Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress
Abstract
1. Introduction
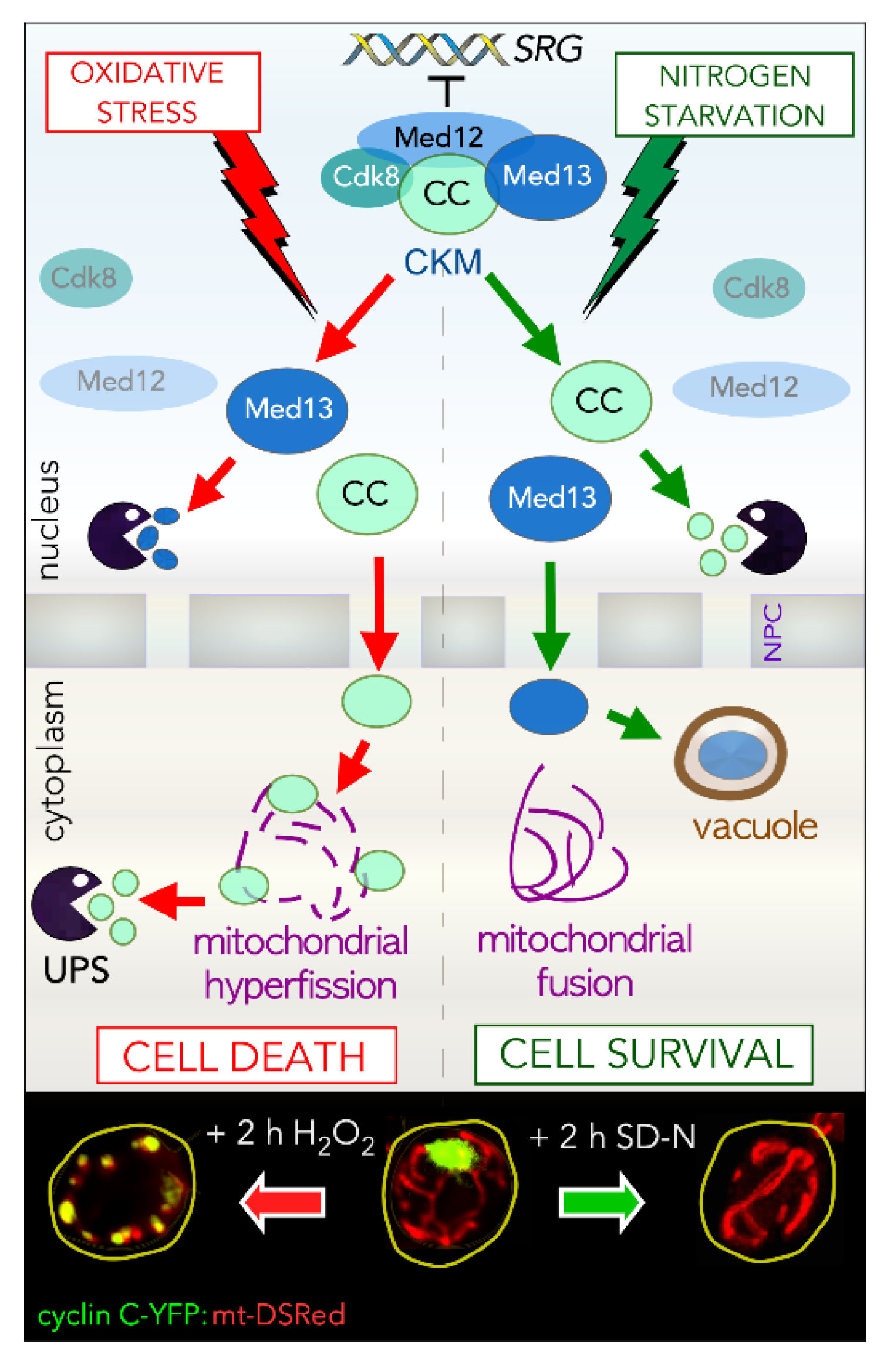
2. The CKM Regulates Cell Fate Decisions
3. The Transcriptional Role of the CKM in Response to Stress
3.1. Association of the CKM with the Core Mediator
3.2. Cdk8 Is a Non-Canonical Cyclin-Dependent Kinase
3.3. Dissociation of the CKM from the Core Mediator
3.4. Disassembly of Cyclin C from Med13
3.5. Genes Regulated by the CKM in Response to Cell Death Cues
3.6. Genes Regulated by the CKM in Response to Cell Survival Cues
4. Upstream Signaling Pathways Link Degradation Machines to Stress
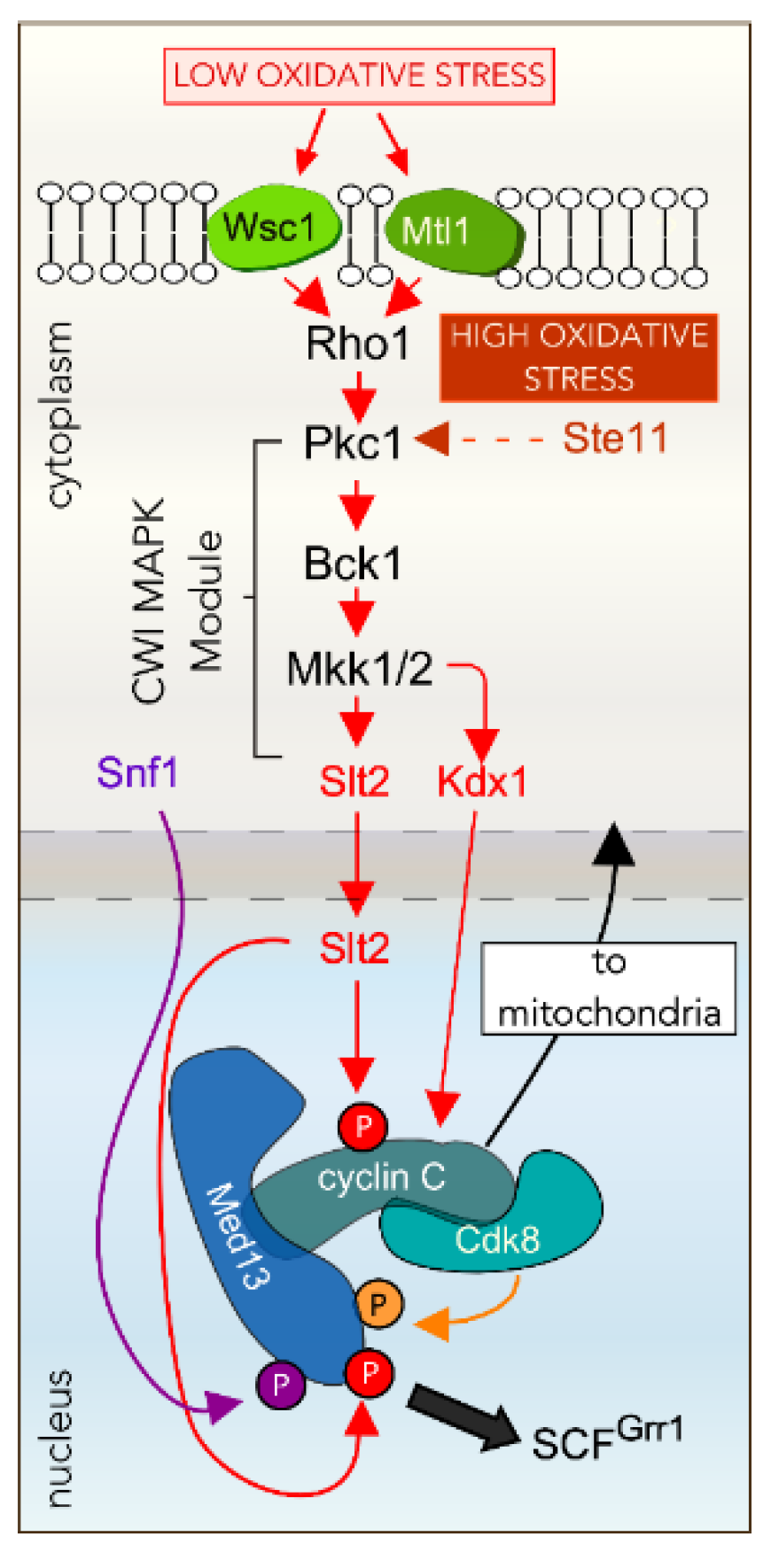
4.1. Oxidative Stress Activates the Cell Wall Integrity Pathway
4.2. The CWI MAPK Slt2 Is Required for Cyclin C Nuclear Release in ROS
4.3. Other Factors Required for ROS-Dependent Cyclin C Nuclear Release
4.4. Cyclin C Nuclear Release Requires Additional MAPK Pathways in Response to More Stringent ROS
4.5. Nitrogen Starvation Inhibits TORC1
4.6. Nitrogen Starvation Activated Slt2 Does Not Directly Phosphorylate Cyclin C
4.7. Cyclin C Destruction in Nitrogen Starvation Is Mediated by the UPS
4.8. TORC1 Inhibition Results in Med13 Degradation by Snx4-Assisted Autophagy
4.9. The Role of Snx4 in Autophagy
5. The Relationship between Mitochondria, the CKM, and Cell Fate Decisions
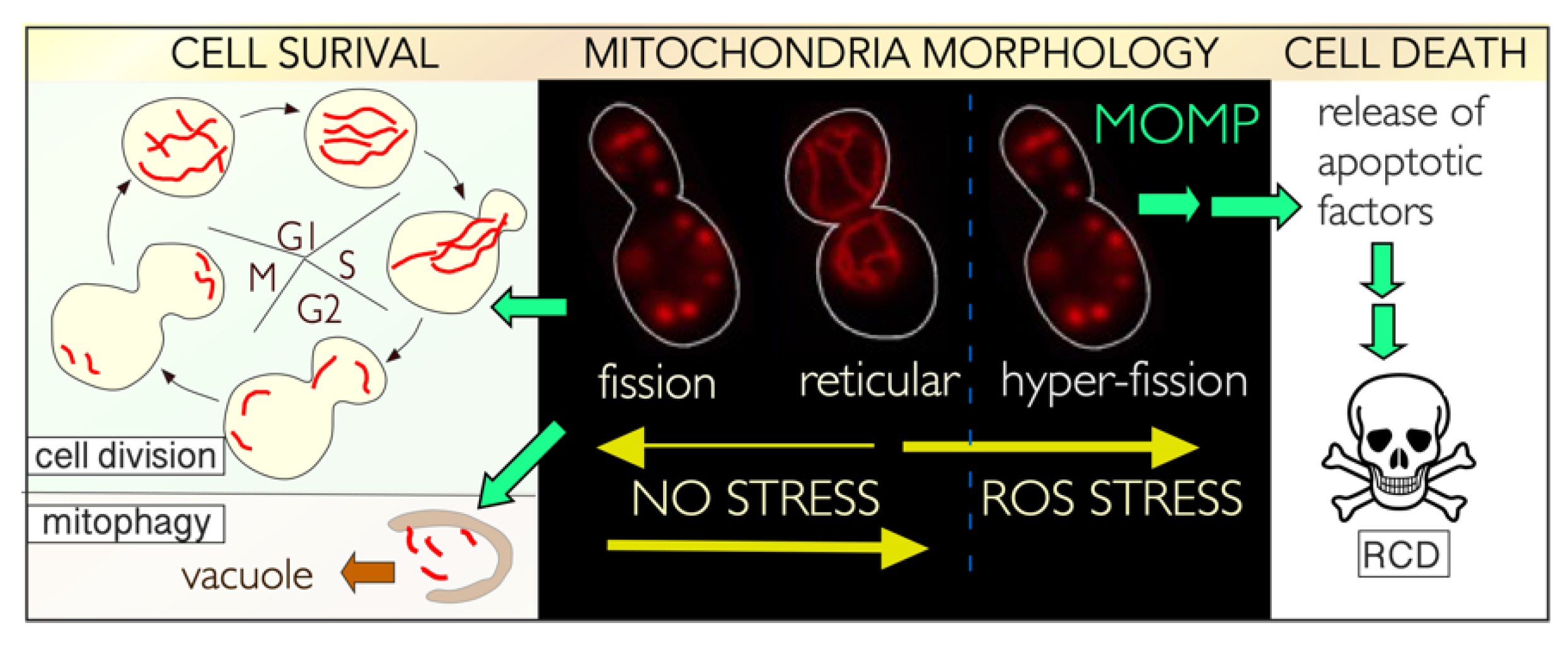
5.1. Control of Mitochondrial Dynamics
5.2. Mitochondrial Dynamics in Unstressed Cells
5.3. Stress-Induced Mitochondrial Fission Is Linked to Cell Death Pathways
5.4. Night Job of Cyclin C in Cell Death: A Mitochondrial Response
5.5. Conservation of the Night Job of Cyclin C
5.6. Mitochondrial Dynamics in Nutrient Starved Cells
5.7. A Different Night Shift for Cyclin C in Cell Survival: Preserving Mitochondrial Integrity
6. Conclusions: The CKM Is at the Crossroads of Different Cell Fates
| Function | S. cerevisiae | Mammals |
|---|---|---|
| Predominantly negatively regulates SRG’s. Positively regulates a few genes. | [18,20,23,68,241,242] | No |
| Equally regulates positive and negative SRGs. | No | [13,14,28,243] |
| Cyclin C -Cdk8 phosphorylate the C-terminal domain of the largest subunit of RNA pol II in vitro, to inhibit transcriptional initiation. | [68,85,242] | [244] |
| Cyclin C -Cdk8 directly phosphorylates other Mediator subunit to negatively regulate transcription. | [245] | [246] |
| Cyclin C -Cdk8 phosphorylates transcription factors and other targets. | [247,248] | [241,246,249,250] |
| MAPK mediated phosphorylation of Med13 mediates its nuclear release following cell death cues. | [35] | unknown |
| Nuclear release of cyclin C requires Med13 degradation by UPS following cell death cues. | [36,40] | unknown |
| Med13 is regulated by SCFGrr1/Fbw7. | [36] | [81] |
| Cytoplasmic cyclin C mediates mitochondrial hyper-fission by binding to mitochondrial fission complex after ROS stress. | [34] | [41] |
| Cytoplasmic cyclin C promotes oligomerization of the GTPase DNM1/Drp1. | unknown | [43] |
| Cyclin C nuclear release is required for RCD (intrinsic pathway). | [31,34] | [41] |
| Cyclin C binds to BAX at mitochondria to induce MOMP following ROS stress. | No BAX | [38] |
| Cyclin C null mutants are deficient in stress-induced mitochondrial fission. | [34] | [41] |
| Cyclin C nuclear release after ROS stress promotes the release of pro-apoptotic factors. | Assumed [34] | [38] |
| Cyclin C is destroyed by the UPS in the cytoplasm in ROS stress after mediating mitochondrial hyper-fission. | [30,33] | No |
| ROS induced night job of cyclin C is independent of Cdk8. | [32] | [41] |
| CKM negatively regulates a subset of ATG genes. | [37,42] | [13,14] |
| Cyclin C is destroyed by the UPS following TORC1 inhibition | [37] | unknown |
| Destruction of cyclin C following TORC1 inhibition promotes cell survival. | [37] | unknown |
| Destruction of cyclin C following TORC1 inhibition promotes mitochondrial fusion. | [37] | unknown |
| Med13 is destroyed by Snx4-assisted autophagy following TORC1 inhibition. | [42] | unknown |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Å | Angstrom |
| AGC-kinase | group of kinases related to PKA PKG and PKC |
| AMPK | adenosine monophosphate-activated protein kinase |
| ATP | adenosine triphosphate |
| CAK | CDK-activating kinase |
| CC | cyclin C |
| CDK | Cyclin-dependent kinase |
| CDKI | cyclin-dependent protein inhibitor |
| Cryo-EM | Cryogenic electron microscopy |
| EGOC | Gtr1, Gtr2, Ego1, and Ego3 complex that can activate TORC1 |
| ERK5 | extracellular signal-regulated kinase 5 |
| GTP | guanosine triphosphate |
| HDAC | Rpd3–Sin3–Ume6 histone deacetylase |
| HOG | high osmolarity glycerol |
| IDR | intrinsic disordered region |
| IMM | inner mitochondrial membrane |
| MAPK | mitogen-activated protein kinase |
| MAPK | mitogen-activated protein kinase kinase |
| MAPKKK | mitogen-activated protein kinase kinase kinase |
| MEFs | mouse embryonic fibroblasts |
| MFF | mitochondrial fission factor |
| MOMP | mitochondrial outer membrane permeabilization |
| mtDNA | mitochondrial DNA |
| NPC | nuclear pore complex |
| OMM | outer mitochondrial membrane |
| OXPHOS | oxidative phosphorylation |
| PAS | pre-autophagosomal structure |
| PIC | pre-initiation complex |
| Pkc1 | protein kinase C |
| PKA | protein kinase A |
| RNA Pol II | RNA Polymerase II |
| ROS | reactive oxygen species |
| SAA | Snx4-assisted autophagy |
| SCF | Skp1-Cullin-F-box E3 ligase complex |
| SRG’s | stress response genes |
| SEACIT | Sea1–Npr2–Npr3 complex that can inhibit TORC1 |
| Ser | serine |
| TORC1 | target of rapamycin kinase complex 1 |
| Thr | threonine |
| TF | Transcription factor |
| UAS | upstream activating sequences |
| UPS | ubiquitin proteasome system |
| YFP | yellow fluorescence protein |
References
- Towers, C.G.; Wodetzki, D.; Thorburn, A. Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations. J. Cell Biol. 2020, 219, e201909033. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Frohlich, E.; Ligr, M.; Grey, M.; Sigrist, S.J.; Wolf, D.H.; Frohlich, K.U. Oxygen stress: A regulator of apoptosis in yeast. J. Cell Biol. 1999, 145, 757–767. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Cooper, K.F. Till Death Do Us Part: The Marriage of Autophagy and Apoptosis. Oxid. Med. Cell. Longev. 2018, 2018, 4701275. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Aguilera, A.; Austriaco, N.; Ayscough, K.; Balzan, R.; Bar-Nun, S.; Barrientos, A.; Belenky, P.; et al. Guidelines and recommendations on yeast cell death nomenclature. Microb. Cell 2018, 5, 4–31. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Tsai, K.L.; Sato, S.; Tomomori-Sato, C.; Conaway, R.C.; Conaway, J.W.; Asturias, F.J. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013, 20, 611–619. [Google Scholar] [CrossRef]
