Microbial Communities Involved in Methane, Sulfur, and Nitrogen Cycling in the Sediments of the Barents Sea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Sampling
2.2. Chemical Analysis
2.3. The Total Abundance of Microorganisms
2.4. Radiotracer Measurements
2.5. 16S rRNA Amplicon Sequencing and Analysis
2.6. Sequencing and Analysis of pmoA Gene Sequences
3. Results
3.1. Characterization of the Sampling Sites and Microbial Processes
3.2. Diversity and Composition of Microbial Communities
3.3. Aerobic Methanotrophs Revealed by pmoA Gene Profiling
4. Discussion
4.1. Methane Cycle
4.2. Sulfur Cycle
4.3. Nitrogen Cycle
4.4. Organic Matter Decomposition in Anoxic Sediments
4.5. Microbial Processes of Transformation of Organic Matter in Sediments of the Arctic Seas: The Role of Methane
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edwards, A.; Cameron, K.A.; Cook, J.M.; Debbonaire, A.R.; Furness, E.; Hay, M.C.; Rassner, S.M.E. Microbial genomics amidst the Arctic crisis. Microb Genom. 2020, 6, e000375. [Google Scholar] [CrossRef] [PubMed]
- James, R.H.; Bousquet, P.; Bussmann, I.; Haeckel, M.; Kipfer, R.; Leifer, I.; Niemann, H.; Ostrovsky, I.; Piskozub, J.; Rehder, G.; et al. Effects of climate change on methane emissions from seafloor sediments in the Arctic Ocean: A review. Limnol. Oceanogr. 2016, 61, 283–299. [Google Scholar] [CrossRef] [Green Version]
- Hunter, S.J.; Goldobin, D.S.; Haywood, A.M.; Ridgewell, A.; Rees, J.G. Sensitivity of the global submarine hydrate inventory to scenarios of future climate change. Earth Planet. Sci. Lett. 2013, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Reagan, M.T.; Moridis, G.J.; Elliott, S.M.; Maltrud, M. Contribution of oceanic gas hydrate dissociation to the formation of Arctic Ocean methane plumes. J. Geophys. Res. 2011, 116. [Google Scholar] [CrossRef] [Green Version]
- Reagan, M.T.; Moridis, G.J. Large-scale simulation of methane hydrate dissociation along the West Spitsbergen Margin. Geophys. Res. Lett. 2009, 36. [Google Scholar] [CrossRef] [Green Version]
- Biastoch, A.T.; Treude, L.H.; Rüpke, U.; Riebesell, C.; Roth, E.B.; Burwicz, W.; Park, M.; Latif, C.W.; Böning, G.; Wallmann, M.K. Rising Arctic Ocean temperatures cause gas hydrate destabilization and ocean acidification. Geophys. Res. Lett. 2011, 38. [Google Scholar] [CrossRef] [Green Version]
- Thatcher, K.E.; Westbrook, G.K.; Sarkar, S.; Minshull, T.A. Methane release from warming-induced hydrate dissociation in the West Svalbard continental margin: Timing, rates, and geological controls. J. Geophys. Res. 2013, 118, 22–38. [Google Scholar] [CrossRef]
- Loeng, H. Features of the physical oceanographic conditions of the Barents Sea. In Proceedings of the Pro Mare Symposium on Polar Marine Ecology, Trondheim, Norway, 12–16 May 1990; Sakshaug, E., Hopkins, C., Britsland, N.A., Eds.; Norwegian Polar Institute: Tromsø, Norway, 1991; Volume 10, pp. 5–18. [Google Scholar] [CrossRef] [Green Version]
- Oziel, L.; Sirven, J.; Gascard, J.C. The Barents Sea frontal zones and water masses variability (1980–2011). Ocean Sci. 2016, 12, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Sakshaug, E. Primary and Secondary Production in the Arctic Seas. In The Organic Carbon Cycle in the Arctic Ocean; Stein, R., MacDonald, R.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 57–81. [Google Scholar] [CrossRef]
- Dobrovolskiy, A.D.; Zalogin, B.S. Seas of the USSR; Moscow State University: Moscow, Russia, 1982; p. 349. [Google Scholar]
- Slagstad, D.; Downing, K.; Carlotti, F.; Hirche, H.J. Modelling the carbon export and air-sea flux of CO2 in the Greenland Sea. Deep Sea Res. II 1999, 46, 1511–1530. [Google Scholar] [CrossRef]
