Abstract
Marine Streptomyces species are underexplored for their pigment molecules and genes. In this study, we report the genome of the undecylprodigiosin biosynthesizing gene cluster carrying Streptomyces sp. strain BSE6.1, displaying antioxidant, antimicrobial, and staining properties. This Gram-positive obligate aerobic bacterium was isolated from the coastal sediment of the Andaman and Nicobar Islands, India. Pink to reddish pigmented colonies with whitish powdery spores on both agar and broth media are the important morphological characteristics of this bacterium. Growth tolerance to NaCl concentrations was 2 to 7%. The assembled genome of Streptomyces sp. BSE6.1 contains one linear chromosome 8.02 Mb in length with 7157 protein-coding genes, 82 tRNAs, 3 rRNAs and at least 11 gene clusters related to the synthesis of various secondary metabolites, including undecylprodigiosin. This strain carries type I, type II, and type III polyketide synthases (PKS) genes. Type I PKS gene cluster is involved in the biosynthesis of red pigment undecylprodigiosin of BSE6.1, similar to the one found in the S. coelicolor A3(2). This red pigment was reported to have various applications in the food and pharmaceutical industries. The genome of Streptomyces sp. BSE6.1 was submitted to NCBI with a BioProject ID of PRJNA514840 (Sequence Read Archive ID: SRR10849367 and Genome accession ID: CP085300).
1. Introduction
In recent years, marine pigmented bacteria have been gaining more research interest due to the potential applications of pigment molecules in the food and drug industries [1,2,3]. Among a wide array of pigmented microbes in terrestrial and marine environments, Streptomyces species have gained enormous attention in biotechnological applications. Although Streptomyces species are well known to produce a wide range of pigments, including blue, yellow, red, orange, pink, purple, blue-green, brown, and black [1,2], prodigiosin molecules, which are red in color, are not well studied amongst the Streptomyces species distributed in marine milieus.
Streptomyces species are known to contain a 5.1–10.1 Mbp size linear chromosome that carries core and adaptive genes [4,5]. They are spore formers with higher G+C contents (69–78%) than other Gram-positive bacteria. Streptomyces species are known to possess 21–45 secondary metabolites biosynthesizing gene clusters [4]. However, most of the gene clusters remain unexplored in this genus, which could have potential applications in the drug and food industry [4]. One such gene cluster is the prodigiosin biosynthetic gene cluster. Although more than 364 Streptomyces species are currently known [6], very few of them, such as Streptomyces spectabilis, Streptomyces pentaticus subsp. jenensis [7], Streptoverticillium rubrireticuli, Streptomyces longispororuber 100-19 (formerly Streptomyces longisporus ruber) [8], S. spectabilis BCC4785 [9], Streptomyces fusant NRCF69 [10], Streptomyces sp. Y-42 [11], Streptomyces sp. WMA-LM31 [12], S. griseoviridis [13], S. lividans [14], Streptomyces sp. CP1130 [15], S. variegatus [16], and S. coelicolor [17,18] are known to produce prodiginine pigments in addition to several well-studied non-actinomycetes bacteria [1].
The biosynthetic pathway of prodigiosin has been well understood in Serratia marcescens [19,20] and among many other prodigiosin-producing bacterial species. S. marcescens synthesizes prodigiosin through 33 genes, whereas S. coelicolor uses only 23 genes to synthesize prodigiosin derivatives [19,21]. The red gene cluster biosynthesizes prodiginines in Streptomyces species. Both Serratia and Streptomyces utilize 4-methoxy-2,2′-bipyrrole-5-carbaldehyde to synthesize prodigiosin and undecylprodigiosin, correspondingly [19,20]. Although the genome contents of several Streptomyces species have been reported in the last decade [4,22], the genomes of red pigment-producing Streptomyces species, especially marine Streptomyces, have remain largely uninvestigated, leaving a gap in the understanding of their evolutionary significances and drug discovery potential. Therefore, we intended to analyze and understand the genome of prodigiosin-producing Streptomyces BSE6.1 isolated from a coastal sediment sample.
Prodigiosin pigments are well known for their antimicrobial, anticancer, and cytotoxic properties [1,2,21,23]. Application of dried prodigiosin as a food-grade colorant in the development of prodigiosin coated microcapsules [24] and agar jellies [25] has been demonstrated from the extractions of S. marcescens [24], Zooshikella sp., and Streptomyces sp. [25]. Prodigiosin extracted from Streptomyces species has demonstrated promising antimicrobial activities against several pathogenic microbes such as Corynebacterium bovis, Mycobacterium smegmatis, Nocardia asteroids [7], and Staphylococcus aureus [7,25]. It is thought that the combined activity of antimicrobial and food colorant applications of prodigiosin would facilitate a synergistic effect in disease treatment. The present study introduces a novel species of a red-pigmented Streptomyces strain isolated from Andaman Islands, India’s marine environment, and its genome for industrial and biotechnological applications. The preliminary studies on prodigiosin-producing Streptomyces have demonstrated antimicrobial [7] and staining properties [8,25]. Although several Streptomyces species are known to produce a wide range of pigment compounds [1,2], the production of prodiginine derivatives by a limited number of Streptomyces species encouraged us to investigate the corresponding gene clusters in this Streptomyces sp. and compare it with other bacterial species.
Andaman and Nicobar Islands are a chain of 836 Islands, including islands, islets and rocky outcrops, that are pristine and unexplored for microbial resources. Bio-prospecting of microbial pigments from this environment was initiated very recently [1,2,26]. The erratic weather conditions observed in this geographically distinct location appear to favor many novel pigmented microbes with potential biotechnological applications. Therefore, the present study explored the pigmented bacterial resources available in the Andaman Islands and found a potential Streptomyces sp. strain BSE6.1 with antibacterial and dye activity. As Andaman waters are still underexplored, we aimed to investigate the novelty of Streptomyces sp. strain BSE6.1 through whole-genome analysis, predict the pigment gene clusters, and compare them with those of other Streptomyces species genomes available in the public nucleotide databases.
2. Materials and Methods
A red-pigmented bacterial isolate designated as BSE6.1 was isolated from a marine sediment sample collected from Burmanallah coast (11°33′52.24″ N, 92°44′ 01.51″ E), South Andaman Islands, India. A serially diluted sediment sample was inoculated onto marine agar 2216 (Himedia, Mumbai) plates and incubated at 28 °C. After a couple of weeks, red-pigmented colonies grown were sub-cultured either on freshly prepared marine agar plates or 2% nutrient agar. Pure cultures were stored as glycerol suspensions (30%, w/v) at −20 °C for further analysis. Salt tolerance was tested on marine agar plates supplemented with various percentages of NaCl (1 to 10%), followed by streaking a pure culture, incubating at 28 °C, and measuring growth after two days. Catalase and oxidase activities were performed according to standard microbial biochemical tests [27].
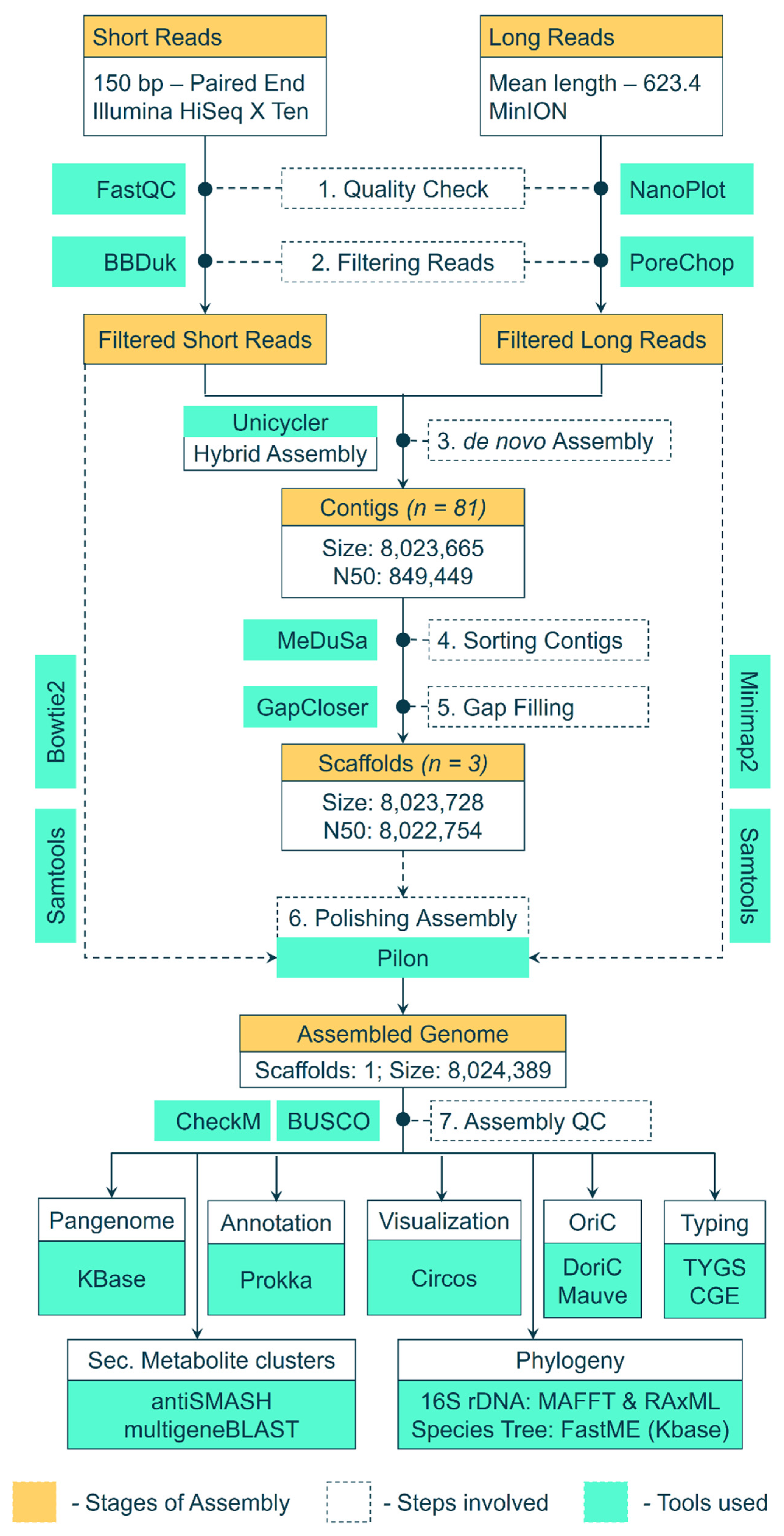
Genomic DNA of Streptomyces BSE6.1 was extracted using the Cetyl Trimethyl Ammonium Bromide (CTAB) and phenol–chloroform method. Extracted DNA was treated with RNase A and purified. DNA was quantified by measuring its absorbance at A260 and A280 in a NanoDrop. The Illumina Hiseq X Ten sequencing system was used to obtain 150 bp short-read paired-end raw data. In addition to these short reads, long reads were obtained using the MinIoN platform. The workflow used to assemble these raw reads and analyze the genome assembly is depicted in Figure 1. The paired-end data quality of short reads was checked using FASTQC v0.11.8 [28]. BBDuk (BBmap v38.93) was used to filter low-quality reads and adaptor sequences [29], whereas the long reads were checked with NanoPlot v1.38.1 [30] and filtered with PoreChop v0.4.8 [31]. The filtered high-quality short and long reads were assembled into contigs using a hybrid de novo assembler Unicycler v0.4.8 [32], in a de novo fashion. The 16S rRNA genes were extracted from the assembled scaffolds using Barrnap [33] and were aligned against the non-redundant nucleotide database at NCBI. The complete genome of the nearest neighbor (Streptomyces sp. KPB2—Accession ID: CP034353.1) [34], was used as a reference. The contigs were sorted and merged into scaffolds with the help of a reference genome using MeDusa v1.6 [35]. A gap-filling step was performed using GapCloser v1.12 [36] to generate a draft genome assembly. Furthermore, the genome assembly was polished with Pilon v1.24 [37] by mapping filtered short reads (Bowtie2 v2.4.4. [38]) and filtered long reads (minimap2 [39]) against the assembly and sorting the alignments with samtools v1.13 [40].