- Kuchin, S.; Yeghiayan, P.; Carlson, M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 1995, 92, 4006–4010. [Google Scholar] [CrossRef]
- Jeronimo, C.; Robert, F. The Mediator Complex: At the Nexus of RNA Polymerase II Transcription. Trends Cell Biol. 2017, 27, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.J.; Trnka, M.J.; Pellarin, R.; Greenberg, C.H.; Bushnell, D.A.; Davis, R.; Burlingame, A.L.; Sali, A.; Kornberg, R.D. Molecular architecture of the yeast Mediator complex. Elife 2015, 4, e08719. [Google Scholar] [CrossRef]
- van de Peppel, J.; Kettelarij, N.; van Bakel, H.; Kockelkorn, T.T.; van Leenen, D.; Holstege, F.C. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 2005, 19, 511–522. [Google Scholar] [CrossRef]
- Law, M.J.; Ciccaglione, K. Fine-Tuning of Histone H3 Lys4 Methylation During Pseudohyphal Differentiation by the CDK Submodule of RNA Polymerase II. Genetics 2015, 199, 435–453. [Google Scholar] [CrossRef]
- Stieg, D.C.; Chang, K.T.; Cooper, K.F.; Strich, R. Cyclin C Regulated Oxidative Stress Responsive Transcriptome in Mus musculus Embryonic Fibroblasts. G3 Genes Genomes Genet. 2019, 9, 1901–1908. [Google Scholar] [CrossRef]
- Stieg, D.C.; Cooper, K.F.; Strich, R. The extent of cyclin C promoter occupancy directs changes in stress-dependent transcription. J. Biol. Chem. 2020, 295, 16280–16291. [Google Scholar] [CrossRef]
- Allen, B.L.; Taatjes, D.J. The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Yarrington, R.M.; Yu, Y.; Yan, C.; Bai, L.; Stillman, D.J. A Role for Mediator Core in Limiting Coactivator Recruitment in Saccharomyces cerevisiae. Genetics 2020, 215, 407–420. [Google Scholar] [CrossRef]
- Strich, R.; Slater, M.R.; Esposito, R.E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc. Natl. Acad. Sci. USA 1989, 86, 10018–10022. [Google Scholar] [CrossRef]
- Holstege, F.C.; Jennings, E.G.; Wyrick, J.J.; Lee, T.I.; Hengartner, C.J.; Green, M.R.; Golub, T.R.; Lander, E.S.; Young, R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 1998, 95, 717–728. [Google Scholar] [CrossRef]
- Ansari, S.A.; Ganapathi, M.; Benschop, J.J.; Holstege, F.C.; Wade, J.T.; Morse, R.H. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J. 2012, 31, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Hirst, K.; Fisher, F.; McAndrew, P.C.; Godding, C.R. The transcription factor, the Cdk, its cyclin and their regulator: Directing the transcriptional response to a nutritional signal. EMBO 1994, 13, 5410–5420. [Google Scholar] [CrossRef]
- Hirst, M.; Kobor, M.S.; Kuriakose, N.; Greenblatt, J.; Sadowski, I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 1999, 3, 673–678. [Google Scholar] [CrossRef]
- Vincent, O.; Kuchin, S.; Hong, S.P.; Townley, R.; Vyas, V.K.; Carlson, M. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 5790–5796. [Google Scholar] [CrossRef]
- Larschan, E.; Winston, F. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 2005, 25, 114–123. [Google Scholar] [CrossRef]
- Xu, W.; Ji, J.Y. Dysregulation of CDK8 and Cyclin C in tumorigenesis. J Genet Genom. 2011, 38, 439–452. [Google Scholar] [CrossRef]
- Nemet, J.; Jelicic, B.; Rubelj, I.; Sopta, M. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 2014, 97, 22–27. [Google Scholar] [CrossRef]
- Berk, A.J. Yin and yang of mediator function revealed by human mutants. Proc. Natl. Acad. Sci. USA 2012, 109, 19519–19520. [Google Scholar] [CrossRef]
- Adegbola, A.; Musante, L.; Callewaert, B.; Maciel, P.; Hu, H.; Isidor, B.; Picker-Minh, S.; Le Caignec, C.; Delle Chiaie, B.; Vanakker, O.; et al. Redefining the MED13L syndrome. Eur. J. Hum. Genet. 2015, 23, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, M.D.; Allen, M.A.; Bensard, C.L.; Wang, X.; Schwinn, M.K.; Qin, B.; Long, H.W.; Daniels, D.L.; Hahn, W.C.; Dowell, R.D.; et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 2013, 153, 1327–1339. [Google Scholar] [CrossRef]
- Dannappel, M.V.; Sooraj, D.; Loh, J.J.; Firestein, R. Molecular and in vivo Functions of the CDK8 and CDK19 Kinase Modules. Front. Cell Dev. Biol. 2019, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.F.; Mallory, M.J.; Smith, J.B.; Strich, R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 1997, 16, 4665–4675. [Google Scholar] [CrossRef] [PubMed]
- Krasley, E.; Cooper, K.F.; Mallory, M.J.; Dunbrack, R.; Strich, R. Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics 2006, 172, 1477–1486. [Google Scholar] [CrossRef]
- Cooper, K.F.; Scarnati, M.S.; Krasley, E.; Mallory, M.J.; Jin, C.; Law, M.J.; Strich, R. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J. Cell Sci. 2012, 125, 1015–1026. [Google Scholar] [CrossRef]
- Cooper, K.F.; Mallory, M.J.; Strich, R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol. Cell. Biol. 1999, 19, 3338–3348. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.F.; Khakhina, S.; Kim, S.K.; Strich, R. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev. Cell 2014, 28, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Strich, R.; Cooper, K.F. Slt2p phosphorylation induces cyclin C nuclear-to-cytoplasmic translocation in response to oxidative stress. Mol. Biol. Cell 2014, 25, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Stieg, D.C.; Willis, S.D.; Ganesan, V.; Ong, K.L.; Scuorzo, J.; Song, M.; Grose, J.; Strich, R.; Cooper, K.F. A complex molecular switch directs stress-induced cyclin C nuclear release through SCF(Grr1)-mediated degradation of Med13. Mol. Biol. Cell 2018, 29, 363–375. [Google Scholar] [CrossRef]
- Willis, S.D.; Hanley, S.E.; Beishke, T.; Tati, P.D.; Cooper, K.F. Ubiquitin-proteasome-mediated cyclin C degradation promotes cell survival following nitrogen starvation. Mol. Biol. Cell 2020, 31, 1015–1031. [Google Scholar] [CrossRef]
- Jezek, J.; Chang, K.T.; Joshi, A.M.; Strich, R. Mitochondrial translocation of cyclin C stimulates intrinsic apoptosis through Bax recruitment. EMBO Rep. 2019, 20, e47425. [Google Scholar] [CrossRef]
- Jezek, J.; Cooper, K.F.; Strich, R. Reactive Oxygen Species and Mitochondrial Dynamics: The Yin and Yang of Mitochondrial Dysfunction and Cancer Progression. Antioxidants 2018, 7, 13. [Google Scholar] [CrossRef]