- Wassmann, P.; Slagstad, D.; Ellingsen, I. Primary production and climatic variability in the European sector of the Arctic Ocean prior to 2007: Preliminary results. Polar Biol. 2010, 33, 1641–1650. [Google Scholar] [CrossRef]
- Wassmann, P.; Slagstad, D.; Riser, C.; Reigstad, M. Modelling the ecosystem dynamics of the Barents Sea including the marginal ice zone II. Carbon flux and interannual variability. J. Mar. Syst. 2006, 59, 1–24. [Google Scholar] [CrossRef]
- Knies, J.; Martinez, P. Organic matter sedimentation in the western Barents Sea region: Terrestrial and marine contribution based on isotopic composition and organic nitrogen content. Nor. J. Geol. 2009, 89, 79–89. [Google Scholar]
- Vetrov, A.; Romankevich, E. The Barents Sea: Distribution, sources, variability and burial of organic carbon. In The Organic Carbon Cycle in the Arctic Ocean; Stein, R., Macdonald, R.W., Eds.; Springer: Berlin, Germany, 2004; pp. 266–278. [Google Scholar]
- Vetrov, A.A.; Romankevich, E.A. Distibution, fluxes and balance of particular organic carbon in the Arctic Ocean. Oceanology 2019, 59, 491–499. [Google Scholar] [CrossRef]
- Winkelmann, D.; Knies, J. Recent distribution and accumulation of organic carbon on the continental margin west off Spitsbergen. Geochem. Geophys. Geosyst. 2005, 6, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Pathirana, I.; Knies, J.; Felix, M.; Mann, U. Towards an improved organic carbon budget for the western Barents Sea shelf. Clim. Past 2014, 10, 569–587. [Google Scholar] [CrossRef] [Green Version]
- Pimenov, N.; Savvichev, A.; Rusanov, I.; Lein, A.; Egorov, A.; Gebruk, A.; Moskalev, L.; Vogt, P. Microbial processes of carbon cycle as the base of food chain of Haakon Mosby Mud Volcano benthic community. Geo-Marine Lett. 1999, 19, 89–96. [Google Scholar] [CrossRef]
- Niemann, H.; Lösekann, T.; de Beer, D.; Elvert, M.; Nadalig, T.; Knittel, K.; Amann, R.; Sauter, E.J.; Schlüter, M.; Klages, M.; et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 2006, 443, 854–858. [Google Scholar] [CrossRef]
- Lösekann, T.; Knittel, K.; Nadalig, T.; Fuchs, B.; Niemann, H.; Boetius, A.; Amann, R. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 2007, 10, 3348–3362. [Google Scholar] [CrossRef] [Green Version]
- Argentino, C.; Waghorn, K.A.; Vadakkepuliyambatta, S.; Polteau, S.; Bünz, S.; Panieri, G. Dynamic and history of methane seepage in the SW Barents Sea: New insights from Leirdjupet Fault Complex. Sci. Rep. 2021, 11, 4373. [Google Scholar] [CrossRef] [PubMed]
- Gründger, F.; Carrier, V.; Svenning, M.M.; Panieri, G.; Vonnahme, T.R.; Klasek, S.; Niemann, H. Methane-fuelled biofilms predominantly composed of methanotrophic ANME-1 in Arctic gas hydrate-related sediments. Sci. Rep. 2019, 1, 9725. [Google Scholar] [CrossRef]
- Pimenov, N.V.; Savvichev, A.S.; Rusanov, I.I.; Lein, A.Y.; Ivanov, M.V. Microbiological processes of the carbon and sulfur cycles at cold methane seeps of the North Atlantic. Microbiology 2000, 6, 709–720. [Google Scholar] [CrossRef]
- Grünke, S.; Lichtschlag, A.; de Beer, D.; Felden, J.; Salman, V.; Ramette, A.; Schulz-Vogt, H.N.; Boetius, A. Mats of psychrophilic thiotrophic bacteria associated with cold seeps of the Barents Sea. Biogeosciences 2012, 9, 2947–2960. [Google Scholar] [CrossRef] [Green Version]
- Shirokolobova, T.I.; Zhichkin, A.P.; Venger, M.P.; Vodopyanova, V.; Moiseev, D.V. Bacteria and viruses of the ice-free aquatic area of the Barents Sea at the beginning of polar night. Dokl. Biol. Sci. 2016, 1, 182–186. [Google Scholar] [CrossRef]
