The Effect of Lithium on the Budding Yeast Saccharomyces cerevisiae upon Stress Adaptation
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
| Strains used in this study. | ||
| Name of the Strain | Genotype | Source |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3∆ | Hohmann collection |
| Hsp104-GFP | HSP104-GFP::HIS3 in BY4741 | Nyström collection |
| crGE | pRS303_TEF1pr_CrGE::HIS3 in BY4741 | Boersma Lab [17] |
2.2. Growth of Yeast Cells under Salt Stress Conditions
2.3. Western Blotting
2.4. Cell Volume Recovery Assessment
2.5. Aggregate Formation upon Salt Stress
2.6. Crowding Measurements
2.7. Microscopy of Aggregates Formed in Cells Exposed to Different Stressors after Pre-adaptation to Salt
2.8. Cell Density Correlation and Survival Assay
3. Results
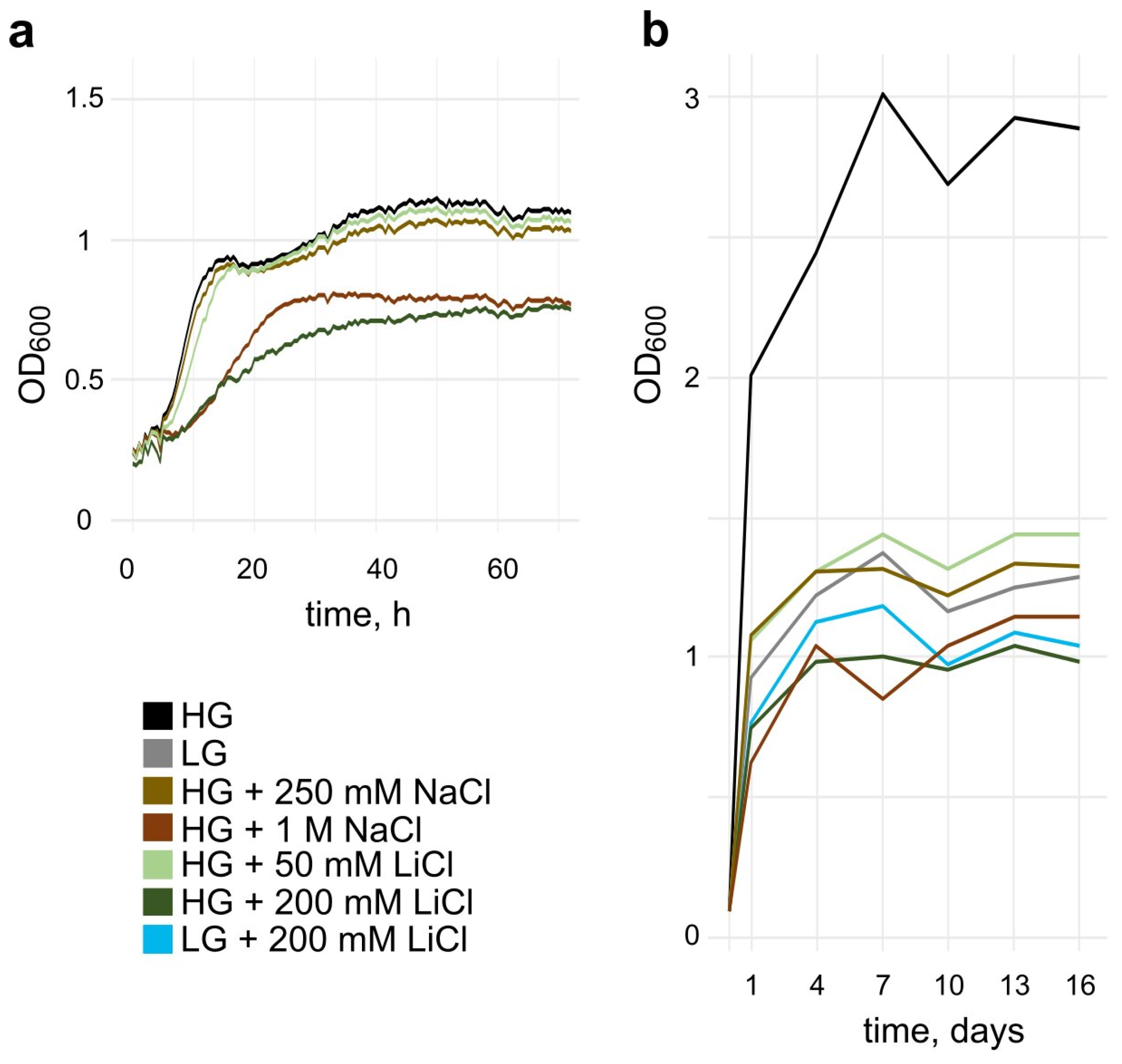
3.1. Salt Stress Reduces the Growth Rate and Maximal Cell Density
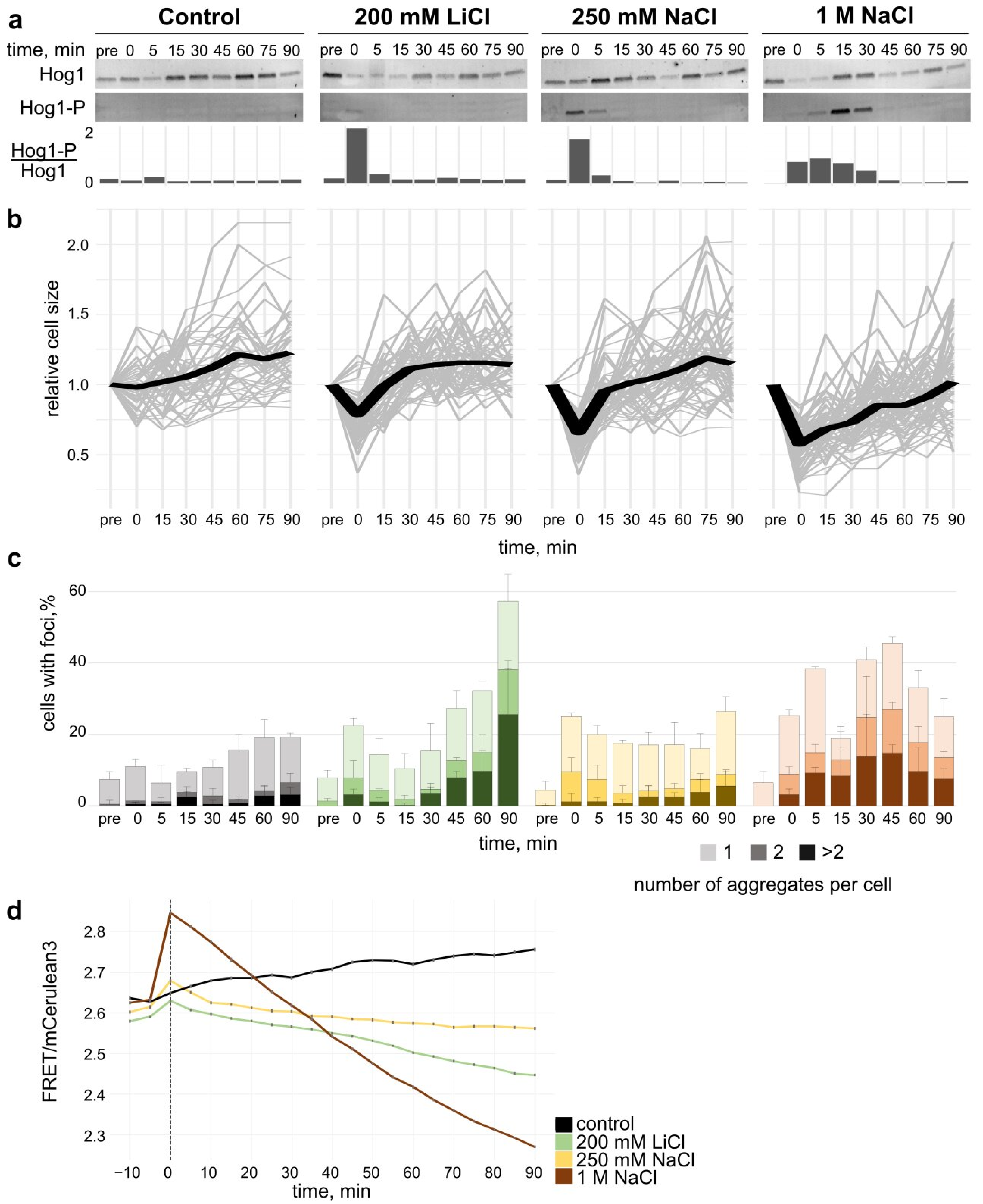
3.2. Cellular Response to Salt Stress
3.2.1. Hog1 Phosphorylation upon Salt Stress
