blasco, M.D.; Filip, R.; Coman, G.; Naas, T.; Nordmann, P. Regional Occurrence of Plasmid-Mediated Carbapenem-Hydrolyzing Oxacillinase OXA-58 in Acinetobacter spp. in Europe. J. Clin. Microbiol. 2005, 43, 4885–4888. [Google Scholar] [CrossRef] [Green Version]
- Pillonetto, M.; Arend, L.; Vespero, E.C.; Pelisson, M.; Chagas, T.P.G.; Carvalho-Assef, A.P.D.; Asensi, M.D. First Report of NDM-1-Producing Acinetobacter baumannii Sequence Type 25 in Brazil. Antimicrob. Agents Chemother. 2014, 58, 7592–7594. [Google Scholar] [CrossRef] [Green Version]
- Almuzara, M.N.; Barberis, C.M.; Rodríguez, C.H.; Famiglietti, A.M.R.; Ramirez, M.S.; Vay, C.A. First Report of an Extensively Drug-Resistant VIM-2 Metallo-β-Lactamase-Producing Brevundimonas diminuta Clinical Isolate. J. Clin. Microbiol. 2012, 50, 2830–2832. [Google Scholar] [CrossRef] [Green Version]
- Docquier, J.-D.; Pantanella, F.; Giuliani, F.; Thaller, M.C.; Amicosante, G.; Galleni, M.; Frère, J.-M.; Bush, K.; Rossolini, G.M. CAU-1, a Subclass B3 Metallo-β-Lactamase of Low Substrate Affinity Encoded by an Ortholog Present in the Caulobacter crescentus Chromosome. Antimicrob. Agents Chemother. 2002, 46, 1823–1830. [Google Scholar] [CrossRef] [Green Version]
- Gudeta, D.D.; Bortolaia, V.; Amos, G.; Wellington, E.M.H.; Brandt, K.K.; Poirel, L.; Nielsen, J.B.; Westh, H.; Guardabassi, L. The Soil Microbiota Harbors a Diversity of Carbapenem-Hydrolyzing β-Lactamases of Potential Clinical Relevance. Antimicrob. Agents Chemother. 2016, 60, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Gudeta, D.D.; Pollini, S.; Docquier, J.-D.; Bortolaia, V.; Rossolini, G.M.; Guardabassi, L. Biochemical Characterization of CPS-1, a Subclass B3 Metallo-β-Lactamase from a Chryseobacterium piscium Soil Isolate. Antimicrob. Agents Chemother. 2016, 60, 1869–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellais, S.; Naas, T.; Nordmann, P. Genetic and Biochemical Characterization of CGB-1, an Ambler Class B Carbapenem-Hydrolyzing β-Lactamase from Chryseobacterium gleum. Antimicrob. Agents Chemother. 2002, 46, 2791–2796. [Google Scholar] [CrossRef] [Green Version]
- Mammeri, H.; Bellais, S.; Nordmann, P. Chromosome-Encoded β-Lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (Formerly Flavobacterium odoratum), New Members of the Lineage of Molecular Subclass B1 Metalloenzymes. Antimicrob. Agents Chemother. 2002, 46, 3561–3567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montaña, S.; Schramm, S.T.J.; Traglia, G.M.; Chiem, K.; Parmeciano Di Noto, G.; Almuzara, M.; Barberis, C.; Vay, C.; Quiroga, C.; Tolmasky, M.E. The Genetic Analysis of an Acinetobacter johnsonii Clinical Strain Evidenced the Presence of Horizontal Genetic Transfer. PLoS ONE 2016, 11, e0161528. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, C.F.M.; Silva, D.M.; Carneiro, M.T.; Ribeiro, S.; Fontana-Maurell, M.; Alvarez, P.; Asensi, M.D.; Zahner, V.; Carvalho-Assef, A.P.D. Detection of Carbapenemase Genes in Aquatic Environments in Rio de Janeiro, Brazil. Antimicrob. Agents Chemother. 2016, 60, 4380–4383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campana, E.H.; Montezzi, L.F.; Paschoal, R.P.; Picão, R.C. NDM-producing Klebsiella pneumoniae ST11 goes to the beach. Int. J. Antimicrob. Agents 2016, 49, 119–121. [Google Scholar] [CrossRef]