- Stevenson, M.A.; Faust, J.C.; Andrade, L.L.; Freitas, F.S.; Gray, N.D.; Tait, K.; Hendry, K.R.; Hilton, R.G.; Henley, S.F.; Tessin, A.; et al. Transformation of organic matter in a Barents Sea sediment profile: Coupled geochemical and microbiological processes. Phil. Trans. R. Soc. A 2020, 378, 20200223. [Google Scholar] [CrossRef] [PubMed]
- Klyuvitkin, A.A.; Politova, N.V.; Novigatsky, A.N.; Kravchishina, M.D. Studies of the European Arctic on Cruise 80 of the R/V Akademik Mstislav Keldysh. Oceanology 2021, 61, 139–141. [Google Scholar] [CrossRef]
- McAuliffe, C. Gas chromatographic determination of solutes by multiple phase equilibrium. Chem. Technol. 1971, 1, 46–51. [Google Scholar]
- Magen, C.; Lapham, L.L.; Pohlman, J.; Marshall, K.; Bossman, S.; Chanton, J.P. A simple headspace equilibration method for measuring dissolved methane. Limnol. Oceanogr. Meth. 2014, 12, 637–650. [Google Scholar] [CrossRef]
- Lapham, L.; Marshall, K.; Magen, C.; Lyubchich, V.; Cooper, L.W.; Grebmeier, J.M. Dissolved methane concentrations in the water column and surface sediments of Hanna Shoal and Barrow Canyon, Northern Chukchi Sea. Deep-Sea Res. II-Topical Stud. Oceanogr. 2017, 144, 92–103. [Google Scholar] [CrossRef]
- Hobbie, J.T.; Daley, R.J.; Jasper, S. Use of Nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 1977, 33, 1225–1228. [Google Scholar] [CrossRef] [Green Version]
- Pimenov, N.V.; Bonch-Osmolovskaya, E.A. In situ activity studies in thermal environments. Methods Microbiol. 2006, 35, 29–53. [Google Scholar]
- Savvichev, A.; Rusanov, I.; Dvornikov, Y.; Kadnikov, V.; Kallistova, A.; Veslopolova, E.; Chetverova, A.; Leibman, M.; Sigalevich, P.; Pimenov, N.; et al. The water column of the Yamal tundra lakes as a microbial filter preventing methane emission. Biogeosciences 2021, 9, 2791–2807. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, 018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoc, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4. [Google Scholar] [CrossRef]
- Luesken, F.A.; Zhu, B.; van Alen, T.A.; Butler, M.K.; Diaz, M.R.; Song, B.; Op den Camp, H.J.; Jetten, M.S.; Ettwig, K.F. pmoA pimers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 2011, 11, 3877–3880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Niemann, H.; Panieri, G. Multi-proxy approach to unravel methane emission history of an Arctic cold seep. Quat. Sci. Rev. 2020, 244, 106490. [Google Scholar] [CrossRef]
- Yao, H.; Panieri, G.; Lehmann, M.F.; Himmler, T.; Niemann, H. Biomarker and isotopic composition of seep carbonates record environmental conditions in two arctic methane seeps. Front. Earth Sci. 2021, 8, 570742. [Google Scholar] [CrossRef]
- Lein, A.Y.; Makkaveev, P.N.; Savvichev, A.S.; Kravchishina, M.D.; Belyaev, N.A.; Dara, O.M.; Ponyaev, M.S.; Zakharova, E.E.; Rozanov, A.G.; Ivanov, M.V. Transformation of suspended particulate matter into sediment in the Kara Sea in September of 2011. Oceanology 2013, 53, 570–606. [Google Scholar] [CrossRef]
- Savvichev, A.S.; Kadnikov, V.V.; Kravchishina, M.D.; Galkin, S.V.; Novigatskii, A.N.; Sigalevich, P.A.; Merkel, A.Y.; Ravin, N.V.; Pimenov, N.V.; Flint, M.V. Methane as an organic matter source and the trophic basis of a Laptev sea cold seep microbial community. Geomicrobiol. J. 2018, 35, 411–423. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Könneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lüke, C.; Krause, S.; Cavigiolo, S.; Greppi, D.; Lupotto, E.; Frenzel, P. Biogeography of wetland rice methanotrophs. Environ. Microbiol. 