3.2.2. Cell Volume Recovery upon Salt Stress
3.2.3. Formation of Hsp104 Aggregates upon Hyperosmolar Salt Shift
3.2.4. Crowding Modulation upon Salt Stress
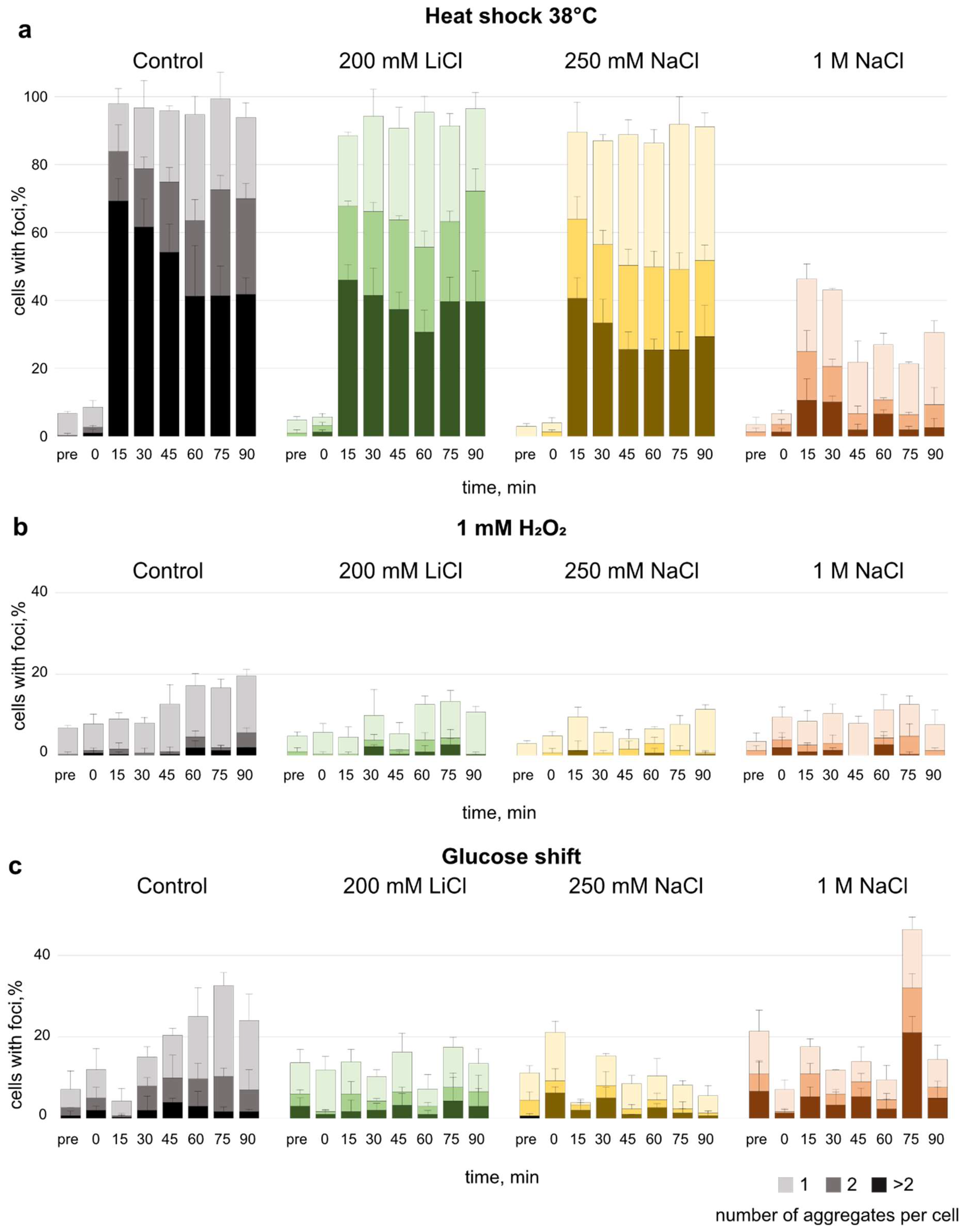
3.3. Effect of Salt Pre-Adaptation on Formation of Protein Aggregates
3.4. Effect of Lithium Chloride on Long-Term Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lithium Cation: Uses, Interactions, Mechanism of Action|DrugBank Online. Available online: https://go.drugbank.com/drugs/DB01356 (accessed on 8 November 2021).