- Khakhina, S.; Cooper, K.F.; Strich, R. Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol. Biol. Cell 2014, 25, 2807–2816. [Google Scholar] [CrossRef]
- Wang, K.; Yan, R.; Cooper, K.F.; Strich, R. Cyclin C mediates stress-induced mitochondrial fission and apoptosis. Mol. Biol. Cell 2015, 26, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Hanley, S.E.; Willis, S.D.; Cooper, K.F. Snx4-assisted vacuolar targeting of transcription factors defines a new autophagy pathway for controlling ATG expression. Autophagy 2021. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, V.; Willis, S.D.; Chang, K.T.; Beluch, S.; Cooper, K.F.; Strich, R. Cyclin C directly stimulates Drp1 GTP affinity to mediate stress-induced mitochondrial hyperfission. Mol. Biol. Cell 2019, 30, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Katz, S.G. Non-apoptotic functions of BCL-2 family proteins. Cell Death Differ. 2017, 24, 1348–1358. [Google Scholar] [CrossRef]
- Shamas-Din, A.; Kale, J.; Leber, B.; Andrews, D.W. Mechanisms of action of Bcl-2 family proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a008714. [Google Scholar] [CrossRef] [PubMed]
- Bourbon, H.M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008, 36, 3993–4008. [Google Scholar] [CrossRef]
- Nagulapalli, M.; Maji, S.; Dwivedi, N.; Dahiya, P.; Thakur, J.K. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res. 2016, 44, 1591–1612. [Google Scholar] [CrossRef]
- Malik, S.; Guermah, M.; Yuan, C.X.; Wu, W.; Yamamura, S.; Roeder, R.G. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 2004, 24, 8244–8254. [Google Scholar] [CrossRef]
- Taatjes, D.J.; Tjian, R. Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol. Cell 2004, 14, 675–683. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Jeronimo, C.; Langelier, M.F.; Bataille, A.R.; Pascal, J.M.; Pugh, B.F.; Robert, F. Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Mol. Cell 2016, 64, 455–466. [Google Scholar] [CrossRef]
- Petrenko, N.; Jin, Y.; Wong, K.H.; Struhl, K. Mediator Undergoes a Compositional Change during Transcriptional Activation. Mol. Cell 2016, 64, 443–454. [Google Scholar] [CrossRef]
- Osman, S.; Mohammad, E.; Lidschreiber, M.; Stuetzer, A.; Bazso, F.L.; Maier, K.C.; Urlaub, H.; Cramer, P. The Cdk8 kinase module regulates interaction of the mediator complex with RNA polymerase II. J. Biol. Chem. 2021, 296, 100734. [Google Scholar] [CrossRef]
- Fan, X.; Chou, D.M.; Struhl, K. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006, 13, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Knuesel, M.T.; Meyer, K.D.; Donner, A.J.; Espinosa, J.M.; Taatjes, D.J. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 2009, 29, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Elmlund, H.; Baraznenok, V.; Lindahl, M.; Samuelsen, C.O.; Koeck, P.J.; Holmberg, S.; Hebert, H.; Gustafsson, C.M. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. USA 2006, 103, 15788–15793. [Google Scholar] [CrossRef]
- Miller, C.; Matic, I.; Maier, K.C.; Schwalb, B.; Roether, S.; Strasser, K.; Tresch, A.; Mann, M.; Cramer, P. Mediator phosphorylation prevents stress response transcription during non-stress conditions. J. Biol. Chem. 2012, 287, 44017–44026. [Google Scholar] [CrossRef]
- Donner, A.J.; Szostek, S.; Hoover, J.M.; Espinosa, J.M. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 2007, 27, 121–133. [Google Scholar] [CrossRef]
- Bancerek, J.; Poss, Z.C.; Steinparzer, I.; Sedlyarov, V.; Pfaffenwimmer, T.; Mikulic, I.; Dolken, L.; Strobl, B.; Muller, M.; Taatjes, D.J.; et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 2013, 38, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef]
- Chen, M.; Liang, J.; Ji, H.; Yang, Z.; Altilia, S.; Hu, B.; Schronce, A.; McDermott, M.S.J.; Schools, G.P.; Lim, C.U.; et al. CDK8/19 Mediator kinases potentiate induction of transcription by NFkappaB. Proc. Natl. Acad. Sci. USA 2017, 114, 10208–10213. [Google Scholar] [CrossRef]
- Philip, S.; Kumarasiri, M.; Teo, T.; Yu, M.; Wang, S. Cyclin-Dependent Kinase 8: A New Hope in Targeted Cancer Therapy? J. Med. Chem. 2018, 61, 5073–5092. [Google Scholar] [CrossRef]
- Clark, A.D.; Oldenbroek, M.; Boyer, T.G. Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 393–426. [Google Scholar] [CrossRef]
- Jezek, J.; Wang, K.; Yan, R.; Di Cristofano, A.; Cooper, K.F.; Strich, R. Synergistic repression of thyroid hyperplasia by cyclin C and Pten. J. Cell Sci. 2019, 132, jcs230029. [Google Scholar] [CrossRef]
- Wahi, M.; Johnson, A.D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics 1995, 140, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, D.; Ronne, H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995, 23, 4421–4425. [Google Scholar] [CrossRef]
- Tabtiang, R.K.; Herskowitz, I. Nuclear proteins Nut1p and Nut2p cooperate to negatively regulate a Swi4p-dependent lacZ reporter gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 4707–4718. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.M.; Zhang, J.; Jeffery, D.A.; Koleske, A.J.; Thompson, C.M.; Chao, D.M.; Viljoen, M.; van Vuuren, H.J.; Young, R.A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 1995, 374, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Bourbon, H.M.; Aguilera, A.; Ansari, A.Z.; Asturias, F.J.; Berk, A.J.; Bjorklund, S.; Blackwell, T.K.; Borggrefe, T.; Carey, M.; Carlson, M.; et al. A Unified Nomenclature for Protein Subunits of Mediator Complexes Linking Transcriptional Regulators to RNA Polymerase II. Mol. Cell 2004, 14, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef]
- Echalier, A.; Endicott, J.A.; Noble, M.E. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim. Biophys. Acta 2010, 1804, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.V.; Bottcher, J.; Blaesse, M.; Neumann, L.; Huber, R.; Maskos, K. The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J. Mol. Biol. 2011, 412, 251–266. [Google Scholar] [CrossRef]
- Knuesel, M.T.; Meyer, K.D.; Bernecky, C.; Taatjes, D.J. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009, 23, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Makinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Shen, H.; Spaeth, J.M.; Tolvanen, J.H.; Failor, C.; Knudtson, J.F.; McLaughlin, J.; Halder, S.K.; Yang, Q.; Bulun, S.E.; et al. Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J. Biol. Chem. 2018, 293, 4870–4882. [Google Scholar] [CrossRef]