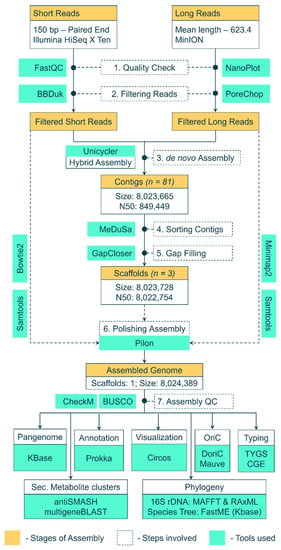
Figure 1.
Workflow and pipeline of tools used to assemble the raw reads into a genome and further analysis of the assembled genome.
Genome assembly was checked for its quality using BUSCO v5.2.2 [41] and CheckM v1.1.3 [42] tools. In silico multi-locus sequence typing (MLST) of the genome was performed using the online webserver at the Centre of Genomic Epidemiology [43]. Type strain identification of the genome was performed at Type(Strain) Genome Server (TYGS) [44]. In addition to the type strain identification, a species tree was constructed with FastME [45] at KBase server [46] using 49 core Clusters of Orthologous Groups (COGs) of 200 related genomes. An additional phylogenetic tree was constructed with the 16s rRNA genes of Streptomyces species available at the Ribosomal RNA database [47]. Duplicate sequences were removed, and multiple sequence alignment (MSA) was performed using default parameters of MAFFT v7.487 for FFT-NS-I refinement method [48]. A maximum-likelihood tree was constructed based on the MSA using default parameters and 1000 bootstraps with RAxML v8.2.12 [49]. The 16s rRNA gene of Staphylococcus aureus (RefSeq ID: GCF_000013425.1) was used as an outgroup. The origin of replication (OriC) was identified using DoriC database [50] and Mauve aligner [51]. Pairwise genomic comparison of strain BSE6.1 was made with 3 other related genomes. Dotplots were constructed with minimap2 based pairwise alignment using D-Genies [52]. Prokka v1.14.6 was used to perform a local de novo annotation [53]. Pan-genome comparison with 100 related genomes (~90% 16S nucleotide identity; ~80% whole-genome aligned fraction identity) was made using the pan-genome tool at KBase server [46]. Gene clusters related to the secondary metabolite biosynthesis were identified using the antiSMASH 5.0 pipeline [54]. The red pigment-producing gene cluster of BSE6.1 was compared with that of S. coelicolor A3(2), Serratia, and Hahella using the multigene BLAST tool [55]. The distribution of various coding sequences (CDS) and gene clusters across the genome was plotted using Circos [56].
3. Results and Discussion
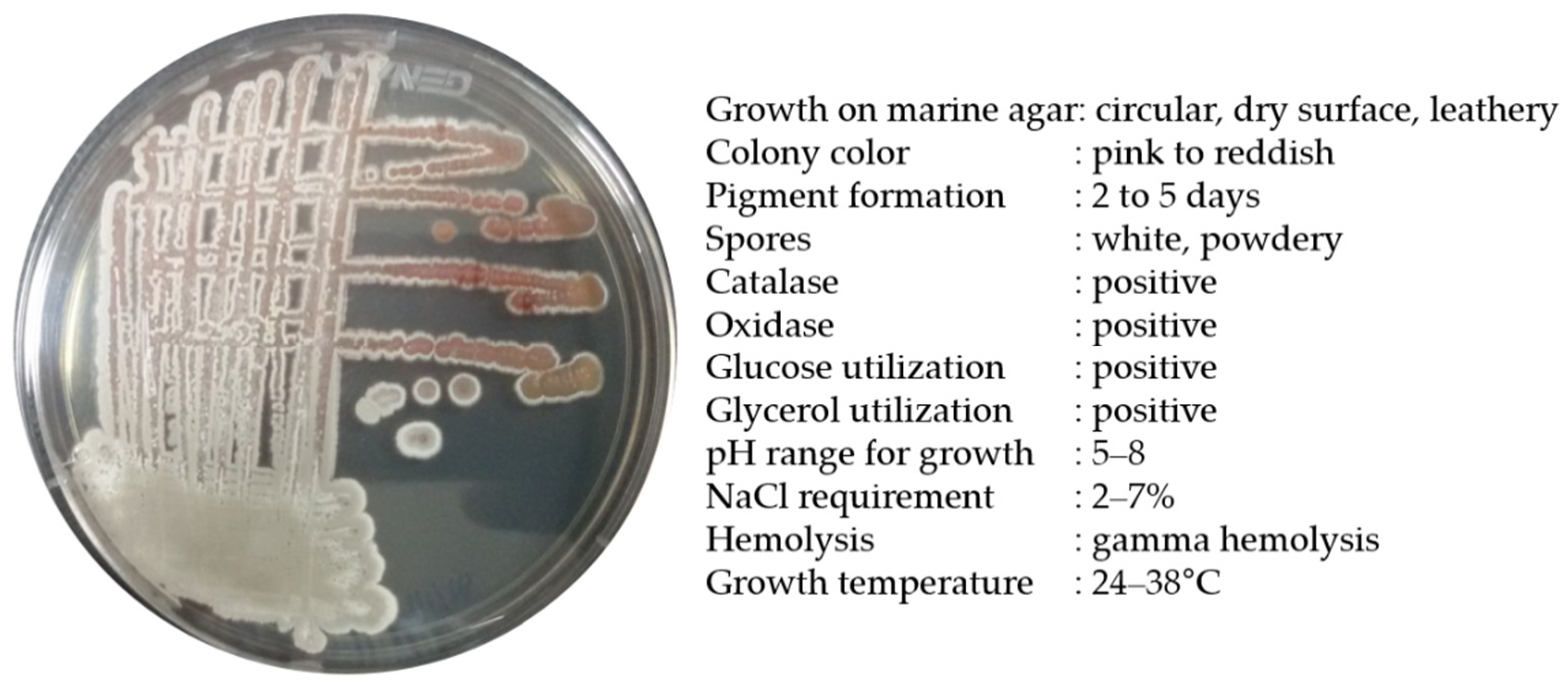
Strain BSE6.1 produced a pink-colored growth in Minimal broth with 2% NaCl and red pigmentation in all other compatible media. Pale pink to reddish colonies with white powdery spores were observed after 7 or 10 days of incubation. Salt tolerance was observed up to a range of 2 to 7%. This bacterium was positive for catalase and oxidase activities. In our earlier study, strain BSE6.1 showed potential antibacterial activity against different human pathogens and also displayed a strong ability to stain epidermis and parenchyma cells of Tridax procumbens stem [25]. The maximum pigment production was observed at 29 °C, and the maximum temperature tolerance for its growth was 38 °C (Figure 2). The peak absorption spectrum of the red pigment of BSE6.1 was observed at 528 nm [25].
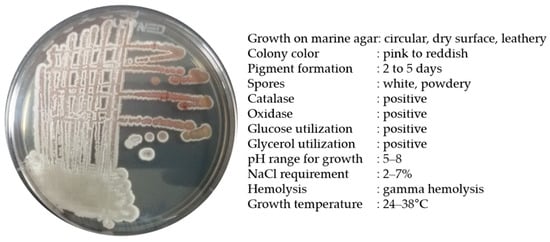
Figure 2.