2010, 4, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Knoblauch, C.; Wagner, D. Methanogenic community composition and anaerobic carbon turnover in submarine permafrost sediments of the Siberian Laptev Sea. Environ. Microbiol. 2009, 3, 657–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchman, D.L.; Cottrell, M.T.; Lovejoy, C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 2010, 5, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.X.; Zhang, F.; He, J.F.; Lee, S.H.; Qiao, Z.Y.; Yu, Y.; Li, H.R. Bacterioplankton community structure in the Arctic waters as revealed by pyrosequencing of 16S rRNA genes. Antonie Van Leeuwenhoek 2013, 6, 1309–1319. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, L.; Liu, Y.; Li, Y. Bacterial and archaeal community structure of pan-Arctic Ocean sediments revealed by pyrosequencing. Acta Oceanol. Sin. 2017, 8, 146–152. [Google Scholar] [CrossRef]
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 2009, 5, 285–292. [Google Scholar] [CrossRef]
- Andreassen, K.; Hubbard, A.; Winsborrow, M.; Patton, H.; Vadakkepuliyambatta, S.; Plaza Faverola, A.; Gudlaugsson, E.; Serov, P.; Deryabin, A.; Mattingsdal, R.; et al. Massive blow-out craters formed by hydrate-controlled methane expulsion from the Arctic seafloor. Science 2017, 356, 948–953. [Google Scholar] [CrossRef] [Green Version]
- Sela-Adler, M.; Ronen, Z.; Herut, B.; Antler, G.; Vigderovich, H.; Eckert, W.; Sivan, O. Co-existence of methanogenesis and sulfate reduction with common substrates in sulfate-rich estuarine sediments. Front. Microbiol. 2017, 8, 766. [Google Scholar] [CrossRef] [Green Version]
- Michaelis, W.; Seifert, R.; Nauhaus, K.; Treude, T.; Thiel, V.; Blumenberg, M.; Knittel, K.; Gieseke, A.; Peterknecht, K.; Pape, T.; et al. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 2002, 5583, 1013–1015. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Ma, A.; Qi, H.; Zhuang, X.; Zhuang, G. Anaerobic oxidation of methane: An “active” microbial process. Microbiol. Open 2015, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Timmers, P.H.; Welte, C.U.; Koehorst, J.J.; Plugge, C.M.; Jetten, M.S.; Stams, A.J. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017, 2017, 1654237. [Google Scholar] [CrossRef]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J.; Rickert, D.; Widdel, F.; Gieseke, A.; Amann, R.; Jørgensen, B.B.; Witte, U.; Pfannkuche, O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Orphan, V.J.; House, C.H.; Hinrichs, K.U.; McKeegan, K.D.; DeLong, E.F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 2002, 99, 7663–7668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmers, P.H.; Suarez-Zuluaga, D.A.; van Rossem, M.; Diender, M.; Stams, A.J.; Plugge, C.M. Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J. 2016, 10, 1400–1412. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, L.; Holler, T.; Knittel, K.; Meyerdierks, A.; Amann, R. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ Microbiol. 2010, 8, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Kleindienst, S.; Ramette, A.; Amann, R.; Knittel, K. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 2012, 10, 2689–2710. [Google Scholar] [CrossRef]
- Wegener, G.; Krukenberg, V.; Ruff, S.E.; Kellermann, M.Y.; Knittel, K. Metabolic Capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 2016, 7, 46. [Google Scholar] [CrossRef] [Green Version]