- Li, Y.C.; Chao, T.C.; Kim, H.J.; Cholko, T.; Chen, S.F.; Li, G.; Snyder, L.; Nakanishi, K.; Chang, C.E.; Murakami, K.; et al. Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Li, N.; Fassl, A.; Chick, J.; Inuzuka, H.; Li, X.; Mansour, M.R.; Liu, L.; Wang, H.; King, B.; Shaik, S.; et al. Cyclin C is a haploinsufficient tumour suppressor. Nat. Cell Biol. 2014, 16, 1080–1091. [Google Scholar] [CrossRef]
- Lynch, C.J.; Bernad, R.; Martinez-Val, A.; Shahbazi, M.N.; Nobrega-Pereira, S.; Calvo, I.; Blanco-Aparicio, C.; Tarantino, C.; Garreta, E.; Richart-Gines, L.; et al. Global hyperactivation of enhancers stabilizes human and mouse naive pluripotency through inhibition of CDK8/19 Mediator kinases. Nat. Cell Biol. 2020, 22, 1223–1238. [Google Scholar] [CrossRef] [PubMed]
- Harper, T.M.; Taatjes, D.J. The complex structure and function of Mediator. J. Biol. Chem. 2018, 293, 13778–13785. [Google Scholar] [CrossRef]
- Davis, M.A.; Larimore, E.A.; Fissel, B.M.; Swanger, J.; Taatjes, D.J.; Clurman, B.E. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 2013, 27, 151–156. [Google Scholar] [CrossRef]
- Klatt, F.; Leitner, A.; Kim, I.V.; Ho-Xuan, H.; Schneider, E.V.; Langhammer, F.; Weinmann, R.; Muller, M.R.; Huber, R.; Meister, G.; et al. A precisely positioned MED12 activation helix stimulates CDK8 kinase activity. Proc. Natl. Acad. Sci. USA 2020, 117, 2894–2905. [Google Scholar] [CrossRef] [PubMed]
- Jezek, J.; Smethurst, D.G.J.; Stieg, D.C.; Kiss, Z.A.C.; Hanley, S.E.; Ganesan, V.; Chang, K.T.; Cooper, K.F.; Strich, R. Cyclin C: The Story of a Non-Cycling Cyclin. Biology 2019, 8, 3. [Google Scholar] [CrossRef]
- Nizon, M.; Laugel, V.; Flanigan, K.M.; Pastore, M.; Waldrop, M.A.; Rosenfeld, J.A.; Marom, R.; Xiao, R.; Gerard, A.; Pichon, O.; et al. Variants in MED12L, encoding a subunit of the mediator kinase module, are responsible for intellectual disability associated with transcriptional defect. Genet. Med. 2019, 21, 2713–2722. [Google Scholar] [CrossRef]
- Cooper, K.F.; Strich, R. Functional analysis of the Ume3p/ Srb11p-RNA polymerase II holoenzyme interaction. Gene Expr. 1999, 8, 43–57. [Google Scholar]
- Moye-Rowley, W.S. Transcription factors regulating the response to oxidative stress in yeast. Antioxid. Redox Signal. 2002, 4, 123–140. [Google Scholar] [CrossRef]
- Taymaz-Nikerel, H.; Cankorur-Cetinkaya, A.; Kirdar, B. Genome-Wide Transcriptional Response of Saccharomyces cerevisiae to Stress-Induced Perturbations. Front. Bioeng. Biotechnol. 2016, 4, 17. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Nataro, J.P.; Corcoran, L.; Zirin, S.; Swink, S.; Taichman, N.; Goin, J.; Harris, M.C. Prospective analysis of coagulase-negative staphylococcal infection in hospitalized infants. J. Pediatr. 1994, 125, 798–804. [Google Scholar] [CrossRef]
- Vomund, S.; Schafer, A.; Parnham, M.J.; Brune, B.; von Knethen, A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef]
- Postnikoff, S.D.; Malo, M.E.; Wong, B.; Harkness, T.A. The yeast forkhead transcription factors fkh1 and fkh2 regulate lifespan and stress response together with the anaphase-promoting complex. PLoS Genet. 2012, 8, e1002583. [Google Scholar] [CrossRef] [PubMed]
- Fuse, Y.; Kobayashi, M. Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time. Molecules 2017, 22, 436. [Google Scholar] [CrossRef]
- Perrone, G.G.; Tan, S.X.; Dawes, I.W. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta 2008, 1783, 1354–1368. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Zdralevic, M.; Antonacci, L.; Passarella, S.; Marra, E.; Giannattasio, S. The role of mitochondria in yeast programmed cell death. Front. Oncol. 2012, 2, 70. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Klionsky, D.J. Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2018, 293, 5396–5403. [Google Scholar] [CrossRef]
- Bartholomew, C.R.; Suzuki, T.; Du, Z.; Backues, S.K.; Jin, M.; Lynch-Day, M.A.; Umekawa, M.; Kamath, A.; Zhao, M.; Xie, Z.; et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. USA 2012, 109, 11206–11210. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; He, D.; Backues, S.K.; Freeberg, M.A.; Liu, X.; Kim, J.K.; Klionsky, D.J. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr. Biol. 2014, 24, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Kadosh, D.; Struhl, K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 1998, 18, 5121–5127. [Google Scholar] [CrossRef]
- Vlahakis, A.; Lopez Muniozguren, N.; Powers, T. Stress-response transcription factors Msn2 and Msn4 couple TORC2-Ypk1 signaling and mitochondrial respiration to ATG8 gene expression and autophagy. Autophagy 2017, 13, 1804–1812. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Orgilles, B.; Saldana-Meyer, R.; Wang, E.; Trompouki, E.; Fassl, A.; Lau, S.; Mullenders, J.; Rocha, P.P.; Raviram, R.; Guillamot, M.; et al. MED12 Regulates HSC-Specific Enhancers Independently of Mediator Kinase Activity to Control Hematopoiesis. Cell Stem Cell 2016, 19, 784–799. [Google Scholar] [CrossRef]
- El Khattabi, L.; Zhao, H.; Kalchschmidt, J.; Young, N.; Jung, S.; Van Blerkom, P.; Kieffer-Kwon, P.; Kieffer-Kwon, K.R.; Park, S.; Wang, X.; et al. A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 2019, 178, 1145–1158 e1120. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Jung, U.S.; Piotrowski, J.; Levin, D.E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995, 9, 1559–1571. [Google Scholar] [CrossRef] [PubMed]
- Vilella, F.; Herrero, E.; Torres, J.; de la Torre-Ruiz, M.A. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 2005, 280, 9149–9159. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Parshin, A.V.; Daly, I.; Strich, R.; Cooper, K.F. The cell wall sensors Mtl1, Wsc1, and Mid2 are required for stress-induced nuclear to cytoplasmic translocation of cyclin C and programmed cell death in yeast. Oxid. Med. Cell. Longev. 2013, 2013, 320823. [Google Scholar] [CrossRef]
- Truman, A.W.; Millson, S.H.; Nuttall, J.M.; King, V.; Mollapour, M.; Prodromou, C.; Pearl, L.H.; Piper, P.W. Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2p (Mpk1p) cell integrity stress-activated protein kinase. Eukaryot. Cell 2006, 5, 1914–1924. [Google Scholar] [CrossRef]
- Garcia, R.; Rodriguez-Pena, J.M.; Bermejo, C.; Nombela, C.; Arroyo, J. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 10901–10911. [Google Scholar] [CrossRef]
- Andrews, B.J.; Moore, L.A. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc. Natl. Acad. Sci. USA 1992, 89, 11852–11856. [Google Scholar] [CrossRef]