Morphological and biochemical characteristics of Streptomyces sp. strain BSE6.1.
Identification of the red pigment through thin layer chromatography (TLC), Fourier-transform infrared spectroscopy (FT-IR), and proton nuclear magnetic resonance (1H-NMR) analyses revealed the presence of antimicrobial pigment –prodiginine derivatives in Streptomyces sp. BSE6.1 [25]. However, the genome analysis of strain BSE6.1 reveals the presence of an undecylprodigiosin gene cluster which is responsible for undecylprodigiosin production. Therefore, the other red fraction of Streptomyces strain BSE6.1 [25] is yet to be elucidated and identified through LC-MS, 13C NMR, HSQC, HMBC, and COSY data to confirm the production of undecylprodigiosin or related derivatives.
Previous studies reported that Streptomyces longisporus, Streptomyces spectabilis [7,57], and Streptomyces variegatus produce prodigiosin [16] (Table 1). However, some strains of Streptomyces coelicolor produce either undecylprodigiosin [17,20,58] or a mixture of prodiginine derivatives [59] (Table 1). Similar to S. coelicolor [17,20,58,59], the first fraction of red pigment eluted from Streptomyces strain BSE6.1 through TLC revealed the presence of methyl-3-propyl prodiginine and 2-methyl-3-butyl prodiginine in mass spectrometry analysis but identified it as prodigiosin in 1H NMR analysis [25]. Methyl-3-propyl prodiginine and 2-methyl-3-butyl prodiginine were also identified in actinomycetes [60], non-actinomycetes bacteria such as Pseudoalteromonas rubra [61], and Serratia marcescens [62]. These studies suggest that some strains of Streptomyces produce either prodigiosin or undecylprodigiosin, whereas some produce a mixture of prodiginine analogs.

Table 1.
Details of various Streptomyces species capable of producing prodiginine derivatives.
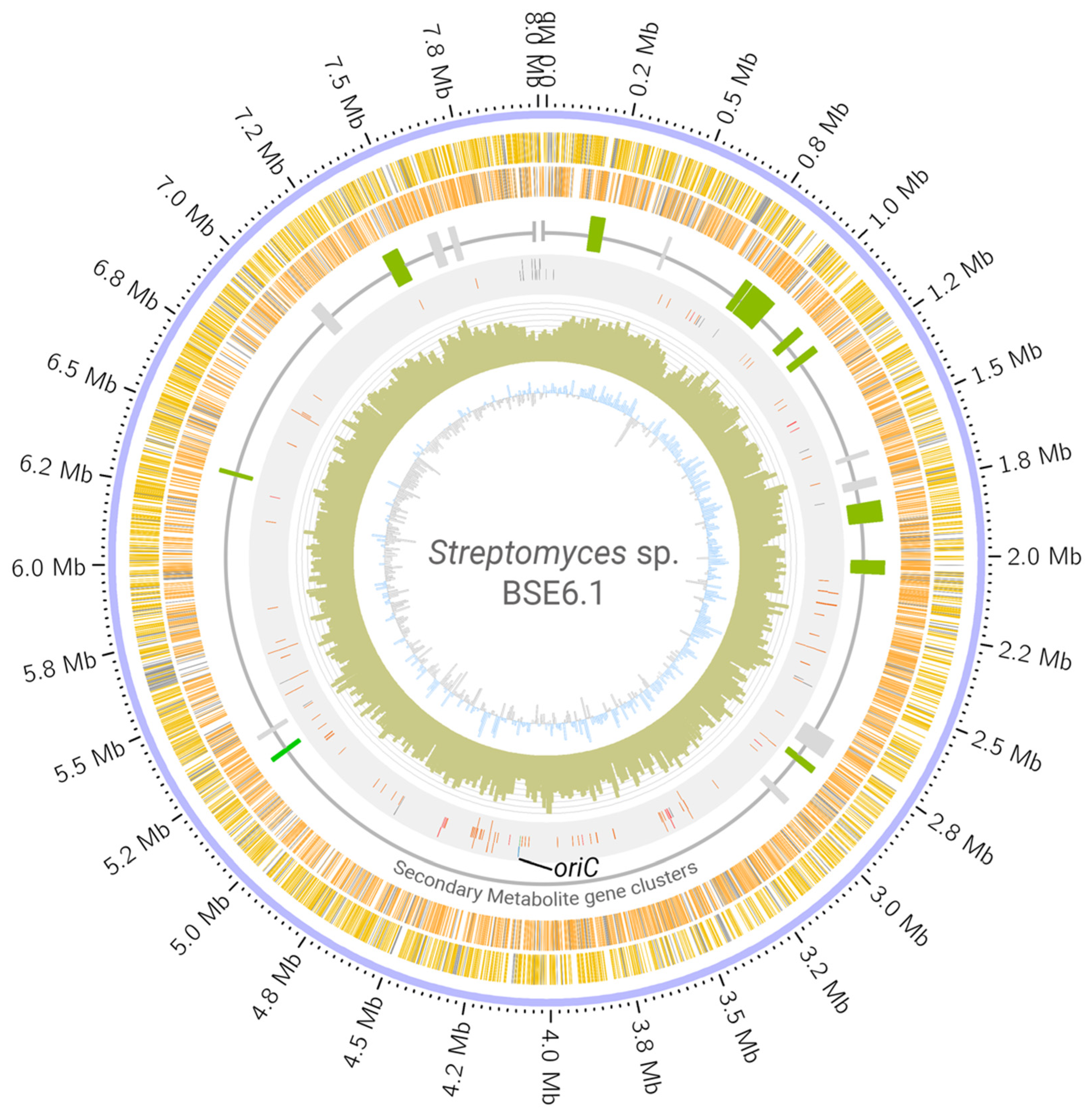
Whole-genome sequencing of strain BSE6.1 produced a total of 7,528,288 reads. Assembling these raw reads resulted in a single scaffold of 8.02 Mb with no extra-chromosomal content. Annotating the assembled genome of strain BSE6.1 indicated the presence of at least 7157 protein-coding genes, 82 tRNA coding genes, 3 rRNA coding genes, and 1 responsible for the production of tmRNA (Table 2, Figure 3). Subsystem coverage of the identified CDS was 19%, involving nearly 324 subsystem types (Figure S1). Subsystems with the highest coverage of genes/features include amino acid, carbohydrate, protein, and vitamin metabolic pathways. Furthermore, at least 43 genes were involved in defense mechanisms such as resistance to antibiotics and toxic compounds. In addition, at least 11 gene clusters involved in the synthesis of other secondary metabolites were also identified (Figure S2). Most members of the Streptomyces genus have linear chromosomes [4,5] and strain BSE6.1 is not an exception. There are no overlapping 5′ and 3′ ends in the scaffold, indicating its non-circular configuration. Furthermore, the oriC region and dnaA gene were identified approximately at the center of the scaffold, similar to that of S. coelicolor A3(2) (Figure 3).

Table 2.
Features of Streptomyces sp. strain BSE6.1 genome.
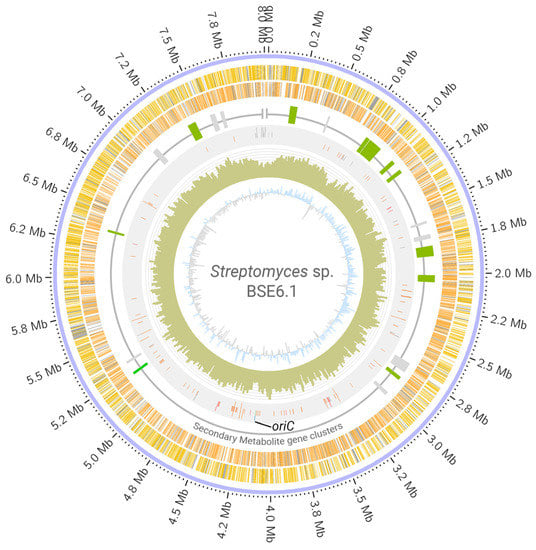
Figure 3.
Circular visualization of Streptomyces sp. strain BSE6.1 genome. The scaffold is represented in the outer circle. The scaffold is followed by coding regions (CDS) in the sense (yellow bands) and anti-sense (orange bands) directions. Grey bands represent hypothetical CDS. The third circle represents the distribution of gene clusters coding for secondary metabolites (green: clusters which are >75% similar to those present in related organisms; grey: <75% similarity). The fourth circle represents the RNA genes (orange), transposases (grey), phage genes (purple) dnaA gene (blue), and oriC region (green and labelled). Histograms in the fifth circle indicate the GC content per 10,000 bases. The innermost circle represents GC skew data per 10,000 bases (blue indicates positive skewness and grey negative skewness).