- Green-Saxena, A.; Dekas, A.E.; Dalleska, N.F.; Orphan, V.J. Nitrate-based niche differentiation by distinct sulfate-reducing bacteria involved in the anaerobic oxidation of methane. ISME J. 2014, 1, 150–163. [Google Scholar] [CrossRef] [Green Version]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D.; et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 2010, 464, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Chistoserdova, L. Methylotrophs in natural habitats: Current insights through metagenomics. Appl. Microbiol. Biotechnol. 2015, 99, 5763–5779. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Ozaki, H.; Hiraoka, S.; Kamagata, Y.; Sakata, S.; Yoshioka, H.; Iwasaki, W. Possible cross-feeding pathway of facultative methylotroph Methyloceanibacter caenitepidi Gela4 on methanotroph Methylocaldum marinum S8. PLoS ONE 2019, 3, e0251538. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Kim, T.G. Development of a novel methanotrophic process with the helper micro-organism Hyphomicrobium sp. NM3. J. Appl. Microbiol. 2019, 2, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Singleton, C.M.; McCalley, C.K.; Woodcroft, B.J.; Boyd, J.A.; Evans, P.N.; Hodgkins, S.B.; Chanton, J.P.; Frolking, S.; Crill, P.M.; Saleska, S.R.; et al. Methanotrophy across a natural permafrost thaw environment. ISME J. 2018, 10, 2544–2558. [Google Scholar] [CrossRef] [Green Version]
- Vekeman, B.; Kerckhof, F.M.; Cremers, G.; de Vos, P.; Vandamme, P.; Boon, N.; Op den Camp, H.J.; Heylen, K. New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ. Microbiol. 2016, 12, 4523–4536. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 3, 203–208. [Google Scholar] [CrossRef]
- Hyman, M.R.; Wood, P.M. Methane oxidation by Nitrosomonas europaea. Biochem. J. 1983, 212, 31–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.D.; Morita, R.Y. Methane Oxidation by Nitrosococcus oceanus and Nitrosomonas europaea. Appl. Environ. Microbiol. 1983, 2, 401–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taher, E.; Chandran, K. High-rate, high-yield production of methanol by ammonia-oxidizing bacteria. Environ. Sci. Technol. 2013, 7, 3167–3173. [Google Scholar] [CrossRef]
- Jørgensen, B.B.; Findlay, A.J.; Pellerin, A. The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 2019. [Google Scholar] [CrossRef]
- Holmes, D.E.; Bond, D.R.; Lovley, D.R. Electron transfer by Desulfobulbus propionicus to Fe(III) and graphite electrodes. Appl. Environ. Microbiol. 2004, 2, 234–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuseler, K.; Cypionka, H. Elemental sulfur as an intermediate of sulfide oxidation with oxygen by Desulfobulbus propionicus. Arch. Microbiol. 1995, 164, 104–109. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Panteleeva, A.N.; Muyzer, G. Desulfonatronobacter acidivorans gen. nov., sp. nov. and Desulfobulbus alkaliphilus sp. nov., haloalkaliphilic heterotrophic sulfate-reducing bacteria from soda lakes. Int. J. Syst. Evol. Microbiol. 2012, 62, 2107–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandfeld, T.; Marzocchi, U.; Petro, C.; Schramm, A.; Risgaard-Petersen, N. Electrogenic sulfide oxidation mediated by cable bacteria stimulates sulfate reduction in freshwater sediments. ISME J. 2020, 5, 1233–1246. [Google Scholar] [CrossRef]
- Kjeldsen, K.U.; Schreiber, L.; Thorup, C.A.; Boesen, T.; Bjerg, J.T.; Yang, T.; Dueholm, M.S.; Larsen, S.; Risgaard-Petersen, N.; Nierychlo, M.; et al. On the evolution and physiology of cable bacteria. Proc. Natl. Acad. Sci. USA 2019, 38, 19116–19125. [Google Scholar] [CrossRef] [Green Version]
- Buongiorno, J.; Herbert, L.C.; Wehrmann, L.M.; Michaud, A.B.; Laufer, K.; Røy, H.; Jørgensen, B.B.; Szynkiewicz, A.; Faiia, A.; Yeager, K.M.; et al. Complex microbial communities drive iron and sulfur cycling in Arctic fjord sediments. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Fang, A.; Yu, X.; Zhang, K.; He, Z.; Wang, C.; Peng, Y.; Xiao, F.; Yang, T.; Zhang, W.; et al. Microbially-driven sulfur cycling microbial communities in different mangrove sediments. Chemosphere 2021, 273, 128597. [Google Scholar] [CrossRef]