- Nash, P.; Tang, X.; Orlicky, S.; Chen, Q.; Gertler, F.B.; Mendenhall, M.D.; Sicheri, F.; Pawson, T.; Tyers, M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 2001, 414, 514–521. [Google Scholar] [CrossRef]
- Ang, X.L.; Wade Harper, J. SCF-mediated protein degradation and cell cycle control. Oncogene 2005, 24, 2860–2870. [Google Scholar] [CrossRef] [PubMed]
- Willis, S.D.; Stieg, D.C.; Ong, K.L.; Shah, R.; Strich, A.K.; Grose, J.H.; Cooper, K.F. Snf1 cooperates with the CWI MAPK pathway to mediate the degradation of Med13 following oxidative stress. Microb. Cell 2018, 5, 357–370. [Google Scholar] [CrossRef]
- Hong, S.P.; Carlson, M. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 2007, 282, 16838–16845. [Google Scholar] [CrossRef] [PubMed]
- Cohen, T.J.; Lee, K.; Rutkowski, L.H.; Strich, R. Ask10p mediates the oxidative stress-induced destruction of the Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p. Eukaryot. Cell 2003, 2, 962–970. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garcia, R.; Pulido, V.; Orellana-Munoz, S.; Nombela, C.; Vazquez de Aldana, C.R.; Rodriguez-Pena, J.M.; Arroyo, J. Signalling through the yeast MAPK Cell Wall Integrity pathway controls P-body assembly upon cell wall stress. Sci. Rep. 2019, 9, 3186. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Kim, S.K.; Willis, S.D.; Cooper, K.F. The MAPKKKs Ste11 and Bck1 jointly transduce the high oxidative stress signal through the cell wall integrity MAP kinase pathway. Microb. Cell 2015, 2, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Moye-Rowley, W.S. Feelin’ it: Differential oxidative stress sensing mediated by Cyclin C. Microb. Cell 2015, 2, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Li, S.P.; Westermarck, J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008, 22, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Saito, H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 2010, 13, 677–683. [Google Scholar] [CrossRef]
- Van Drogen, F.; Dard, N.; Pelet, S.; Lee, S.S.; Mishra, R.; Srejic, N.; Peter, M. Crosstalk and spatiotemporal regulation between stress-induced MAP kinase pathways and pheromone signaling in budding yeast. Cell Cycle 2020, 19, 1707–1715. [Google Scholar] [CrossRef]
- Gonzalez, S.; Rallis, C. The TOR Signaling Pathway in Spatial and Temporal Control of Cell Size and Growth. Front. Cell Dev. Biol. 2017, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Albert, V.; Hall, M.N. mTOR signaling in cellular and organismal energetics. Curr. Opin. Cell Biol. 2015, 33, 55–66. [Google Scholar] [CrossRef]
- Sullivan, A.; Wallace, R.L.; Wellington, R.; Luo, X.; Capaldi, A.P. Multilayered regulation of TORC1-body formation in budding yeast. Mol. Biol. Cell 2019, 30, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Reinke, A.; Anderson, S.; McCaffery, J.M.; Yates, J., 3rd; Aronova, S.; Chu, S.; Fairclough, S.; Iverson, C.; Wedaman, K.P.; Powers, T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 14752–14762. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Kaiser, C.A. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006, 8, 657–667. [Google Scholar] [CrossRef]
- Binda, M.; Peli-Gulli, M.P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; De Virgilio, C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 2009, 35, 563–573. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Kim, J.S.; Ro, S.H.; Kim, M.; Park, H.W.; Semple, I.A.; Park, H.; Cho, U.S.; Wang, W.; Guan, K.L.; Karin, M.; et al. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 2015, 5, 9502. [Google Scholar] [CrossRef] [PubMed]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 2015, 4, e09181. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, R.; Peli-Gulli, M.P.; Hu, Z.; Jaquenoud, M.; Garcia Osuna, G.M.; Sardu, A.; Dengjel, J.; De Virgilio, C. Spatially Distinct Pools of TORC1 Balance Protein Homeostasis. Mol. Cell 2019, 73, 325–338 e328. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, V.; Klionsky, D.J. Spatially distinct pools of TORC1 balance protein homeostasis. Autophagy 2019, 15, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef]
- Kamada, Y.; Yoshino, K.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef]
- Torres, J.; Di Como, C.J.; Herrero, E.; De La Torre-Ruiz, M.A. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 2002, 277, 43495–43504. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Jaquenoud, M.; De Virgilio, C. TORC1 controls G1-S cell cycle transition in yeast via Mpk1 and the greatwall kinase pathway. Nat. Commun. 2015, 6, 8256. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Jaquenoud, M.; Peli-Gulli, M.P.; Nicastro, R.; De Virgilio, C. TORC1 coordinates the conversion of Sic1 from a target to an inhibitor of cyclin-CDK-Cks1. Cell Discov. 2017, 3, 17012. [Google Scholar] [CrossRef]
- Gonzalez-Rubio, G.; Sellers-Moya, A.; Martin, H.; Molina, M. Differential Role of Threonine and Tyrosine Phosphorylation in the Activation and Activity of the Yeast MAPK Slt2. Int. J. Mol. Sci. 2021, 22, 1110. [Google Scholar] [CrossRef]
- Yurko, N.; Liu, X.; Yamazaki, T.; Hoque, M.; Tian, B.; Manley, J.L. MPK1/SLT2 Links Multiple Stress Responses with Gene Expression in Budding Yeast by Phosphorylating Tyr1 of the RNAP II CTD. Mol. Cell 2017, 68, 913–925 e913. [Google Scholar] [CrossRef]
- Zhu, J.; Deng, S.; Lu, P.; Bu, W.; Li, T.; Yu, L.; Xie, Z. The Ccl1-Kin28 kinase complex regulates autophagy under nitrogen starvation. J. Cell Sci. 2016, 129, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.H.; Shi, Y.; Zhang, N.; de Poot, S.A.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, T.; Ziv, I.; Rosenzweig, R.; Matiuhin, Y.; Bronner, V.; Glickman, M.H.; Fushman, D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol. Cell 2009, 36, 1018–1033. [Google Scholar] [CrossRef]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Noble, K.N.; Tran, E.J.; Alcazar-Roman, A.R.; Hodge, C.A.; Cole, C.N.; Wente, S.R. The Dbp5 cycle at the nuclear pore complex during mRNA export II: Nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011, 25, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.J.; Zhou, Y.; Corbett, A.H.; Wente, S.R. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 2007, 28, 850–859. [Google Scholar] [CrossRef]
- Aryanpur, P.P.; Regan, C.A.; Collins, J.M.; Mittelmeier, T.M.; Renner, D.M.; Vergara, A.M.; Brown, N.P.; Bolger, T.A. Gle1 Regulates RNA Binding of the DEAD-Box Helicase Ded1 in Its Complex Role in Translation Initiation. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef]