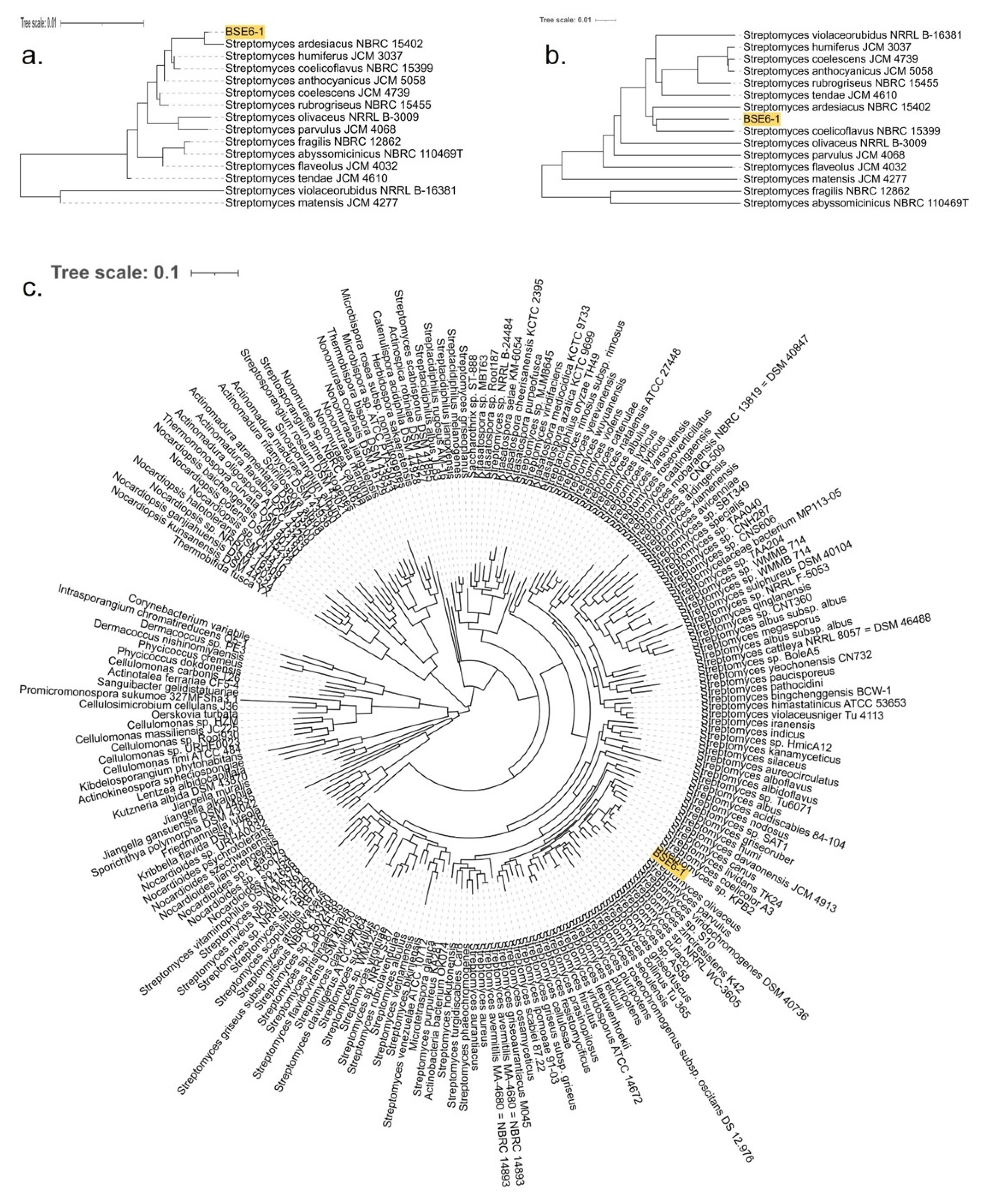
BLAST analysis based on the 16s rRNA sequences suggested that strain BSE6.1 had a 99.71% similarity with various unclassified Streptomyces species available in the GenBank. The most similar strains include Streptomyces sp. NA03103 (isolated from marine sediment in China) (GenBank: CP054920), Streptomyces sp. strain HB-N217 (isolated from a marine sponge, Forcepia sp. in the USA) [77], Streptomyces sp. CCM_MD2014 (soil isolate from the USA) [78], Streptomyces sp. KPB2 (isolated from the pollen of kiwi fruit from South Korea) [34], Streptomyces sp. PM-R01 (isolated from Durian fruit, Durio zibethinus, in Thailand) (GenBank: LC381944), and Streptomyces sp. IT-M01 (isolated from a sea crab, Thalamita crenata, in Thailand) (GenBank: LC386952). Furthermore, 16S rRNA genes of BSE6.1 and 208 Streptomyces species were used to construct a phylogenetic tree (Figure S3). The strain typing of BSE6.1 at TYGS indicated no available type strain, which is closely related to the query genome. The highest pairwise digital DNA–DNA hybridization similarity (dDDH, d4 value corresponding to the sum of all identities found in HSPs divided by overall HSP length) was 48.7% with type strain Streptomyces coelicoflavus NBRC 15399 (Sup. Data 1). A genome blast distance phylogenetic (GBDP) tree was constructed for BSE6.1 and the related type strains using 16S rRNA gene and complete genome data (Figure 4a,b). In addition to detecting the closest type strain, a species tree was constructed using 49 core COGs in related genomes [46] (Sup. Data 2). In the species tree, BSE6.1 clustered with the strains viz. Streptomyces sp. KPB2, S. coelicolor A3(2), S. lividans TK24, S. olivaceus, S. parvulus, etc (Figure 4c).
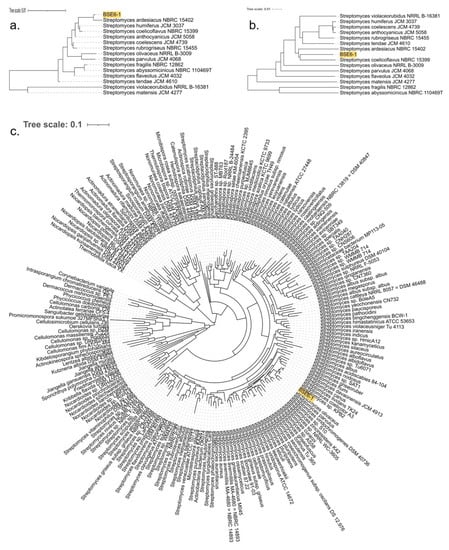
Figure 4.
GBDP tree with 100 bootstraps for (a) 16S rRNA genes and (b) genomes of strain BSE6.1 along with 14 type strains with highest dDDH (d4) similarity. (c) Species tree constructed using 49 core/conservative COGs of strain BSE6.1 and 200 related/homologous genomes with at least 90% 16S nucleotide identity and ~80% whole-genome aligned fraction identity.
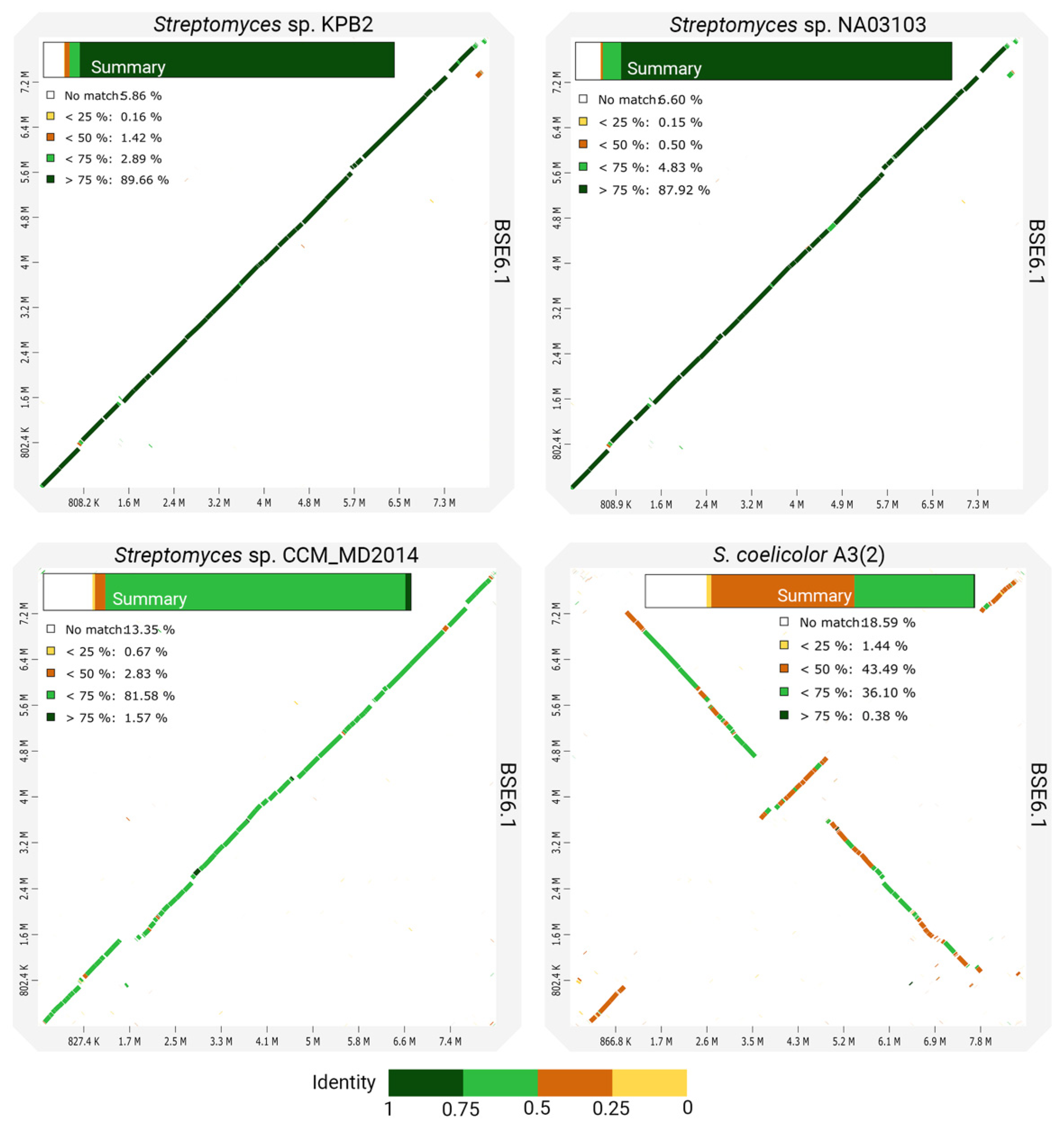
However, the whole-genome comparison of BSE6.1 with other closely related species shows many variations in its genomic content (Figure 5). In concordance with the phylogenetic distances, the genomes of strain KPB2 and strain NA03103 have the most similar genomic regions with BSE6.1. Comparatively less identical homologous regions were observed while comparing BSE6.1 with strain CCM_MD2014. Another comparison of BSE6.1 with one of the well-studied pigment-producing bacteria, S. coelicolor A3(2) [70], presented the least identical synteny among the four comparisons. Furthermore, the in silico MLST analysis of the BSE6.1 genome revealed the presence of a novel allelic profile—16S_99, atpD_185, gyrB_124, recA_156, rpoB_175 and trpB_190 (Table 3). All the in silico analyses suggested that the strain BSE6.1 could be a novel species of Streptomyces. However, further phenotypic characterizations are needed to confirm its novelty.