- Feng, S.; Lin, X.; Tong, Y.; Huang, X.; Yang, H. Biodesulfurization of sulfide wastewater for, elemental sulfur recovery by isolated Halothiobacillus neapolitanus in an internal airlift loop reactor. Bioresour. Technol. 2018, 264, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Zhang, M.; Jing, R.; Bai, L.; Zhou, B.; Zhao, M.; Chen, G.H. Thiosulfate as the electron acceptor in sulfur bioconversion-associated process (SBAP) for sewage treatment. Water Res. 2019, 163, 114850. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Ekama, G.A.; Wang, Y.; Dai, J.; Biswal, B.K.; Chen, G.; Wu, D. Advances in sulfur conversion-associated enhanced biological phosphorus removal in sulfate-rich wastewater treatment: A review. Bioresour. Technol. 2019, 285, 121303. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Galinski, E.A.; Muyzer, G.; Kuenen, J.G. Thiohalorhabdus denitrificans gen. nov., sp. nov., an extremely halophilic, sulfur-oxidizing, deep-lineage gammaproteobacterium from hypersaline habitats. Int. J. Syst. Evol. Microbiol. 2008, 58, 2890–2897. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.L.; Zhang, J.; Wang, M.X.; Cao, L.; Du, Z.F.; Sun, Y.Y.; Liu, S.Q.; Li, C.L.; Sun, L. High-throughput sequencing reveals a potentially novel Sulfurovum species dominating the microbial communities of the seawater-sediment interface of a deep-sea cold seep in south China Sea. Microorganisms 2020, 5, 687. [Google Scholar] [CrossRef]
- Mori, K.; Yamaguchi, K.; Hanada, S. Sulfurovum denitrificans sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int. J. Syst. Evol. Microbiol. 2018, 68, 2183–2187. [Google Scholar] [CrossRef] [PubMed]
- Mulder, A.; van de Graaf, A.A.; Robertson, L.A.; Kuenen, J.G. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 1995, 16, 177–183. [Google Scholar] [CrossRef]
- Lam, P.; Kuypers, M.M. Microbial nitrogen cycling processes in oxygen minimum zones. Ann. Rev. Mar. Sci. 2011, 3, 317–345. [Google Scholar] [CrossRef]
- Matthew, D.P.; Micha, J.A.; Rijkenberg, P.J.; Statham, M.C.; Stinchcombe, E.P.; Achterberg, M.M. Determination of nitrate and phosphate in seawater at nanomolar concentrations. Trends Anal. Chem. 2008, 27, 169–182. [Google Scholar]
- Sedlacek, C.J.; McGowan, B.; Suwa, Y.; Sayavedra-Soto, L.; Laanbroek, H.J.; Stein, L.Y.; Norton, J.M.; Klotz, M.G.; Bollmann, A. A physiological and genomic comparison of Nitrosomonas cluster 6a and 7 ammonia-oxidizing bacteria. Microb. Ecol. 2019, 4, 985–994. [Google Scholar] [CrossRef]
- Taylor, H.B.; Kurtz, H.D. Composition, diversity, and activity of aerobic ammonia-oxidizing Bacteria and Archaea in the intertidal sands of a grand strand South Carolina beach. Microbiol. Open 2020, 9, e1011. [Google Scholar] [CrossRef] [Green Version]
- Van Kessel, M.A.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 7583, 555–559. [Google Scholar] [CrossRef] [Green Version]
- Daims, H.; Wagner, M. Nitrospira. Trends Microbiol. 2018, 5, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.G.; Schreiber, L.; Petersen, D.G.; Kjeldsen, K.U.; Lever, M.A.; Steen, A.D.; Stepanauskas, R.; Richter, M.; Kleindienst, S.; Lenk, S.; et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 2013, 496, 215–218. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Lloyd, K.G.; Pan, J.; Yang, Y.; Gu, J.D.; Li, M. Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). ISME J. 2019, 4, 885–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farag, I.F.; Davis, J.P.; Youssef, N.H.; Elshahed, M.S. Global patterns of abundance, diversity and community structure of the Aminicenantes (candidate phylum OP8). PLoS ONE 2014, 3, e92139. [Google Scholar] [CrossRef]