- Mahon, B.M.; Brehony, C.; McGrath, E.; Killeen, J.; Cormican, M.; Hickey, P.; Keane, S.; Hanahoe, B.; Dolan, A.; Morris, D. Indistinguishable NDM-producing Escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Eurosurveillance 2017, 22, 30513. [Google Scholar] [CrossRef] [Green Version]
- Xin, R.; Zhang, K.; Wu, N.; Zhang, Y.; Niu, Z. The pollution level of the blaOXA-58 carbapenemase gene in coastal water and its host bacteria characteristics. Environ. Pollut. 2019, 244, 66–71. [Google Scholar] [CrossRef]
- Mahon, B.M.; Brehony, C.; Cahill, N.; McGrath, E.; O’Connor, L.; Varley, A.; Cormican, M.; Ryan, S.; Hickey, P.; Keane, S. Detection of OXA-48-like-producing Enterobacterales in Irish recreational water. Sci. Total Environ. 2019, 690, 1–6. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Nordmann, P. Genetic and biochemical characterization of the chromosome-encoded class B β-lactamases from Shewanella livingstonensis (SLB-1) and Shewanella frigidimarina (SFB-1). J. Antimicrob. Chemother. 2005, 55, 680–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristiansen, A.; Grgic, M.; Altermark, B.; Leiros, I. Properties and distribution of a metallo-β-lactamase (ALI-1) from the fish pathogen Aliivibrio salmonicida LFI1238. J. Antimicrob. Chemother. 2015, 70, 766–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatosy, S.M.; Martiny, A.C.; Nojiri, H. The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Pruden, A.; Virta, M.; Zhang, T. Antibiotic Resistance in Aquatic Systems. Front. Microbiol. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almuzara, M.N.; Vazquez, M.; Tanaka, N.; Turco, M.; Ramirez, M.S.; Lopez, E.L.; Pasteran, F.; Rapoport, M.; Procopio, A.; Vay, C.A. First Case of Human Infection Due to Pseudomonas fulva, an Environmental Bacterium Isolated from Cerebrospinal Fluid. J. Clin. Microbiol. 2010, 48, 660–664. [Google Scholar] [CrossRef] [Green Version]
- Rizek, C.; Fu, L.; Dos Santos, L.C.; Leite, G.; Ramos, J.; Rossi, F.; Guimaraes, T.; Levin, A.S.; Costa, S.F. Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Buehrle, D.J.; Shields, R.K.; Clarke, L.G.; Potoski, B.A.; Clancy, C.J.; Nguyen, M.H. Carbapenem-Resistant Pseudomonas aeruginosa Bacteremia: Risk Factors for Mortality and Microbiologic Treatment Failure. Antimicrob. Agents Chemother. 2017, 61, e01243-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, T.R.; Toleman, M.A.; Hryniewicz, W.; Bennett, P.M.; Jones, R.N. Evolution of an integron carrying blaVIM-2 in Eastern Europe: Report from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2003, 52, 116–119. [Google Scholar] [CrossRef] [Green Version]
- Yong, D.; Toleman, M.A.; Bell, J.; Ritchie, B.; Pratt, R.; Ryley, H.; Walsh, T.R. Genetic and Biochemical Characterization of an Acquired Subgroup B3 Metallo-β-Lactamase Gene, blaAIM-1, and Its Unique Genetic Context in Pseudomonas aeruginosa from Australia. Antimicrob. Agents Chemother. 2012, 56, 6154–6159. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Ros, A.; Carricajo, A.; Berthelot, P.; Pozzetto, B.; Bernabeu, S.; Nordmann, P. Extremely Drug-ResistantCitrobacter freundiiIsolate Producing NDM-1 and Other Carbapenemases Identified in a Patient Returning from India. Antimicrob. Agents Chemother. 2011, 55, 447–448. [Google Scholar] [CrossRef] [Green Version]