- Manji, H.K.; Lenox, R.H. Lithium: A Molecular Transducer of Mood-Stabilization in the Treatment of Bipolar Disorder. Neuropsychopharmacology 1998, 19, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.T.; Chuang, D.M. Neuroprotective Action of Lithium in Disorders of the Central Nervous System. J. Cent. South Univ. Medical Sci. 2011, 36, 461–475. [Google Scholar] [CrossRef]
- EEML—Electronic Essential Medicines List. Available online: https://list.essentialmeds.org/medicines/248 (accessed on 8 November 2021).
- Sarkar, S.; Floto, R.A.; Berger, Z.; Imarisio, S.; Cordenier, A.; Pasco, M.; Cook, L.J.; Rubinsztein, D.C. Lithium Induces Autophagy by Inhibiting Inositol Monophosphatase. J. Cell Biol. 2005, 170, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Sanchez-Lopez, E.; Karin, M. Autophagy, Inflammation, and Immunity: A Troika Governing Cancer and Its Treatment. Cell 2016, 166, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Jakobsson, E. Systems Biology Understanding of the Effects of Lithium on Affective and Neurodegenerative Disorders. Front. Neurosci. 2018, 12, 296. [Google Scholar] [CrossRef]
- Ruiz, A.; González, A.; García-Salcedo, R.; Ramos, J.; Ariño, J. Role of Protein Phosphatases 2C on Tolerance to Lithium Toxicity in the Yeast Saccharomyces Cerevisiae. Mol. Microbiol. 2006, 62, 263–277. [Google Scholar] [CrossRef]
- Masuda, C.A.; Xavier, M.A.; Mattos, K.A.; Galina, A.; Montero-Lomelí, M. Phosphoglucomutase Is an in Vivo Lithium Target in Yeast. J. Biol. Chem. 2001, 276, 37794–37801. [Google Scholar] [CrossRef]
- Lopez, F.; Leube, M.; Gil-Mascarell, R.; Navarro-Aviñó, J.P.; Serrano, R. The Yeast Inositol Monophosphatase Is a Lithium- and Sodium-Sensitive Enzyme Encoded by a Non-Essential Gene Pair. Mol. Microbiol. 1999, 31, 1255–1264. [Google Scholar] [CrossRef]
- Forlenza, O.V.; De Paula, V.J.; MacHado-Vieira, R.; Diniz, B.S.; Gattaz, W.F. Does Lithium Prevent Alzheimer’s Disease? Drugs Aging 2012, 29, 335–342. [Google Scholar] [CrossRef]
- Damri, O.; Shemesh, N.; Agam, G. Is There Justification to Treat Neurodegenerative Disorders by Repurposing Drugs? The Case of Alzheimer’s Disease, Lithium, and Autophagy. Int. J. Mol. Sci. 2020, 22, 189. [Google Scholar] [CrossRef]
- Medeiros, R.; Baglietto-Vargas, D.; Laferla, F.M. The Role of Tau in Alzheimer’s Disease and Related Disorders. CNS Neurosci. Ther. 2011, 17, 514. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.; Planel, E.; Zehr, C.; Olm, V.; Meyerson, J.; Suleman, F.; Gaynor, K.; Wang, L.; LaFrancois, J.; Feinstein, B.; et al. Inhibition of Glycogen Synthase Kinase-3 by Lithium Correlates with Reduced Tauopathy and Degeneration in Vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 6990–6995. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Sofola-Adesakin, O.; Castillo-Quan, J.I.; Rallis, C.; Tain, L.S.; Bjedov, I.; Rogers, I.; Li, L.; Martinez, P.; Khericha, M.; Cabecinha, M.; et al. Lithium Suppresses Aβ Pathology by Inhibiting Translation in an Adult Drosophila Model of Alzheimer’s Disease. Front. Aging Neurosci. 2014, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Boersma, A.J.; Zuhorn, I.S.; Poolman, B. A Sensor for Quantification of Macromolecular Crowding in Living Cells. Nat. Methods 2015, 12, 227–229. [Google Scholar] [CrossRef]