- Alcazar-Roman, A.R.; Tran, E.J.; Guo, S.; Wente, S.R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006, 8, 711–716. [Google Scholar] [CrossRef]
- Weirich, C.S.; Erzberger, J.P.; Flick, J.S.; Berger, J.M.; Thorner, J.; Weis, K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006, 8, 668–676. [Google Scholar] [CrossRef]
- Mao, K.; Chew, L.H.; Inoue-Aono, Y.; Cheong, H.; Nair, U.; Popelka, H.; Yip, C.K.; Klionsky, D.J. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc. Natl. Acad. Sci. USA 2013, 110, E2875–E2884. [Google Scholar] [CrossRef]
- Hollenstein, D.M.; Gomez-Sanchez, R.; Ciftci, A.; Kriegenburg, F.; Mari, M.; Torggler, R.; Licheva, M.; Reggiori, F.; Kraft, C. Vac8 spatially confines autophagosome formation at the vacuole in S. cerevisiae. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Millen, J.I.; Krick, R.; Prick, T.; Thumm, M.; Goldfarb, D.S. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy 2009, 5, 75–81. [Google Scholar] [CrossRef]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef]
- Papandreou, M.E.; Tavernarakis, N. Nucleophagy: From homeostasis to disease. Cell Death Differ. 2019, 26, 630–639. [Google Scholar] [CrossRef]
- Hanley, S.E.; Cooper, K.F. Sorting Nexins in Protein Homeostasis. Cells 2020, 10, 17. [Google Scholar] [CrossRef]
- Haft, C.R.; de la Luz Sierra, M.; Barr, V.A.; Haft, D.H.; Taylor, S.I. Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol. Cell. Biol. 1998, 18, 7278–7287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hu, B.; You, Y.; Yang, Z.; Liu, L.; Tang, H.; Bao, W.; Guan, Y.; Shen, X. Sorting nexin 10 acts as a tumor suppressor in tumorigenesis and progression of colorectal cancer through regulating chaperone mediated autophagy degradation of p21(Cip1/WAF1). Cancer Lett. 2018, 419, 116–127. [Google Scholar] [CrossRef]
- Hu, Y.B.; Dammer, E.B.; Ren, R.J.; Wang, G. The endosomal-lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 2015, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Stanishneva-Konovalova, T.B.; Derkacheva, N.I.; Polevova, S.V.; Sokolova, O.S. The Role of BAR Domain Proteins in the Regulation of Membrane Dynamics. Acta Nat. 2016, 8, 60–69. [Google Scholar] [CrossRef]
- Popelka, H.; Damasio, A.; Hinshaw, J.E.; Klionsky, D.J.; Ragusa, M.J. Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc. Natl. Acad. Sci. USA 2017, 114, E10112–E10121. [Google Scholar] [CrossRef] [PubMed]
- Bean, B.D.; Davey, M.; Conibear, E. Cargo selectivity of yeast sorting nexins. Traffic 2017, 18, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Nemec, A.A.; Howell, L.A.; Peterson, A.K.; Murray, M.A.; Tomko, R.J., Jr. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J. Biol. Chem. 2017, 292, 21466–21480. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Kawamata, T.; Iwasaki, S.; Ohsumi, Y. Selectivity of mRNA degradation by autophagy in yeast. Nat. Commun. 2021, 12, 2316. [Google Scholar] [CrossRef]
- Ma, M.; Burd, C.G.; Chi, R.J. Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic 2017, 18, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Burd, C.G. Retrograde trafficking and plasma membrane recycling pathways of the budding yeast Saccharomyces cerevisiae. Traffic 2020, 21, 45–59. [Google Scholar] [CrossRef]
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010, 11, 872–884. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020, 15, 235–259. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Atkins, K.; Dasgupta, A.; Chen, K.H.; Mewburn, J.; Archer, S.L. The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: Implications for human disease. Clin. Sci. 2016, 130, 1861–1874. [Google Scholar] [CrossRef]
- Frohlich, C.; Grabiger, S.; Schwefel, D.; Faelber, K.; Rosenbaum, E.; Mears, J.; Rocks, O.; Daumke, O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013, 32, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Otsuga, D.; Keegan, B.R.; Brisch, E.; Thatcher, J.W.; Hermann, G.J.; Bleazard, W.; Shaw, J.M. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998, 143, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Bleazard, W.; McCaffery, J.M.; King, E.J.; Bale, S.; Mozdy, A.; Tieu, Q.; Nunnari, J.; Shaw, J.M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999, 1, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Mozdy, A.D.; McCaffery, J.M.; Shaw, J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000, 151, 367–380. [Google Scholar] [CrossRef]
- Tieu, Q.; Nunnari, J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000, 151, 353–366. [Google Scholar] [CrossRef]
- Cerveny, K.L.; Jensen, R.E. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 2003, 14, 4126–4139. [Google Scholar] [CrossRef] [PubMed]
- Motley, A.M.; Ward, G.P.; Hettema, E.H. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 2008, 121, 1633–1640. [Google Scholar] [CrossRef]
- Naylor, K.; Ingerman, E.; Okreglak, V.; Marino, M.; Hinshaw, J.E.; Nunnari, J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 2006, 281, 2177–2183. [Google Scholar] [CrossRef]
- Bhar, D.; Karren, M.A.; Babst, M.; Shaw, J.M. Dimeric Dnm1-G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 2006, 281, 17312–17320. [Google Scholar] [CrossRef]
- Anton, F.; Fres, J.M.; Schauss, A.; Pinson, B.; Praefcke, G.J.; Langer, T.; Escobar-Henriques, M. Ugo1 and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer membrane fusion. J. Cell Sci. 2011, 124, 1126–1135. [Google Scholar] [CrossRef]
- Sesaki, H.; Jensen, R.E. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 2001, 152, 1123–1134. [Google Scholar] [CrossRef]
- Hoppins, S.; Lackner, L.; Nunnari, J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007, 76, 751–780. [Google Scholar] [CrossRef]
- Chan, D.C. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012, 46, 265–287. [Google Scholar] [CrossRef]
- Kraus, F.; Roy, K.; Pucadyil, T.J.; Ryan, M.T. Function and regulation of the divisome for mitochondrial fission. Nature 2021, 590, 57–66. [Google Scholar] [CrossRef]
- Liesa, M.; Palacin, M.; Zorzano, A. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef]
- Liesa, M.; Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Kanki, T.; Wang, K.; Baba, M.; Bartholomew, C.R.; Lynch-Day, M.A.; Du, Z.; Geng, J.; Mao, K.; Yang, Z.; Yen, W.L.; et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 2009, 20, 4730–4738. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Westermann, B. Mitochondrial dynamics in model organisms: What yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin. Cell Dev. Biol. 2010, 21, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.C.; Scorrano, L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim. Biophys. Acta 2013, 1833, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Kroemer, G.; Kepp, O. Mitophagy: An Emerging Role in Aging and Age-Associated Diseases. Front. Cell Dev. Biol. 2020, 8, 200. [Google Scholar] [CrossRef]