- Goren, M.G.; Chmelnitsky, I.; Carmeli, Y.; Navon-Venezia, S. Plasmid-encoded OXA-48 carbapenemase in Escherichia coli from Israel. J. Antimicrob. Chemother. 2011, 66, 672–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maravić, A.; Skočibušić, M.; Cvjetan, S.; Šamanić, I.; Fredotović, Ž.; Puizina, J. Prevalence and diversity of extended-spectrum-β-lactamase-producing Enterobacteriaceae from marine beach waters. Mar. Pollut. Bull. 2015, 90, 60–67. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Park, S.E.; Kang, S.-J.; Oh, T.-K. Rheinheimera aquimaris sp. nov., isolated from seawater of the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 2007, 57, 1386–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanenko, L.A.; Uchino, M.; Falsen, E.; Zhukova, N.V.; Mikhailov, V.V.; Uchimura, T. Rheinheimera pacifica sp. nov., a novel halotolerant bacterium isolated from deep sea water of the Pacific. Int. J. Syst. Evol. Microbiol. 2003, 53, 1973–1977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kümmerer, K. Antibiotics in the aquatic environment—A review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, G.; Zou, S.; Li, X.; Liu, Y. Determination of selected antibiotics in the Victoria Harbour and the Pearl River, South China using high-performance liquid chromatography-electrospray ionization tandem mass spectrometry. Environ. Pollut. 2007, 145, 672–679. [Google Scholar] [CrossRef] [Green Version]
- Genilloud, O.O. Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm? Antibiotics 2018, 7, 85. [Google Scholar] [CrossRef] [Green Version]
- Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics. Mar. Drugs 2016, 14, 137. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.S.; Nicholson, B.; Teisan, S.; Lam, K.S.; Potts, B.C.M. Aureoverticillactam, a Novel 22-Atom Macrocyclic Lactam from the Marine Actinomycete Streptomyces aureoverticillatus. J. Nat. Prod. 2004, 67, 1400–1402. [Google Scholar] [CrossRef]
- Manam, R.R.; Teisan, S.; White, D.J.; Nicholson, B.; Grodberg, J.; Neuteboom, S.T.C.; Lam, K.S.; Mosca, D.A.; Lloyd, G.K.; Potts, B.C.M. Lajollamycin, a Nitro-tetraene Spiro-β-lactone-γ-lactam Antibiotic from the Marine Actinomycete Streptomyces nodosus. J. Nat. Prod. 2005, 68, 240–243. [Google Scholar] [CrossRef]
- Cayô, R.; San Segundo, L.Y.; del Molino Bernal, I.C.P.; de la Fuente, C.G.; Rodríguez, M.A.B.; Calvo, J.; Martínez-Martínez, L. Bloodstream infection caused by Acinetobacter junii in a patient with acute lymphoblastic leukaemia after allogenic haematopoietic cell transplantation. J. Med. Microbiol. 2011, 60, 375–377. [Google Scholar] [CrossRef]