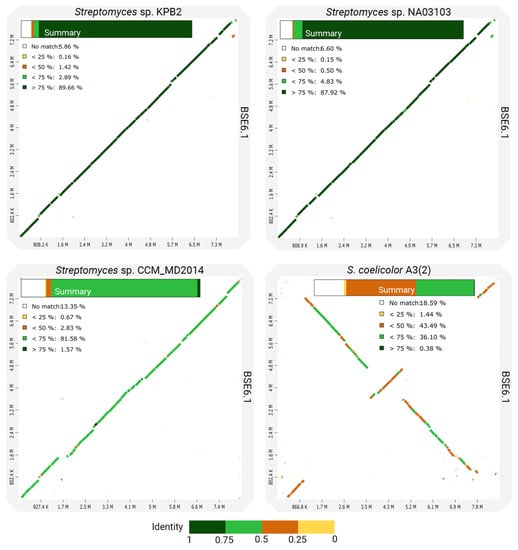
Figure 5.
Dotplots showing the pairwise comparisons of strain BSE6.1 genome and three closely related genomes. A summary of each comparison is shown within the corresponding plot.

Table 3.
MLST profile of Streptomyces sp. strain BSE6.1 genome.
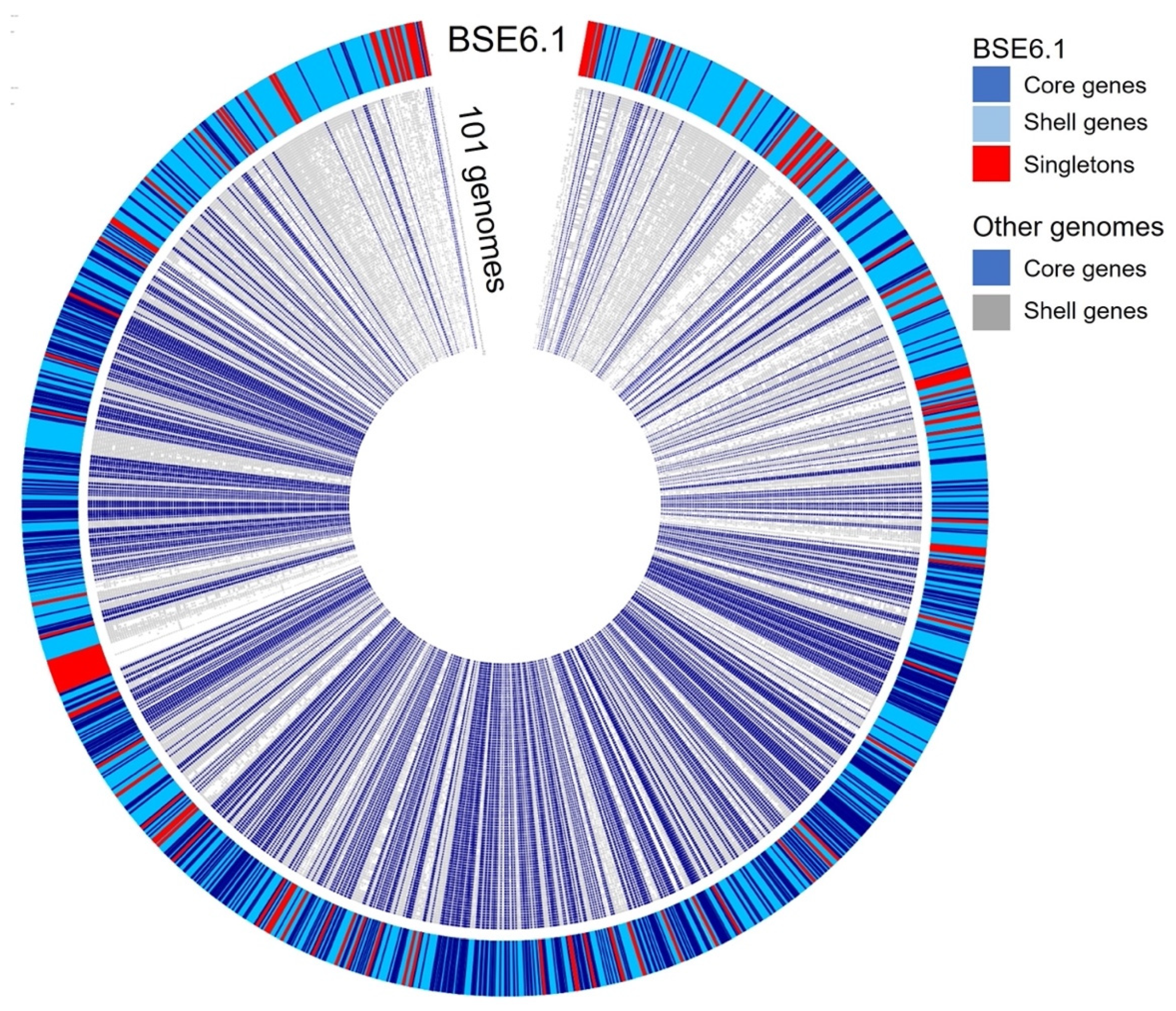
A pan-genomic comparison was made between 101 related genomes belonging to the Streptomycetaceae family and that of strain BSE6.1 (Figure 6). A total of 720,604 translated genes belong to 123,491 homologous gene families were identified. Out of these, 726 families were conserved across the genomes, 41,274 were shell gene families, and 81,497 were singletons. Strain BSE6.1 has 7157 genes, of which 902 belong to the core gene cluster, 6016 genes belonging to the shell gene cluster, and 239 genes are unique to BSE6.1. The genes confined to strain BSE6.1 are mostly hypothetical (184 out of 239 genes), apart from some interesting genes viz. serine protease genes (perform physiological roles), MarR family (responsible for multiple antibiotic resistance), SsgA sporulation regulator, etc (Sup. Data 3).
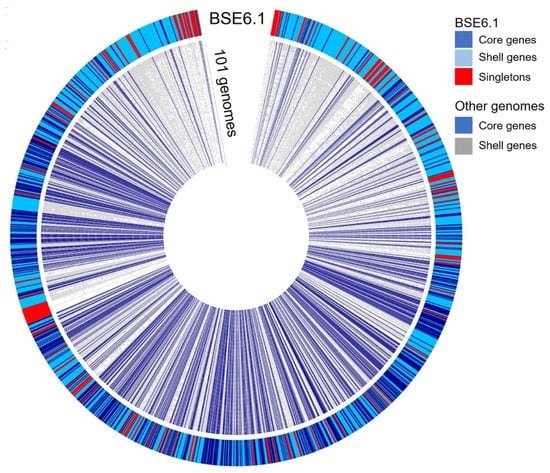
Figure 6.
Pangenome comparison of strain BSE6.1 and 101 related genomes (Sup. Data 4) of Streptomycetaceae family. The genome of strain BSE6.1 has 12.6% of conserved genes, 84.1% of shared or shell genes, and 3.3% of unique genes.
Streptomyces species are ubiquitous in nature, with more than 500 Streptomyces species reported from various environments such as terrestrial, coastal, deep-sea, deserts, and polar regions [6]. Under unfavorable conditions, these species produce external hyphae, which divide into spores. Streptomyces species possess antibiotic resistance genes; thus, they display potential bioactive properties. Many species of Streptomyces are known to produce secondary metabolites, antibiotics [79,80], and very few Streptomyces species are known to produce pigments such as prodigiosin derivatives having antimicrobial and anticancer properties [1,6,19]. The genome analysis of BSE6.1 revealed the presence of 23 gene clusters responsible for the production of ectoine, polyketides, etc (Figure S2). Out of these 23 clusters, at least 11 showed >75% similarity with existing gene clusters of different strains (Figures S4 and S5). The information about all the other gene clusters and their similarity to the other Streptomyces may be accessed through anti-smash (Sup. Data 5).
The genome of BSE6.1 contains three types of PKSs, namely type I, type II, and type III. Strain BSE6.1 has two copies of type III polyketide synthase (PKS) genes observed in clusters 20 and 21, coding for herboxidiene, an antitumor molecule reported in Streptomyces sp. [81], and germicidin, which is responsible for the development of spore formation and aerial hyphae elongation [82], respectively. The type III PKS genes in Streptomyces species are known to produce red to brownish pigments with potential antimicrobial and antioxidant activities [83,84]. Cluster 13 represents a type II PKS, which is responsible for grey-pink spore pigmentation in Streptomyces species [85,86].