- Robbins, S.J.; Evans, P.N.; Parks, D.H.; Golding, S.D.; Tyson, G.W. Genome-centric analysis of microbial populations enriched by hydraulic fracture fluid additives in a coal bed methane production well. Front. Microbiol. 2016, 7, 731. [Google Scholar] [CrossRef]
- Kadnikov, V.V.; Mardanov, A.V.; Beletsky, A.V.; Karnachuk, O.V.; Ravin, N.V. Genome of the candidate phylum Aminicenantes bacterium from a deep subsurface thermal aquifer revealed its fermentative saccharolytic lifestyle. Extremophiles 2019, 2, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Hwang, K.; Lee, J.I.; Kim, M.; Hwang, C.Y.; Noh, H.J.; Choi, H.; Lee, H.K.; Chun, J.; Hong, S.G.; et al. Genomic insight into the predominance of candidate phylum Atribacteria JS1 lineage in marine sediments. Front. Microbiol. 2018, 9, 2909. [Google Scholar] [CrossRef]
- Nobu, M.K.; Dodsworth, J.A.; Murugapiran, S.K.; Rinke, C.; Gies, E.A.; Webster, G.; Schwientek, P.; Kille, P.; Parkes, R.J.; Sass, H.; et al. Phylogeny and physiology of candidate phylum ‘Atribacteria’ (OP9/JS1) inferred from cultivation-independent genomics. ISME J. 2016, 10, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Müller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63, 625–635. [Google Scholar] [CrossRef] [Green Version]
- Parkes, R.; Cragg, J.B.; Roussel, E.; Webster, G.; Weightman, A.; Sass, H. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar. Geol. 2014, 352, 409–425. [Google Scholar] [CrossRef]
- Yamada, T.; Sekiguchi, Y.; Hanada, S.; Imachi, H.; Ohashi, A.; Harada, H.; Kamagata, Y. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2006, 56, 1331–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fincker, M.; Huber, J.A.; Orphan, V.J.; Rappé, M.S.; Teske, A.; Spormann, A.M. Metabolic strategies of marine subseafloor Chloroflexi inferred from genome reconstructions. Environ. Microbiol. 2020, 8, 3188–3204. [Google Scholar] [CrossRef] [PubMed]
- Futagami, T.; Morono, Y.; Terada, T.; Kaksonen, A.H.; Inagaki, F. Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl. Environ. Microbiol. 2009, 21, 6905–6909. [Google Scholar] [CrossRef] [Green Version]
- Savvichev, A.S.; Rusanov, I.I.; Kadnikov, V.V.; Beletskii, A.V.; Ravin, N.V.; Pimenov, N.V. Microbial Community Composition and Rates of the Methane Cycle Microbial Processes in the Upper Sediments of the Yamal Sector of the Southwestern Kara Sea. Microbiology 2018, 87, 238–248. [Google Scholar] [CrossRef]
- Bussmann, I.; Hackbusch, S.; Schaal, P.; Wichels, A. Methane distribution and oxidation around the Lena Delta in summer 2013. Biogeosciences 2017, 14, 4985–5002. [Google Scholar] [CrossRef] [Green Version]
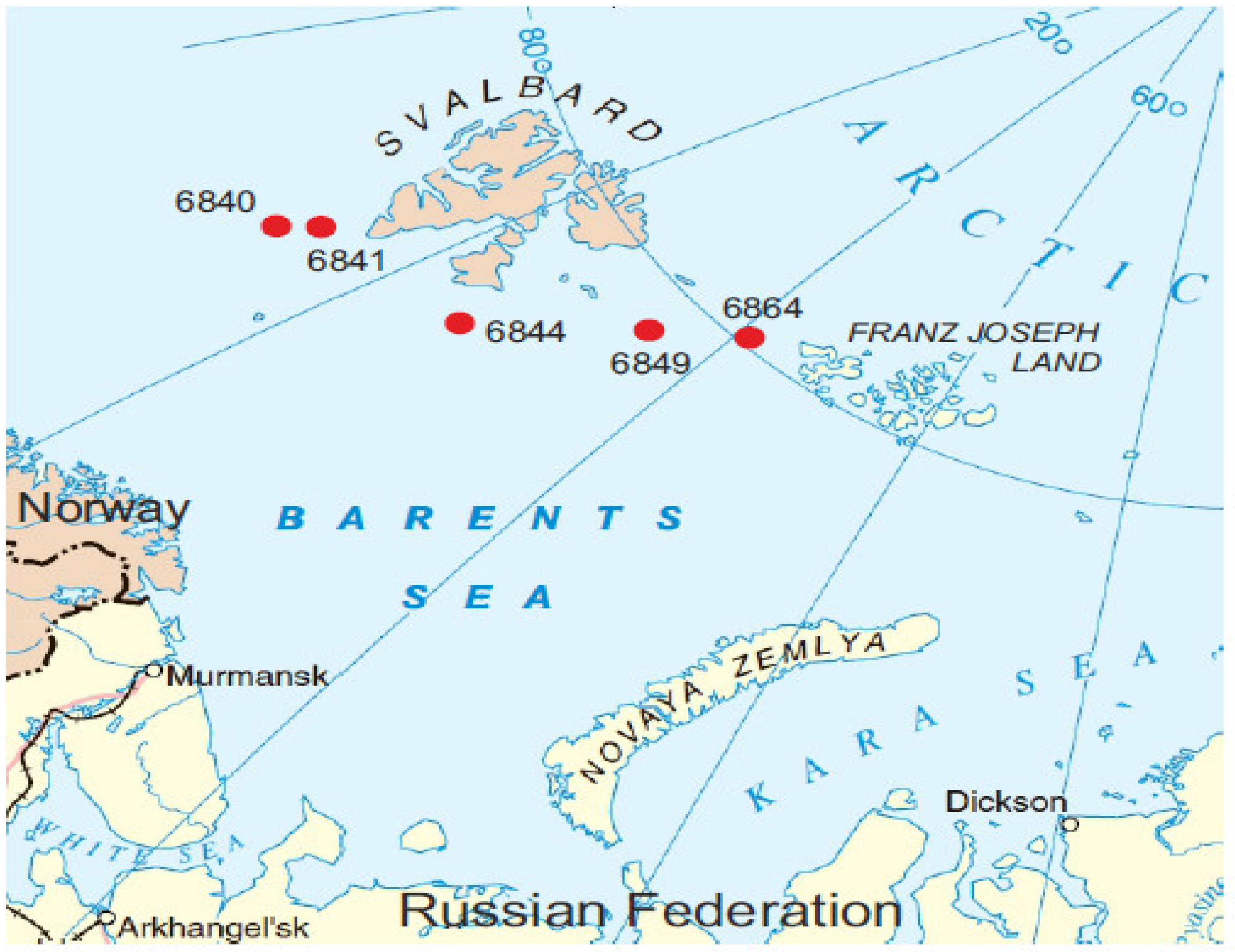




| Sampling Station | Sea Depth (m) | Coordinates | Sediment Depth (cm) | 16S rRNA Gene Sequences * |
|---|---|---|---|---|
| 6840 | 1514 | 75.21990 N 13.11843 E | 0–1 | 10,140 |
| 6841 | 385 | 76.06437 N 15.57961 E | 0–5 | 19,649 |
| 6–7 | 16,146 | |||
| 16–19 | 17,395 | |||
| 6844 | 101 | 77.03582 N 25.58852 E | 0–1 | 174,986 |
| 6849 | 307 | 78.59960 N 35.39939 E | 0–1 | 23,286 |
| 6864 | 584 | 80.59010 N 40.45922 E | 0–1 | 22,583 |
| Station | Sediment Depth, cm | Eh (mV) | Alk (mM) | Methane (μM) | MO (nmol L−1 day−1) | Sulfate (mM) | SR (µmol L−1 day−1) | DCA (µmol L−1 day−1) | TMC (×106 cells ml−1) |