- Talemi, S.R.; Tiger, C.-F.; Andersson, M.; Babazadeh, R.; Welkenhuysen, N.; Klipp, E.; Hohmann, S.; Schaber, J. Systems Level Analysis of the Yeast Osmo-Stat. Sci. Rep. 2016, 6, 30950. [Google Scholar] [CrossRef]
- Liu, B.; Mavrova, S.N.; Van Den Berg, J.; Kristensen, S.K.; Mantovanelli, L.; Veenhoff, L.M.; Poolman, B.; Boersma, A.J. Influence of Fluorescent Protein Maturation on FRET Measurements in Living Cells. ACS Sensors 2018, 3, 1735–1742. [Google Scholar] [CrossRef]
- Mouton, S.N.; Thaller, D.J.; Crane, M.M.; Rempel, I.L.; Steen, A.; Kaeberlein, M.; Lusk, C.P.; Boersma, A.J.; Veenhoff, L.M. A Physicochemical Roadmap of Yeast Replicative Aging. bioRxiv 2019. [Google Scholar] [CrossRef]
- Laidlaw, K.; Bisinski, D.; Shashkova, S.; Paine, K.; Veillon, M.; Leake, M.; MacDonald, C. A Glucose Starvation Response Governs Endocytic Trafficking and Eisosomal Retention of Surface Cargoes in Budding Yeast. J. Cell Sci. 2020, 2, jcs257733. [Google Scholar] [CrossRef]
- Lu, A.X.; Zarin, T.; Hsu, I.S.; Moses, A.M. YeastSpotter: Accurate and Parameter-Free Web Segmentation for Microscopy Images of Yeast Cells. Bioinformatics 2019, 35, 4525–4527. [Google Scholar] [CrossRef]
- Huh, G.H.; Damsz, B.; Matsumoto, T.K.; Reddy, M.P.; Rus, A.M.; Ibeas, J.I.; Narasimhan, M.L.; Bressan, R.A.; Hasegawa, P.M. Salt Causes Ion Disequilibrium-Induced Programmed Cell Death in Yeast and Plants. Plant J. 2002, 29, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Babazadeh, R.; Adiels, C.B.; Smedh, M.; Petelenz-Kurdziel, E.; Goksor, M.; Hohmann, S. Osmostress-Induced Cell Volume Loss Delays Yeast Hog1 Signaling by Limiting Diffusion Processes and by Hog1-Specific Effects. PLoS ONE 2013, 8, e80901. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Control of High Osmolarity Signalling in the Yeast Saccharomyces Cerevisiae. FEBS Lett. 2009, 583, 4025–4029. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic Stress Signaling and Osmoadaptation in Yeasts Osmotic. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Klipp, E.; Nordlander, B.; Krüger, R.; Gennemark, P.; Hohmann, S. Integrative Model of the Response of Yeast to Osmotic Shock. Nat. Biotechnol. 2005, 23, 975–982. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Rivas, G.; Minton, A.P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys. 2008, 37, 375–397. [Google Scholar] [CrossRef]
- Spitzer, J.; Poolman, B. The Role of Biomacromolecular Crowding, Ionic Strength, and Physicochemical Gradients in the Complexities of Life’s Emergence. Microbiol. Mol. Biol. Rev. 2009, 73, 371–388. [Google Scholar] [CrossRef]
- Zhou, C.; Slaughter, B.D.; Unruh, J.R.; Eldakak, A.; Rubinstein, B.; Li, R. Motility and Segregation of Hsp104-Associated Protein Aggregates in Budding Yeast. Cell 2011, 147, 1186–1196. [Google Scholar] [CrossRef]