- Chakravorty, A.; Jetto, C.T.; Manjithaya, R. Dysfunctional Mitochondria and Mitophagy as Drivers of Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 2019, 11, 311. [Google Scholar] [CrossRef]
- Jezek, J.; Cooper, K.F.; Strich, R. The Impact of Mitochondrial Fission-Stimulated ROS Production on Pro-Apoptotic Chemotherapy. Biology 2021, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Subramanian, M.; Yurdagul, A., Jr.; Barbosa-Lorenzi, V.C.; Cai, B.; de Juan-Sanz, J.; Ryan, T.A.; Nomura, M.; Maxfield, F.R.; Tabas, I. Mitochondrial Fission Promotes the Continued Clearance of Apoptotic Cells by Macrophages. Cell 2017, 171, 331–345.e322. [Google Scholar] [CrossRef]
- Pena-Blanco, A.; Garcia-Saez, A.J. Bax, Bak and beyond-mitochondrial performance in apoptosis. FEBS J. 2018, 285, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Karbowski, M.; Lee, Y.J.; Gaume, B.; Jeong, S.Y.; Frank, S.; Nechushtan, A.; Santel, A.; Fuller, M.; Smith, C.L.; Youle, R.J. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 2002, 159, 931–938. [Google Scholar] [CrossRef]
- Tsai, C.J.; Liu, S.; Hung, C.L.; Jhong, S.R.; Sung, T.C.; Chiang, Y.W. BAX-induced apoptosis can be initiated through a conformational selection mechanism. Structure 2015, 23, 139–148. [Google Scholar] [CrossRef][Green Version]
- Antonsson, B.; Montessuit, S.; Sanchez, B.; Martinou, J.C. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J. Biol. Chem. 2001, 276, 11615–11623. [Google Scholar] [CrossRef]
- Saito, M.; Korsmeyer, S.J.; Schlesinger, P.H. BAX-dependent transport of cytochrome c reconstituted in pure liposomes. Nat. Cell Biol. 2000, 2, 553–555. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Saez, A.J. The BCL-2 family saga. Nat. Rev. Mol. Cell Biol. 2020, 21, 564–565. [Google Scholar] [CrossRef] [PubMed]
- Rathore, R.; McCallum, J.E.; Varghese, E.; Florea, A.M.; Busselberg, D. Overcoming chemotherapy drug resistance by targeting inhibitors of apoptosis proteins (IAPs). Apoptosis 2017, 22, 898–919. [Google Scholar] [CrossRef]
- Breckenridge, D.G.; Stojanovic, M.; Marcellus, R.C.; Shore, G.C. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 2003, 160, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Estaquier, J.; Arnoult, D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007, 14, 1086–1094. [Google Scholar] [CrossRef]
- Parone, P.A.; James, D.I.; Da Cruz, S.; Mattenberger, Y.; Donze, O.; Barja, F.; Martinou, J.C. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol. Cell. Biol. 2006, 26, 7397–7408. [Google Scholar] [CrossRef]
- Strich, R. Programmed Cell Death Initiation and Execution in Budding Yeast. Genetics 2015, 200, 1003–1014. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Eisenberg, T.; Buttner, S.; Meisinger, C.; Kroemer, G.; Madeo, F. Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 2010, 17, 763–773. [Google Scholar] [CrossRef]
- Chaves, S.R.; Rego, A.; Martins, V.M.; Santos-Pereira, C.; Sousa, M.J.; Corte-Real, M. Regulation of Cell Death Induced by Acetic Acid in Yeasts. Front. Cell Dev. Biol. 2021, 9, 642375. [Google Scholar] [CrossRef]
- Falcone, C.; Mazzoni, C. External and internal triggers of cell death in yeast. Cell. Mol. Life Sci. 2016, 73, 2237–2250. [Google Scholar] [CrossRef]
- Frank, S.; Gaume, B.; Bergmann-Leitner, E.S.; Leitner, W.W.; Robert, E.G.; Catez, F.; Smith, C.L.; Youle, R.J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 2001, 1, 515–525. [Google Scholar] [CrossRef]
- Demetrick, D.J.; Matsumoto, S.; Hannon, G.J.; Okamoto, K.; Xiong, Y.; Zhang, H.; Beach, D.H. Chromosomal mapping of the genes for the human cell cycle proteins cyclin C (CCNC), cyclin E (CCNE), p21 (CDKN1) and KAP (CDKN3). Cytogenet. Cell Genet. 1995, 69, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Ohata, N.; Ito, S.; Yoshida, A.; Kunisada, T.; Numoto, K.; Jitsumori, Y.; Kanzaki, H.; Ozaki, T.; Shimizu, K.; Ouchida, M. Highly frequent allelic loss of chromosome 6q16-23 in osteosarcoma: Involvement of cyclin C in osteosarcoma. Int. J. Mol. Med. 2006, 18, 1153–1158. [Google Scholar] [CrossRef]
- Liang, J.; Chen, M.; Hughes, D.; Chumanevich, A.A.; Altilia, S.; Kaza, V.; Lim, C.U.; Kiaris, H.; Mythreye, K.; Pena, M.M.; et al. CDK8 Selectively Promotes the Growth of Colon Cancer Metastases in the Liver by Regulating Gene Expression of TIMP3 and Matrix Metalloproteinases. Cancer Res. 2018, 78, 6594–6606. [Google Scholar] [CrossRef] [PubMed]
- Bragelmann, J.; Klumper, N.; Offermann, A.; von Massenhausen, A.; Bohm, D.; Deng, M.; Queisser, A.; Sanders, C.; Syring, I.; Merseburger, A.S.; et al. Pan-Cancer Analysis of the Mediator Complex Transcriptome Identifies CDK19 and CDK8 as Therapeutic Targets in Advanced Prostate Cancer. Clin. Cancer. Res. 2017, 23, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.S.; Chumanevich, A.A.; Lim, C.U.; Liang, J.; Chen, M.; Altilia, S.; Oliver, D.; Rae, J.M.; Shtutman, M.; Kiaris, H.; et al. Inhibition of CDK8 mediator kinase suppresses estrogen dependent transcription and the growth of estrogen receptor positive breast cancer. Oncotarget 2017, 8, 12558–12575. [Google Scholar] [CrossRef]
- Roninson, I.B.; Gyorffy, B.; Mack, Z.T.; Shtil, A.A.; Shtutman, M.S.; Chen, M.; Broude, E.V. Identifying Cancers Impacted by CDK8/19. Cells 2019, 8, 821. [Google Scholar] [CrossRef] [PubMed]
- Fant, C.B.; Taatjes, D.J. Regulatory functions of the Mediator kinases CDK8 and CDK19. Transcription 2019, 10, 76–90. [Google Scholar] [CrossRef]
- Rambold, A.S.; Kostelecky, B.; Elia, N.; Lippincott-Schwartz, J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 10190–10195. [Google Scholar] [CrossRef]
- Rambold, A.S.; Kostelecky, B.; Lippincott-Schwartz, J. Together we are stronger: Fusion protects mitochondria from autophagosomal degradation. Autophagy 2011, 7, 1568–1569. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Prudent, J.; Basu, K.; Goyon, V.; Katsumura, S.; Hulea, L.; Pearl, D.; Siddiqui, N.; Strack, S.; McGuirk, S.; et al. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell 2017, 67, 922–935 e925. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef]