- Linde, H.-J.; Hahn, J.; Holler, E.; Reischl, U.; Lehn, N. Septicemia Due to Acinetobacter junii. J. Clin. Microbiol. 2002, 40, 2696–2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, K.; Yu, X.; Li, B.; Cao, B. Identification and control of a Pseudomonas spp (P. fulva and P. putida) bloodstream infection outbreak in a teaching hospital in Beijing, China. Int. J. Infect. Dis. 2014, 23, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, H.; Strate, A.; Schulze, A.; Pulverer, G. Vascular Catheter--Related Bloodstream Infection Due to Acinetobacter johnsonii (Formerly Acinetobacter calcoaceticus var. lwoffii): Report of 13 Cases. Clin. Infect. Dis. 1993, 17, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Vijayakrishnan, R.; Rapose, A. Fatal Enterococcus durans aortic valve endocarditis: A case report and review of the literature. Case Rep. 2012, 2012, bcr0220125855. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Kastoris, A.C.; Vouloumanou, E.K.; Dimopoulos, G. Community-acquired Stenotrophomonas maltophilia infections: A systematic review. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 719–730. [Google Scholar] [CrossRef]
- Sakhnini, E.; Weissmann, A.; Oren, I. Fulminant Stenotrophomonas maltophilia Soft Tissue Infection in Immunocompromised Patients: An Outbreak Transmitted via Tap Water. Am. J. Med. Sci. 2002, 323, 269–272. [Google Scholar] [CrossRef]
- Turenne, C.; Chedore, P.; Wolfe, J.; Jamieson, F.; Broukhanski, G.; May, K.; Kabani, A. Mycobacterium lacus sp. nov., a novel slowly growing, non-chromogenic clinical isolate. Int. J. Syst. Evol. Microbiol. 2002, 52, 2135–2140. [Google Scholar] [CrossRef] [Green Version]
- Kanderi, T.; Shrimanker, I.; Mansoora, Q.; Shah, K.; Yumen, A.; Komanduri, S. Stenotrophomonas maltophilia: An Emerging Pathogen of the Respiratory Tract. Am. J. Case Rep. 2020, 21, e921466-1. [Google Scholar] [CrossRef]
- Mauger, T.F.; Kuennen, R.A.; Smith, R.H.; Sawyer, W. Acanthamoeba and Stenotrophomonas maltophilia keratitis with fungal keratitis in the contralateral eye. Clin. Ophthalmol. 2010, 4, 1207–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prashanth, K.; Ranga, M.P.M.; Rao, V.A.; Kanungo, R. Corneal perforation due to Acinetobacter junii: A case report. Diagn. Microbiol. Infect. Dis. 2000, 37, 215–217. [Google Scholar] [CrossRef]
- Pandey, P.K.; Kass, P.H.; Soupir, M.L.; Biswas, S.; Singh, V.P. Contamination of water resources by pathogenic bacteria. AMB Express 2014, 4, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yau, V.; Wade, T.J.; De Wilde, C.K.; Colford, J.M. Skin-related symptoms following exposure to recreational water: A systematic review and meta-analysis. Water Qual. Expo. Health 2009, 1, 79–103. [Google Scholar] [CrossRef] [Green Version]
- Pitart, C.; Solé, M.; Roca, I.; Román, A.; Moreno, A.; Vila, J.; Marco, F. Molecular Characterization of blaNDM-5 Carried on an IncFII Plasmid in an Escherichia coli Isolate from a Nontraveler Patient in Spain. Antimicrob. Agents Chemother. 2015, 59, 659–662. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.A.; Islam, M.; Hasan, R.; Hossain, M.I.; Nabi, A.; Rahman, M.; Goessens, W.H.F.; Endtz, H.P.; Boehm, A.B.; Faruque, S.M. Environmental Spread of New Delhi Metallo-β-Lactamase-1-Producing Multidrug-Resistant Bacteria in Dhaka, Bangladesh. Appl. Environ. Microbiol. 2017, 83, e00793-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, Y.; Akeda, Y.; Hagiya, H.; Sakamoto, N.; Takeuchi, D.; Shanmugakani, R.K.; Motooka, D.; Nishi, I.; Zin, K.N.; Aye, M.M.; et al. Spreading Patterns of NDM-Producing Enterobacteriaceae in Clinical and Environmental Settings in Yangon, Myanmar. Antimicrob. Agents Chemother. 2019, 63, e01924-18. [Google Scholar] [CrossRef] [Green Version]