- Schneider, K.L.; Wollman, A.J.M.; Nyström, T.; Shashkova, S. Comparison of Endogenously Expressed Fluorescent Protein Fusions Behaviour for Protein Quality Control and Cellular Ageing Research. Sci. Rep. 2021, 11, 12819. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending Healthy Life Span-from Yeast to Humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Andalis, A.A.; Fink, G.R.; Guarente, L. High Osmolarity Extends Life Span in Saccharomyces Cerevisiae by a Mechanism Related to Calorie Restriction. Mol. Cell. Biol. 2002, 22, 8056–8066. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie Restriction Extends Saccharomyces Cerevisiae Lifespan by Increasing Respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Delaney, J.R.; Murakami, C.; Chou, A.; Carr, D.; Schleit, J.; Sutphin, G.L.; An, E.H.; Castanza, A.S.; Fletcher, M.; Goswami, S.; et al. Dietary Restriction and Mitochondrial Function Link Replicative and Chronological Aging in Saccharomyces Cerevisiae. Exp. Gerontol. 2013, 48, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Quan, J.I.; Li, L.; Kinghorn, K.J.; Ivanov, D.K.; Tain, L.S.; Slack, C.; Kerr, F.; Nespital, T.; Thornton, J.; Hardy, J.; et al. Lithium Promotes Longevity through GSK3/NRF2-Dependent Hormesis. Cell Rep. 2016, 15, 638–650. [Google Scholar] [CrossRef]
- Maxwell, P.H. Growth Conditions That Increase or Decrease Lifespan in Saccharomyces Cerevisiae Lead to Corresponding Decreases or Increases in Rates of Interstitial Deletions and Non-Reciprocal Translocations. BMC Genet. 2016, 17, 140. [Google Scholar] [CrossRef]
- Zimkus, A.; Misiūnas, A.; Chaustova, L. Li + Effect on the Cell Wall of the Yeast Saccharomyces Cerevisiae as Probed by FT-IR Spectroscopy. Cent. Eur. J. Biol. 2013, 8, 724–729. [Google Scholar] [CrossRef][Green Version]
- Gietz, R.D.; Schiestl, R.H. Frozen Competent Yeast Cells That Can Be Transformed with High Efficiency Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 1–4. [Google Scholar] [CrossRef]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The Response to Heat Shock and Oxidative Stress in Saccharomyces Cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef]
- Sanchez, Y.; Taulien, J.; Borkovich, K.A.; Lindquist, S. Hsp104 Is Required for Tolerance to Many Forms of Stress. EMBO J. 1992, 11, 2357–2364. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Reichmann, D.; Voth, W.; Jakob, U. Maintaining a Healthy Proteome during Oxidative Stress. Mol. Cell 2018, 69, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Sathyanarayanan, U.; Musa, M.; Bou Dib, P.; Raimundo, N.; Milosevic, I.; Krisko, A. ATP Hydrolysis by Yeast Hsp104 Determines Protein Aggregate Dissolution and Size In Vivo. Nat. Commun. 2020, 11, 5226. [Google Scholar] [CrossRef]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A Novel Chaperone System That Rescues Previously Aggregated Proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Dudev, T.; Grauffel, C.; Lim, C. How Native and Alien Metal Cations Bind ATP: Implications for Lithium as a Therapeutic Agent. Sci. Rep. 2017, 7, 42377. [Google Scholar] [CrossRef] [PubMed]
- Marek, P.J.; Patsalo, V.; Green, D.F.; Raleigh, D.P. Ionic Strength Effects on Amyloid Formation by Amylin Are a Complicated Interplay among Debye Screening, Ion Selectivity, and Hofmeister Effects. Biochemistry 2012, 51, 8478–8490. [Google Scholar] [CrossRef]
- Zhang, Y.; Cremer, P.S. Interactions between Macromolecules and Ions: The Hofmeister Series. Curr. Opin. Chem. Biol. 2006, 10, 658–663. [Google Scholar] [CrossRef]
- Leirmo, S.; Harrison, C.; Cayley, D.S.; Record, M.T.; Burgess, R.R. Replacement of Potassium Chloride by Potassium Glutamate Dramatically Enhances Protein-DNA Interactions In Vitro. Biochemistry 1987, 26, 2095–2101. [Google Scholar] [CrossRef]