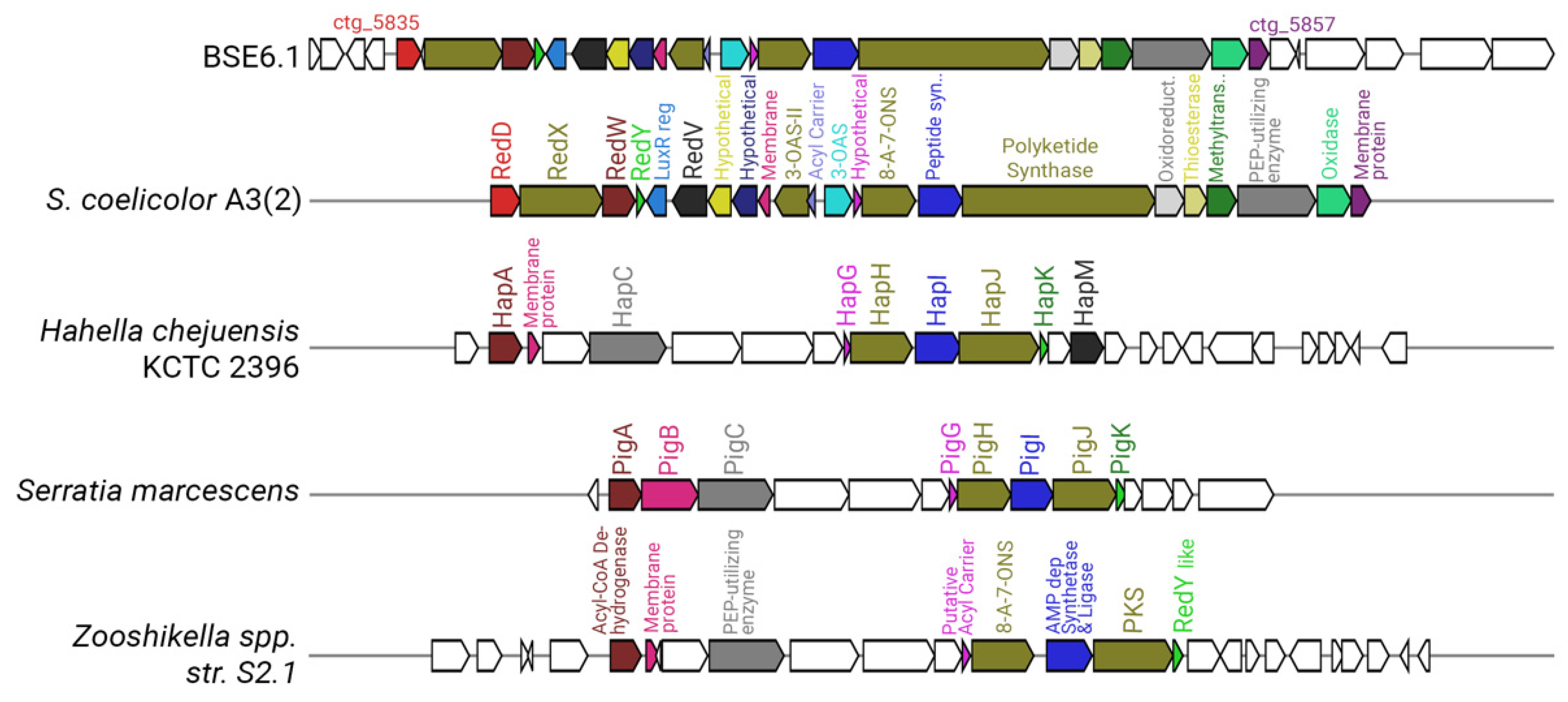
Strain BSE6.1 has a type I PKS system in cluster 10, which is responsible for undecylprodigiosin production. The prodigiosin biosynthesis gene cluster was identified as pig gene cluster in Serratia marcescens [19,87]. Prodigiosin synthesizing genes in Hahella chejuensis KCTC 2396 and Pseudoalteromonas species were identified as hap gene cluster [88], while red gene cluster was identified for undecylprodigiosin biosynthesis in S. coelicolor A3(2) [58]. The prodigiosin biosynthesizing cluster found in the Streptomyces species is the largest cluster, with 23 genes (Figure 7). Cluster 10 of strain BSE6.1 showed 100% similarity with red gene cluster of S. coelicolor A3(2). Cluster 10 has 32 genes, of which 23 genes are responsible for the production of the bioactive red pigment undecylprodigiosin. This cluster comprises regulatory genes, core, and additional genes involved in the biosynthesis of prodigiosin derivatives, similar to that of red gene cluster of S. coelicolor A3(2).
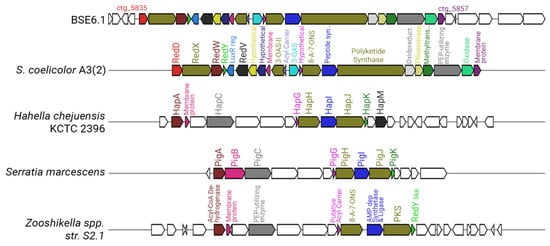
Figure 7.
Comparative representation of the undecylprodigiosin cluster in BSE6.1 (cluster 10 of Supplementary Figure S2) with that of S. coelicolor A3(2), Hahella chejuensis KCTC 2396, Serratia marcescens, and Zooshikella sp. S2.1. (Genes represented without any color in the strain BSE6.1 have unknown functions, whereas those in the other species have no homologs in BSE6.1).
Cluster 19 displayed 100% similarity with ectoine biosynthesizing genes, indicating that BSE6.1 is capable of producing ectoine—an anticancer molecule [89] that regulates osmotic stress [90] and acts as a stress protectant against various environmental stresses [91]. Clusters 7 and 17 displayed siderophore genes that were involved in the production of enduracidin and desferrioxamine B, respectively. Desferrioxamine B is a drug used to treat iron overload disease in humans [92,93], while enduracidin is known to display antibiotic activity [94]. Cluster 14 represents albaflavenone, an antibiotic terpene molecule produced by S. coelicolor A3(2) [95,96]. Chemical molecules produced by S. coelicolor A3(2) such as germicidin [82], ectoine [91], albaflavenone [95], coelichelin [97], hopanoids [98], sapB protein [99], and coelibactin [100] are observed in Streptomyces strain BSE6.1 with a 100% similarity match. Ashimides molecules produced by Streptomyces sp. NA03103 [101] are not detected in S. coelicolor A3(2), but Streptomyces strain BSE6.1 shows 100% similarity with ashimides synthesizing gene.
Interestingly, the genome content of strain BSE6.1 is distinct from other Streptomyces species. It is an important evolutionary aspect that these related and non-related bacterial lineages are capable of producing a variety of prodiginine analogs for their defensive function in the surrounding milieus. As studies on the diversity and distribution of marine pigmented Streptomyces species are scarce, further research on this aspect would provide new insights into the evolutionary spread and species distribution of pigmented Streptomyces in different environments. We infer that pigment gene clusters of microbes such as Streptomyces may serve as an evolutionary marker to address the actual place of origin and spread of prodiginine pigments in the marine or terrestrial milieus during the evolutionary process. The variability in the whole genome content and novel alleles in the MLST profile indicate its status as a novel species. Thus, based on complete genome analysis, we propose strain BSE6.1 as Streptomyces prasanthi sp. nov. This study provides the whole genome of Streptomyces sp. BSE6.1 for further comparative studies with other Streptomyces species on taxonomical, evolutionary, and biotechnological aspects. As it is the first ever mined genome of prodigiosin-producing marine Streptomyces BSE6.1, it would serve as a reference genome for comparative studies to predict the novelty of the genomic contents of other Streptomyces species and non-Streptomyces species.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9112249/s1, Figure S1: Subsystems, Figure S2: Clusters of BSE6.1, Figure S3: 16S rRNA based phylogenetic tree, Figures S4 and S5: Clusters in detail, Sup. Data 1: TYGS summary, Sup. Data 2: Core COGs used in the construction of species tree, Sup. Data 3: Unique genes of BSE6.1, Sup. Data 4: List of genomes, Sup. Data 5: All clusters and their similarity to the other Streptomyces.
Author Contributions
Conceptualization, lab work, data analysis, validation, and manuscript writing were completed by C.R., M.A. worked on bioinformatics and manuscript writing. Supervision, editing, and approval by N.V.V. and R.K., L.D. edited and provided additional information to improve the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science and Engineering Research Board (SERB), New Delhi, under File no: SERB/N-PDF/2016/ 000354.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genome sequence of Streptomyces BSE6.1 is submitted in Sequence Read Archive (SRA) under Bioproject: PRJNA514840. The BioSample accession ID of strain BSE6.1 is SAMN12598824. Genome assembly was submitted and is under process (SUB10526264).
Acknowledgments
Ramesh thanks the Science and Engineering Research Board (SERB), New Delhi, for the National Postdoctoral Research Fellowship award. This the outcome of CSIR-NIO’s project OLP2005 and the contribution number of this publication is 6826.
Conflicts of Interest
Authors declare that there are no conflicts of interest.