|---|---|---|---|---|---|---|---|---|---|
| 6840 | 0–1 | +100 | 2.4 | 0.288 | 6.2 ± 0.28 | 28.0 | 1.73 ± 0.16 | 2.71 ± 0.22 | 1.40 ± 0.12 |
| 6844 | 0–1 | +125 | 2.2 | 0.597 | 8.2 ± 0.40 | 27.3 | 2.16 ± 0.21 | 5.75 ± 0.46 | 1.65 ± 0.17 |
| 6849 | 0–1 | +110 | 2.4 | 0.217 | 6.6 ± 0.29 | 27.6 | 1.12 ± 0.12 | 4.12 ± 0.29 | 1.30 ± 0.15 |
| 6864 | 0–1 | +110 | 2.3 | 0.398 | 2.1 ± 0.7 | 27.9 | 0.27 ± 0.03 | 2.14 ± 0.17 | 0.90 ± 0.1 |
| 6841 | 0–5 | +10 | 2.6 | 2.39 | 22.8 ± 0.95 | 27.1 | 2.77 ± 0.24 | 12.42 ± 1.1 | 1.96 ± 0.21 |
| 6–7 | −80 | 4.0 | 5.08 | 0.9 ± 0.05 | 26.5 | 2.00 ± 0.23 | 0.51 ± 0.04 | 0.60 ± 0.1 | |
| 16–19 | −120 | 4.4 | 9.51 | 0.3 ± 0.03 | 26.0 | 0.97 ± 0.10 | 0.31 ± 0.03 | 0.40 ± 0.1 |
| Station | Sediment Depth, cm | Chao1 | Shannon | ||
|---|---|---|---|---|---|
| Bacteria | Archaea | Bacteria | Archaea | ||
| 6840 | 0–1 | 643.8 | 20 | 4.92 | 1.91 |
| 6844 | 0–1 | 929.3 | 67.1 | 5.37 | 2.96 |
| 6849 | 0–1 | 994.9 | 31.5 | 5.62 | 1.81 |
| 6864 | 0–1 | 1097.9 | 66.1 | 5.6 | 2.52 |
| 6841 | 0–5 | 1241.2 | 63.3 | 5.94 | 1.78 |
| 6841 | 6–7 | 897.9 | 149.1 | 5.43 | 4.08 |
| 6841 | 16–19 | 561.8 | 128.4 | 4.43 | 2.96 |
| Site (Number of Samples) | Eh (mV) | Alkalinity (mM) | Methane (μM) | MO (nmol L−1 day−1) | SR (µmol L−1 day−1) | DCA (µmol L−1 day−1) | Reference |
|---|---|---|---|---|---|---|---|
| Northern part of the Kara Sea (2) | ND | ND | 0.02 ÷ 0.3 | 2.2 ÷ 12 | 0.4 ÷ 4.2 | ND | [111] |
| Southwestern part of the Kara Sea (18) | −160 ÷ +40 | 2.4 ÷ 8.0 | 1.9 ÷ 20.3 | 9.1 ÷ 103 | 0.46 ÷ 2.2 | 2.1 ÷ 11.8 | [111] |
| Laptev Sea, outside the methane seep field (7) | +40 ÷ +180 | 2.2 ÷ 2.8 | <0.012 | <2 | <0.05 | 0.01 ÷ 0.12 | [46] |
| Laptev Sea, methane seep field (5) | −160 ÷ +20 | 5.5 ÷ 18.0 | 19 ÷ 539 | 460 ÷ 3900 | 0.34 ÷ 4.8 | 0.2 ÷ 40.4 | [46] |
| Laptev Sea, near the Lena river delta (19) | ND | ND | 0.4 ÷ 5.4 | <5.7 | ND | ND | [112] |
| HMMV (18) | −350 ÷ −200 | 15.0 ÷ 30.5 | > 2000 | 1500 ÷ 70000 | 0.5 ÷ 394 | 1.5 ÷ 154 | [20,25] |
| Northern part of the Barents Sea (7) | −120 ÷ +125 | 2.2 ÷ 4.4 | 0.2 ÷ 9.5 | 0.3 ÷ 23 | 0.3 ÷ 2.8 | 0.3 ÷ 12.4 | this work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Begmatov, S.; Savvichev, A.S.; Kadnikov, V.V.; Beletsky, A.V.; Rusanov, I.I.; Klyuvitkin, A.A.; Novichkova, E.A.; Mardanov, A.V.; Pimenov, N.V.; Ravin, N.V. Microbial Communities Involved in Methane, Sulfur, and Nitrogen Cycling in the Sediments of the Barents Sea. Microorganisms 2021, 9, 2362. https://doi.org/10.3390/microorganisms9112362
Begmatov S, Savvichev AS, Kadnikov VV, Beletsky AV, Rusanov II, Klyuvitkin AA, Novichkova EA, Mardanov AV, Pimenov NV, Ravin NV. Microbial Communities Involved in Methane, Sulfur, and Nitrogen Cycling in the Sediments of the Barents Sea. Microorganisms. 2021; 9(11):2362. https://doi.org/10.3390/microorganisms9112362
Chicago/Turabian StyleBegmatov, Shahjahon, Alexander S. Savvichev, Vitaly V. Kadnikov, Alexey V. Beletsky, Igor I. Rusanov, Alexey A. Klyuvitkin, Ekaterina A. Novichkova, Andrey V. Mardanov, Nikolai V. Pimenov, and Nikolai V. Ravin. 2021. "Microbial Communities Involved in Methane, Sulfur, and Nitrogen Cycling in the Sediments of the Barents Sea" Microorganisms 9, no. 11: 2362. https://doi.org/10.3390/microorganisms9112362
APA StyleBegmatov, S., Savvichev, A. S., Kadnikov, V. V., Beletsky, A. V., Rusanov, I. I., Klyuvitkin, A. A., Novichkova, E. A., Mardanov, A. V., Pimenov, N. V., & Ravin, N. V. (2021). Microbial Communities Involved in Methane, Sulfur, and Nitrogen Cycling in the Sediments of the Barents Sea. Microorganisms, 9(11), 2362. https://doi.org/10.3390/microorganisms9112362