- Berglund, F.; Marathe, N.P.; Österlund, T.; Bengtsson-Palme, J.; Kotsakis, S.; Flach, C.-F.; Larsson, D.G.J.; Kristiansson, E. Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome 2017, 5, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, D.G.J.; Andremont, A.; Bengtsson-Palme, J.; Brandt, K.K.; de Roda Husman, A.M.; Fagerstedt, P.; Fick, J.; Flach, C.-F.; Gaze, W.H.; Kuroda, M.; et al. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018, 117, 132–138. [Google Scholar] [CrossRef]
- Diene, S.M.; Rolain, J.M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliday, E.; McLellan, S.L.; Amaral-Zettler, L.A.; Sogin, M.L.; Gast, R.J. Comparison of Bacterial Communities in Sands and Water at Beaches with Bacterial Water Quality Violations. PLoS ONE 2014, 9, e90815. [Google Scholar] [CrossRef] [PubMed]

| Carbapenem Resistant Bacteria | Carbapenem Resistance Determinants | Reference |
|---|---|---|
| Vibrio cholerae | Not identified | [24] |
| Rheinheimera spp. | B3-MBL | [25] |
| Variovorax spp. | NDM | |
| Enterobacteriaceae | KPC, OXA | [72] |
| Citrobacter sp., Citrobacter sp., Kluyvera sp., Aeromonas sp. | KPC-2 | [73] |
| Acinetobacter spp. | OXA | [76] |
| Aeromonas spp. | KPC-2, GES-5, GES-16 | |
| Citrobacter sp. | KPC-2, OXA-370 | |
| Enterobacter spp. | KPC-2, KPC-26, GES-5, GES-16 | |
| Klebsiella spp. | KPC-2, KPC-26, GES-16, NDM-1 | |
| Kluyvera spp., Serratia spp. | KPC-2 | |
| Pseudomonas spp. | VIM-2, SPM-1 | |
| Enterobacter asburiae | IMI-2 | [77] |
| Enterobacter bugandensis | IMI-20 | |
| Escherichia coli | OXA-48 | |
| Erythrobacter litoralis | ElBla2 * | [78] |
| Enterobacter cloacae | KPC-2, CTX-M-15, OXA-17 | [79] |
| Pelagibacterium halotolerans | PH-1 * | [80] |
| Aeromonas punctata, Enterobacter asburiae, K. pneumoniae, Enterobacter kobei | KPC, GES-16, OXA-48-like | [91] |
| K. pneumoniae | NDM | [93] |
| K. pneumoniae | NDM-1, OXA-1 | [92] |
| Pseudomonas spp., Rheinheimera spp., Stenotrophomonas sp., Shewanella sp., Raoultella sp., Vibrio sp., Pseudoalteromonas sp., Algoriphagus sp., Bowmanella sp., and Thalassospira sp. | OXA-58 | [94] |
| E. coli, K. pneumoniae | OXA-48 | [95] |
| Shewanella livingstonensis | SLB-1 * | [96] |
| Shewanella frigidimarina | SFB-1 * | |
| Aliivibrio salmonicida | ALI-1 * | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dewi, D.A.P.R.; Thomas, T.; Ahmad Mokhtar, A.M.; Mat Nanyan, N.S.; Zulfigar, S.B.; Salikin, N.H. Carbapenem Resistance among Marine Bacteria—An Emerging Threat to the Global Health Sector. Microorganisms 2021, 9, 2147. https://doi.org/10.3390/microorganisms9102147
Dewi DAPR, Thomas T, Ahmad Mokhtar AM, Mat Nanyan NS, Zulfigar SB, Salikin NH. Carbapenem Resistance among Marine Bacteria—An Emerging Threat to the Global Health Sector. Microorganisms. 2021; 9(10):2147. https://doi.org/10.3390/microorganisms9102147
Chicago/Turabian StyleDewi, Dewa A.P. Rasmika, Torsten Thomas, Ana Masara Ahmad Mokhtar, Noreen Suliani Mat Nanyan, Siti Balqis Zulfigar, and Nor Hawani Salikin. 2021. "Carbapenem Resistance among Marine Bacteria—An Emerging Threat to the Global Health Sector" Microorganisms 9, no. 10: 2147. https://doi.org/10.3390/microorganisms9102147