References
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Multifaceted Applications of Microbial Pigments: Current Knowledge, Challenges and Future Directions for Public Health Implications. Microorganisms 2019, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R. Marine pigmented bacteria: A prospective source of antibacterial compounds. J. Nat. Sci. Biol. Med. 2019, 10, 104. [Google Scholar] [CrossRef]
- Soliev, A.B.; Hosokawa, K.; Enomoto, K. Bioactive Pigments from Marine Bacteria: Applications and Physiological Roles. Evid. -Based Complement. Altern. Med. 2011, 2011, 1–17. [Google Scholar] [CrossRef]
- Lee, N.; Kim, W.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Sci. Data 2020, 7, 1–9. [Google Scholar] [CrossRef]
- Kirby, R.; Sangal, V.; Tucker, N.P.; Zakrzewska-Czerwińska, J.; Wierzbicka, K.; Herron, P.R.; Chu, C.-J.; Chandra, G.; Fahal, A.H.; Goodfellow, M.; et al. Draft Genome Sequence of the Human Pathogen Streptomyces somaliensis, a Significant Cause of Actinomycetoma. J. Bacteriol. 2012, 194, 3544–3545. [Google Scholar] [CrossRef]
- Kämpfer, P.; Glaeser, S.P.; Parkes, L.; Van Keulen, G.; Dyson, P. The family Streptomycetaceae. In The Prokaryotes: Actinobacteria; Rosenberg, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 889–1010. [Google Scholar]
- Gerber, N.N.; Lechevalier, M.P. Prodiginine (prodigiosin-like) pigments from Streptomyces and other aerobic Actinomycetes. Can. J. Microbiol. 1976, 22, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Gerber, N.N.; Stahly, D.P. Prodiginine (prodigiosin-like) pigments from Streptoverticillium rubrireticuli, an organism that causes pink staining of polyvinyl chloride. Appl. Microbiol. 1975, 30, 807–810. [Google Scholar] [CrossRef]
- Isaka, M.; Jaturapat, A.; Kramyu, J.; Tanticharoen, M.; Thebtaranonth, Y. Potent In Vitro Antimalarial Activity of Metacycloprodigiosin Isolated from Streptomyces spectabilis BCC 4785. Antimicrob. Agents Chemother. 2002, 46, 1112–1113. [Google Scholar] [CrossRef]
- El-Bondkly, A.M.A.; El-Gendy, M.M.A.; Bassyouni, R.H. Overproduction and biological activity of prodigiosin-like pigments from recombinant fusant of endophytic marine Streptomyces species. Antonie Van Leeuwenhoek 2012, 102, 719–734. [Google Scholar] [CrossRef]
- Gerber, N.N. A new prodiginne (prodigiosin-like) pigment from Streptomyces. Antimalarial activity of several prodiginnes. J. Antibiot. 1975, 28, 194–199. [Google Scholar] [CrossRef]
- Sajjad, W.; Ahmad, S.; Aziz, I.; Azam, S.S.; Hasan, F.; Shah, A.A. Antiproliferative, antioxidant and binding mechanism analysis of prodigiosin from newly isolated radio-resistant Streptomyces sp. strain WMA-LM31. Mol. Biol. Rep. 2018, 45, 1787–1798. [Google Scholar] [CrossRef]
- Kawasaki, T.; Sakurai, F.; Hayakawa, Y. A Prodigiosin from the Roseophilin Producer Streptomyces griseoviridis. J. Nat. Prod. 2008, 71, 1265–1267. [Google Scholar] [CrossRef]
- Sola-Landa, A.; Moura, R.S.; Martin, J.F. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 2003, 100, 6133–6138. [Google Scholar] [CrossRef] [PubMed]
- Lewer, P.; Chapin, E.L.; Graupner, P.R.; Gilbert, J.R.; Peacock, C. Tartrolone C: A novel insecticidal macrodiolide produced by Streptomyces sp. CP1130. J. Nat. Prod. 2003, 66, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, Y.-S.; Park, S.; Kim, J.; Kang, S.-J.; Lee, M.-H.; Ryu, S.; Choi, J.M.; Oh, T.-K.; Yoon, J.-H. Exceptional Production of both Prodigiosin and Cycloprodigiosin as Major Metabolic Constituents by a Novel Marine Bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 2011, 77, 4967–4973. [Google Scholar] [CrossRef]
- Kang, S.G.; Jin, W.; Bibb, M.; Lee, K.J. Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3(2) grown in continuous culture. FEMS Microbiol. Lett. 1998, 168, 221–226. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, H.; Zheng, G.; Jiang, W.; Lu, Y. Metabolic engineering of Streptomyces coelicolor for enhanced prodigiosins (RED) production. Sci. China Life Sci. 2017, 60, 948–957. [Google Scholar] [CrossRef]
- Williamson, N.R.; Fineran, P.; Leeper, F.J.; Salmond, G.P.C. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Genet. 2006, 4, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Williamson, N.R.; Simonsen, H.T.; Ahmed, R.A.A.; Goldet, G.; Slater, H.; Woodley, L.; Leeper, F.J.; Salmond, G.P.C. Biosynthesis of the red antibiotic, prodigiosin, in Serratia: Identification of a novel 2-methyl-3-n-amylpyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Strept. Mol. Microbiol. 2005, 56, 971–989. [Google Scholar] [CrossRef]
- Darshan, N.; Manonmani, H.K. Prodigiosin and its potential applications. J. Food Sci. Technol. 2015, 52, 5393–5407. [Google Scholar] [CrossRef]
- Harrison, J.; Studholme, D.J. Recently published Streptomyces genome sequences. Microb. Biotechnol. 2014, 7, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L. Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2014, 41, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Namazkar, S.; Ahmad, W.A. Spray-dried Prodigiosin from Serratia marcescens As A colorant. Biosci. Biotechnol. Res. Asia 2013, 10, 69–76. [Google Scholar] [CrossRef]
- Chatragadda, R.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Applications of Prodigiosin Extracted from Marine Red Pigmented Bacteria Zooshikella sp. and Actinomycete Streptomyces sp. Microorganisms 2020, 8, 556. [Google Scholar] [CrossRef]
- Ramesh, C.H.; Mohanraju, R.; Murthy, K.N.; Karthick, P. Molecular characterization of marine pigmented bacteria showing antibacterial activity. Indian J. Mar. Sci. 2017, 46, 2081–2087. [Google Scholar]
- Benson, H.J. Microbiological Applications Laboratory Manual in General Microbiology, 8th ed.; The McGraw−Hill Companies: New York, NY, USA, 2001. [Google Scholar]
- Andrews, S. FastQC: A Quality-Control Tool for High-through Put Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bushnell, B. BBTools Software Package. Available online: http://sourceforge.net/projects/bbmap (accessed on 23 November 2020).
- Coster, W.; De D’Hert, S.; Schultz, D.T.; Cruts, M.; Broeckhoven, C. Van NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.; Gorrie, C.; Holt, K. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Seemann, T. Barrnap 0.7: Rapid Ribosomal RNA Prediction. 2013. Available online: https://github.com/tseemann/barrnap (accessed on 23 November 2020).
- Kim, M.-J.; Chae, D.-H.; Cho, G.; Kim, D.-R.; Kwak, Y.-S. Characterization of Antibacterial Strains against Kiwifruit Bacterial Canker Pathogen. Plant. Pathol. J. 2019, 35, 473–485. [Google Scholar] [CrossRef]
- Bosi, E.; Donati, B.; Galardini, M.; Brunetti, S.; Sagot, M.-F.; Lio, P.; Crescenzi, P.; Fani, R.; Fondi, M. MeDuSa: A multi-draft based scaffolder. Bioinformatics 2015, 31, 2443–2451. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Subgroup, 1000 Genome Project Data Processing The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program: Table 1. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.; Schmidt, T.M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2014, 43, D593–D598. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, C.-T. DoriC: A database of oriC regions in bacterial genomes. Bioinformatics 2007, 23, 1866–1867. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Cabanettes, F.; Klopp, C. D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ 2018, 6, e4958. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Medema, M.H.; Takano, E.; Breitling, R. Detecting Sequence Homology at the Gene Cluster Level with MultiGeneBlast. Mol. Biol. Evol. 2013, 30, 1218–1223. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Variyar, P.S.; Chander, R.; Venkatachalam, S.R.; Bongirwar, D.R. ChemInform Abstract: A New Red Pigment from an Alkalophilic Micrococcus Species. ChemInform 2010, 33. [Google Scholar] [CrossRef]
- Cerdeño, A.M.; Bibb, M.J.; Challis, G.L.; Cerdeño, A.M.; Bibb, M.J.; Challis, G.L.; Cerdeño, A.M.; Bibb, M.J.; Challis, G.L.; Cerdeño, A.M.; et al. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): New mechanisms for chain initiation and termination in modular multienzymes. Chem. Biol. 2001, 8, 817–829. [Google Scholar] [CrossRef]
- Tsao, S.-W.; Rudd, B.A.M.; He, X.-G.; Chang, C.-J.; Floss, H.G. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 1985, 38, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Gerber, N.N.; McInnes, A.G.; Smith, D.G.; Walter, J.A.; Wright, J.L.C.; Vining, L.C. Biosynthesis of prodiginines. 13C resonance assignments and enrichment patterns in nonyl-, cyclononyl-, methylcyclodecyl-, and butylcycloheptylprodiginine produced by actinomycete cultures supplemented with 13C-labeled acetate and 15N-labeled nitrate. Can. J. Chem. 1978, 56, 1155–1163. [Google Scholar] [CrossRef]
- Setiyono, E.; Adhiwibawa, M.; Indrawati, R.; Prihastyanti, M.N.U.; Shioi, Y.; Brotosudarmo, T.H.P. An Indonesian Marine Bacterium, Pseudoalteromonas rubra, Produces Antimicrobial Prodiginine Pigments. ACS Omega 2020, 5, 4626–4635. [Google Scholar] [CrossRef]