- Tondera, D.; Grandemange, S.; Jourdain, A.; Karbowski, M.; Mattenberger, Y.; Herzig, S.; Da Cruz, S.; Clerc, P.; Raschke, I.; Merkwirth, C.; et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009, 28, 1589–1600. [Google Scholar] [CrossRef]
- Egner, A.; Jakobs, S.; Hell, S.W. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. USA 2002, 99, 3370–3375. [Google Scholar] [CrossRef]
- Walden, E.A.; Fong, R.Y.; Pham, T.T.; Knill, H.; Laframboise, S.J.; Huard, S.; Harper, M.E.; Baetz, K. Phenomic screen identifies a role for the yeast lysine acetyltransferase NuA4 in the control of Bcy1 subcellular localization, glycogen biosynthesis, and mitochondrial morphology. PLoS Genet. 2020, 16, e1009220. [Google Scholar] [CrossRef]
- Zemirli, N.; Morel, E.; Molino, D. Mitochondrial Dynamics in Basal and Stressful Conditions. Int. J. Mol. Sci. 2018, 19, 564. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Anbo, H.; Sato, M.; Okoshi, A.; Fukuchi, S. Functional Segments on Intrinsically Disordered Regions in Disease-Related Proteins. Biomolecules 2019, 9, 88. [Google Scholar] [CrossRef]
- Brodsky, S.; Jana, T.; Mittelman, K.; Chapal, M.; Kumar, D.K.; Carmi, M.; Barkai, N. Intrinsically Disordered Regions Direct Transcription Factor In Vivo Binding Specificity. Mol. Cell 2020, 79, 459–471 e454. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Bassel-Duby, R.; Olson, E.N. Heart- and muscle-derived signaling system dependent on MED13 and Wingless controls obesity in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, 9491–9496. [Google Scholar] [CrossRef] [PubMed]
- Tutter, A.V.; Kowalski, M.P.; Baltus, G.A.; Iourgenko, V.; Labow, M.; Li, E.; Kadam, S. Role for Med12 in regulation of Nanog and Nanog target genes. J. Biol. Chem. 2009, 284, 3709–3718. [Google Scholar] [CrossRef]
- Miao, Y.L.; Gambini, A.; Zhang, Y.; Padilla-Banks, E.; Jefferson, W.N.; Bernhardt, M.L.; Huang, W.; Li, L.; Williams, C.J. Mediator complex component MED13 regulates zygotic genome activation and is required for postimplantation development in the mouse. Biol. Reprod. 2018, 98, 449–464. [Google Scholar] [CrossRef]
- Loncle, N.; Boube, M.; Joulia, L.; Boschiero, C.; Werner, M.; Cribbs, D.L.; Bourbon, H.M. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007, 26, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Gillmor, C.S.; Silva-Ortega, C.O.; Willmann, M.R.; Buendia-Monreal, M.; Poethig, R.S. The Arabidopsis Mediator CDK8 module genes CCT (MED12) and GCT (MED13) are global regulators of developmental phase transitions. Development 2014, 141, 4580–4589. [Google Scholar] [CrossRef] [PubMed]
- Keightley, M.C.; Layton, J.E.; Hayman, J.W.; Heath, J.K.; Lieschke, G.J. Mediator subunit 12 is required for neutrophil development in zebrafish. PLoS ONE 2011, 6, e23845. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Makinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, R.; Zweier, M.; Gogoll, L.; Schiffmann, R.; Sticht, H.; Steindl, K.; Rauch, A. Genotype-phenotype evaluation of MED13L defects in the light of a novel truncating and a recurrent missense mutation. Eur. J. Med. Genet. 2017, 60, 451–464. [Google Scholar] [CrossRef]
- van Haelst, M.M.; Monroe, G.R.; Duran, K.; van Binsbergen, E.; Breur, J.M.; Giltay, J.C.; van Haaften, G. Further confirmation of the MED13L haploinsufficiency syndrome. Eur. J. Hum. Genet. 2015, 23, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Tyler, J.K.; Johnson, J.E. The role of autophagy in the regulation of yeast life span. Ann. N. Y. Acad. Sci. 2018, 1418, 31–43. [Google Scholar] [CrossRef]
- Alarcon, C.; Zaromytidou, A.I.; Xi, Q.; Gao, S.; Yu, J.; Fujisawa, S.; Barlas, A.; Miller, A.N.; Manova-Todorova, K.; Macias, M.J.; et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 2009, 139, 757–769. [Google Scholar] [CrossRef]
- Hengartner, C.J.; Thompson, C.M.; Zhang, J.; Chao, D.M.; Liao, S.M.; Koleske, A.J.; Okamura, S.; Young, R.A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995, 9, 897–910. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Donner, A.J.; Hoover, J.M.; Szostek, S.A.; Espinosa, J.M. Stimulus-specific transcriptional regulation within the p53 network. Cell Cycle 2007, 6, 2594–2598. [Google Scholar] [CrossRef]
- Guo, Y.E.; Manteiga, J.C.; Henninger, J.E.; Sabari, B.R.; Dall’Agnese, A.; Hannett, N.M.; Spille, J.H.; Afeyan, L.K.; Zamudio, A.V.; Shrinivas, K.; et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572, 543–548. [Google Scholar] [CrossRef]
- Gonzalez, D.; Hamidi, N.; Del Sol, R.; Benschop, J.J.; Nancy, T.; Li, C.; Francis, L.; Tzouros, M.; Krijgsveld, J.; Holstege, F.C.; et al. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl. Acad. Sci. USA 2014, 111, 2500–2505. [Google Scholar] [CrossRef]
- Poss, Z.C.; Ebmeier, C.C.; Odell, A.T.; Tangpeerachaikul, A.; Lee, T.; Pelish, H.E.; Shair, M.D.; Dowell, R.D.; Old, W.M.; Taatjes, D.J. Identification of Mediator Kinase Substrates in Human Cells using Cortistatin A and Quantitative Phosphoproteomics. Cell Rep. 2016, 15, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Huddleston, M.J.; Zhang, X.; Young, R.A.; Annan, R.S.; Carr, S.A.; Deshaies, R.J. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001, 15, 1078–1092. [Google Scholar] [CrossRef]
- Raithatha, S.; Su, T.C.; Lourenco, P.; Goto, S.; Sadowski, I. Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol. Cell. Biol. 2012, 32, 664–674. [Google Scholar] [CrossRef]
- Zhao, J.; Ramos, R.; Demma, M. CDK8 regulates E2F1 transcriptional activity through S375 phosphorylation. Oncogene 2013, 32, 3520–3530. [Google Scholar] [CrossRef] [PubMed]
- Fryer, C.J.; White, J.B.; Jones, K.A. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 2004, 16, 509–520. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Friedson, B.; Cooper, K.F. Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress. Microorganisms 2021, 9, 2152. https://doi.org/10.3390/microorganisms9102152
Friedson B, Cooper KF. Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress. Microorganisms. 2021; 9(10):2152. https://doi.org/10.3390/microorganisms9102152
Chicago/Turabian StyleFriedson, Brittany, and Katrina F. Cooper. 2021. "Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress" Microorganisms 9, no. 10: 2152. https://doi.org/10.3390/microorganisms9102152
APA StyleFriedson, B., & Cooper, K. F. (2021). Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress. Microorganisms, 9(10), 2152. https://doi.org/10.3390/microorganisms9102152