- Liu, B.; Poolman, B.; Boersma, A.J. Ionic Strength Sensing in Living Cells. ACS Chem. Biol. 2017, 12, 2510–2514. [Google Scholar] [CrossRef]
- Dudev, T.; Mazmanian, K.; Lim, C. Competition between Li+ and Na+ in Sodium Transporters and Receptors: Which Na+-Binding Sites Are “Therapeutic” Li+ Targets? Chem. Sci. 2018, 9, 4093–4103. [Google Scholar] [CrossRef]
- Ni, Y.G.; Chen, J.G.; Androutsellis-Theotokis, A.; Huang, C.J.; Moczydlowski, E.; Rudnick, G. A Lithium-Induced Conformational Change in Serotonin Transporter Alters Cocaine Binding, Ion Conductance, and Reactivity of Cys-109. J. Biol. Chem. 2001, 276, 30942–30947. [Google Scholar] [CrossRef]
- Dudev, T.; Mazmanian, K.; Weng, W.H.; Grauffel, C.; Lim, C. Free and Bound Therapeutic Lithium in Brain Signaling. Acc. Chem. Res. 2019, 52, 2960–2970. [Google Scholar] [CrossRef] [PubMed]
- Harwood, A.J. Lithium and Bipolar Mood Disorder: The Inositol-Depletion Hypothesis Revisited. Mol. Psychiatry 2005, 10, 117–126. [Google Scholar] [CrossRef]
- Shirra, M.K.; Patton-Vogt, J.; Ulrich, A.; Liuta-Tehlivets, O.; Kohlwein, S.D.; Henry, S.A.; Arndt, K.M. Inhibition of Acetyl Coenzyme A Carboxylase Activity Restores Expression of the INO1 Gene in a Snf1 Mutant Strain of Saccharomyces Cerevisiae. Mol. Cell. Biol. 2001, 21, 5710–5722. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, C.; Miller, R.A.; Smith, I.; Bui, T.; Molgó, J.; Müller, M.; Vais, H.; Cheung, K.-H.; Yang, J.; Parker, I.; et al. Essential Regulation of Cell Bioenergetics by Constitutive InsP3 Receptor Ca2+ Transfer to Mitochondria. Cell 2010, 142, 270–283. [Google Scholar] [CrossRef]
- Fu, L.; Miseta, A.; Hunton, D.; Marchase, R.B.; Bedwell, D.M. Loss of the Major Isoform of Phosphoglucomutase Results in Altered Calcium Homeostasis in Saccharomyces Cerevisiae. J. Biol. Chem. 2000, 275, 5431–5440. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reith, P.; Braam, S.; Welkenhuysen, N.; Lecinski, S.; Shepherd, J.; MacDonald, C.; Leake, M.C.; Hohmann, S.; Shashkova, S.; Cvijovic, M. The Effect of Lithium on the Budding Yeast Saccharomyces cerevisiae upon Stress Adaptation. Microorganisms 2022, 10, 590. https://doi.org/10.3390/microorganisms10030590
Reith P, Braam S, Welkenhuysen N, Lecinski S, Shepherd J, MacDonald C, Leake MC, Hohmann S, Shashkova S, Cvijovic M. The Effect of Lithium on the Budding Yeast Saccharomyces cerevisiae upon Stress Adaptation. Microorganisms. 2022; 10(3):590. https://doi.org/10.3390/microorganisms10030590
Chicago/Turabian StyleReith, Patrick, Svenja Braam, Niek Welkenhuysen, Sarah Lecinski, Jack Shepherd, Chris MacDonald, Mark C. Leake, Stefan Hohmann, Sviatlana Shashkova, and Marija Cvijovic. 2022. "The Effect of Lithium on the Budding Yeast Saccharomyces cerevisiae upon Stress Adaptation" Microorganisms 10, no. 3: 590. https://doi.org/10.3390/microorganisms10030590
APA StyleReith, P., Braam, S., Welkenhuysen, N., Lecinski, S., Shepherd, J., MacDonald, C., Leake, M. C., Hohmann, S., Shashkova, S., & Cvijovic, M. (2022). The Effect of Lithium on the Budding Yeast Saccharomyces cerevisiae upon Stress Adaptation. Microorganisms, 10(3), 590. https://doi.org/10.3390/microorganisms10030590