- Eckelmann, D.; Spiteller, M.; Kusari, S. Spatial-temporal profling of prodiginines and serratamolides produced by endophytic Serratia marcescens harbored in Maytenus serrata. Sci. Rep. 2018, 8, 5283. [Google Scholar] [CrossRef]
- Abidin, Z.A.Z.; Ahmad, A.; Latip, J.; Usup, G. MARINE Streptomyces SP. UKMCC_PT15 producing undecylprodigiosin with algicidal activity. J. Teknol. 2016, 78. [Google Scholar] [CrossRef]
- Song, Y.; Liu, G.; Li, J.; Huang, H.; Zhang, X.; Zhang, H.; Ju, J. Cytotoxic and Antibacterial Angucycline- and Prodigiosin- Analogues from the Deep-Sea Derived Streptomyces sp. SCSIO 11594. Mar. Drugs 2015, 13, 1304–1316. [Google Scholar] [CrossRef]
- Meng-Xi, L.I.; Hui-Bin, H.; Jie-Yun, L.; Jing-Xiao, C.A.O.; Zhen-Wang, Z. Antibacterial Performance of a Streptomyces spectabilis Strain Producing Metacycloprodigiosin. Curr. Microbiol. 2021, 78, 2569–2576. [Google Scholar] [CrossRef]
- Stankovic, N.; Radulovic, V.; Petkovic, M.; Vuckovic, I.; Jadranin, M.; Vasiljevic, B.; Nikodinovic-Runic, J. Streptomyces sp. JS520 produces exceptionally high quantities of undecylprodigiosin with antibacterial, antioxidative, and UV-protective properties. Appl. Microbiol. Biotechnol. 2012, 96, 1217–1231. [Google Scholar] [CrossRef]
- Bikash, B.; Vilja, S.; Mitchell, L.; Keith, Y.; Mikael, I.; Mikko, M.-K.; Jarmo, N. Differential regulation of undecylprodigiosin biosynthesis in the yeast-scavenging Streptomyces strain MBK6. FEMS Microbiol. Lett. 2021, 368, fnab044. [Google Scholar] [CrossRef] [PubMed]
- Kramar, A.; Ilic-Tomic, T.; Petkovic, M.; Radulovic, N.; Kostic, M.; Jocic, D.; Nikodinovic-Runic, J. Crude bacterial extracts of two new Streptomyces sp. isolates as bio-colorants for textile dyeing. World J. Microbiol. Biotechnol. 2014, 30, 2231–2240. [Google Scholar] [CrossRef]
- Abraham, J.; Chauhan, R. Profiling of red pigment produced by Streptomyces sp. JAR6 and its bioactivity. 3 Biotech. 2017, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.; Thomson, N.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Wentzel, A.; Bruheim, P.; Øverby, A.; Jakobsen, M.; Sletta, H.; Omara, W.A.M.; A Hodgson, D.; E Ellingsen, T. Optimized submerged batch fermentation strategy for systems scale studies of metabolic switching in Streptomyces coelicolor A3(2). BMC Syst. Biol. 2012, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Sakurai, F.; Nagatsuka, S.-Y.; Hayakawa, Y. Prodigiosin biosynthesis gene cluster in the roseophilin producer Streptomyces griseoviridis. J. Antibiot. 2009, 62, 271–276. [Google Scholar] [CrossRef]
- Kimata, S.; Matsuda, T.; Suizu, Y.; Hayakawa, Y. Prodigiosin R2, a new prodigiosin from the roseophilin producer Streptomyces griseoviridis 2464-S5. J. Antibiot. 2018, 71, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Beppu, T. Production in Large Quantities of Actinorhodin and Undecyl-prodigiosin Induced by afsB in Streptomyces lividans. Agric. Biol. Chem. 1984, 48, 2131–2133. [Google Scholar] [CrossRef][Green Version]
- Gerber, N.N. Prodigiosin-Like Pigments. CRC Crit. Rev. Microbiol. 1975, 3, 469–485. [Google Scholar] [CrossRef]
- Mirzaei, S.A.; Kavishahi, M.S.; Keshavarz, Z.; Elahian, F. Unlike Butylcycloheptylprodigiosin, Isolated Undecylprodigiosin from Streptomyces parvulus Is Not a MDR1 and BCRP Substrate in Multidrug-Resistant Cancers. DNA Cell Biol. 2018, 37, 535–542. [Google Scholar] [CrossRef]
- Xavier, R.K.M.; Xu, D.; McCarthy, P.J.; Yang, S.; Wang, G. Genome Sequence of Streptomyces sp. Strain HB-N217, Isolated from the Marine Sponge Forcepia sp. Microbiol. Resour. Announc. 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Mariita, R.M.; Bhatnagar, S.; Hanselmann, K.; Hossain, M.J.; Korlach, J.; Boitano, M.; Roberts, R.J.; Liles, M.R.; Moss, A.G.; Leadbetter, J.R.; et al. Complete Genome Sequence of Streptomyces sp. Strain CCM_MD2014, Isolated from Topsoil in Woods Hole, Massachusetts. Genome Announc. 2015, 3, e01506-15. [Google Scholar] [CrossRef]
- Sharma, A.; Gautam, S.; Saxena, S. Streptomyces. In Encyclopedia of Food Microbiology; Batt, C.A., Robinson, R.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 3, pp. 560–566. [Google Scholar]
- Quinn, G.A.; Banat, A.M.; Abdelhameed, A.M.; Banat, I.M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol. 2020, 69, 1040–1048. [Google Scholar] [CrossRef]
- Sakai, Y.; Yoshida, T.; Ochiai, K.; Uosaki, Y.; Saitoh, Y.; Tanaka, F.; Akiyama, T.; Akinaga, S.; Mizukami, T. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J. Antibiot. 2002, 55, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Matsumoto, D.; Kawaide, H.; Natsume, M. Physiological role of germicidins in spore germination and hyphal elongation in Streptomyces coelicolor A3(2). J. Antibiot. 2011, 64, 607–611. [Google Scholar] [CrossRef]
- Thanapipatsiri, A.; Claesen, J.; Gomez-Escribano, J.-P.; Bibb, M.J.; Thamchaipenet, A. A Streptomyces coelicolor host for the heterologous expression of Type III polyketide synthase genes. Microb. Cell Factories 2015, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Risdian, C.; Mozef, T.; Wink, J. Biosynthesis of Polyketides in Streptomyces. Microorganisms 2019, 7, 124. [Google Scholar] [CrossRef]
- Novakova, R.; Bistakova, J.; Kormanec, J. Characterization of the polyketide spore pigment cluster whiESa in Streptomyces aureofaciens CCM3239. Arch. Microbiol. 2004, 182, 388–395. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Zhang, X.-S.; Luo, H.-D.; Ren, N.-N.; Jiang, X.-H.; Jiang, H.; Li, Y.-Q. Characterization of Discrete Phosphopantetheinyl Transferases in Streptomyces tsukubaensis L19 Unveils a Complicate Phosphopantetheinylation Network. Sci. Rep. 2016, 6, 24255. [Google Scholar] [CrossRef]
- Harris, A.K.P.; Williamson, N.R.; Slater, H.; Cox, A.; Abbasi, S.; Foulds, I.; Simonsen, H.T.; Leeper, F.J.; Salmond, G.P.C. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 2004, 150, 3547–3560. [Google Scholar] [CrossRef]
- Kim, D.; Park, Y.K.; Lee, J.S.; Kim, J.F.; Jeong, H.; Kim, B.S.; Lee, C.H. Analysis of a prodigiosin biosynthetic gene cluster from the marine bacterium Hahella chejuensis KCTC 2396. J. Microbiol. Biotechnol. 2006, 16, 1912–1918. [Google Scholar]
- Shaikhpour, M.; Sadeghi, A.; Yazdian, F.; Mansoori, A.; Movafagh, A. Anticancer and Apoptotic Effects of Ectoine and Hydroxyectoine on Non-Small Cell Lung Cancer cells: An in-vitro Investigation. Multidiscip. Cancer Investig. 2019, 3, 14–19. [Google Scholar] [CrossRef]
- Sadeghi, A.; Soltani, B.M.; Nekouei, M.K.; Jouzani, G.S.; Mirzaei, H.H.; Sadeghizadeh, M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and Uptake of the Compatible Solutes Ectoine and 5-Hydroxyectoine by Streptomyces coelicolor A3(2) in Response to Salt and Heat Stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef] [PubMed]
- Blondeau, R.; Imbert, M.; Béchet, M. Comparison of the main siderophores produced by some species of Streptomyces. Curr. Microbiol. 1995, 31, 129–133. [Google Scholar] [CrossRef]
- Müller, G.; Raymond, K.N. Specificity and mechanism of ferrioxamine-mediated iron transport in Streptomyces pilosus. J. Bacteriol. 1984, 160, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Lucas, X.; Senger, C.; Erxleben, A.; Grüning, B.; Döring, K.; Mosch, J.; Flemming, S.; Günther, S. StreptomeDB: A resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 2012, 41, D1130–D1136. [Google Scholar] [CrossRef]
- Zhao, B.; Lin, X.; Lei, L.; Lamb, D.C.; Kelly, S.L.; Waterman, M.R.; Cane, D.E. Biosynthesis of the Sesquiterpene Antibiotic Albaflavenone in Streptomyces coelicolor A3(2). J. Biol. Chem. 2008, 283, 8183–8189. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Cane, D.E. Biosynthesis of the Sesquiterpene Antibiotic Albaflavenone in Streptomyces coelicolor. Mechanism and Stereochemistry of the Enzymatic Formation of Epi-isozizaene. J. Am. Chem. Soc. 2009, 131, 6332–6333. [Google Scholar] [CrossRef]
- Challis, G.L.; Ravel, J. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: Structure prediction from the sequence of its non- ribosomal peptide synthetase. FEMS Microbiol. Lett. 2000, 187, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Poralla, K.; Muth, G.; Härtner, T. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2000, 189, 93–95. [Google Scholar] [CrossRef]
- De Jong, W.; Vijgenboom, E.; Dijkhuizen, L.; Wösten, H.A.; Claessen, D. SapB and the rodlins are required for development of Streptomyces coelicolor in high osmolarity media. FEMS Microbiol. Lett. 2012, 329, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Moody, S.C.; Hider, R.C.; Lei, L.; Kelly, S.L.; Waterman, M.R.; Lamb, D.C. Structural Analysis of Cytochrome P450 105N1 Involved in the Biosynthesis of the Zincophore, Coelibactin. Int. J. Mol. Sci. 2012, 13, 8500–8513. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zeng, Y.J.; Zhang, B.; Shao, F.L.; Chen, Y.C.; Xu, X.; Sun, Y.; Xu, Q.; Tan, R.X.; Ge, H.M. Comparative genome mining and heterologous expression of an orphan NRPS gene cluster direct the production of ashimides. Chem. Sci. 2019, 10, 3042–3048. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).