Genetic Characterization of Salmonella Infantis with Multiple Drug Resistance Profiles Isolated from a Poultry-Farm in Chile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Salmonella Isolation
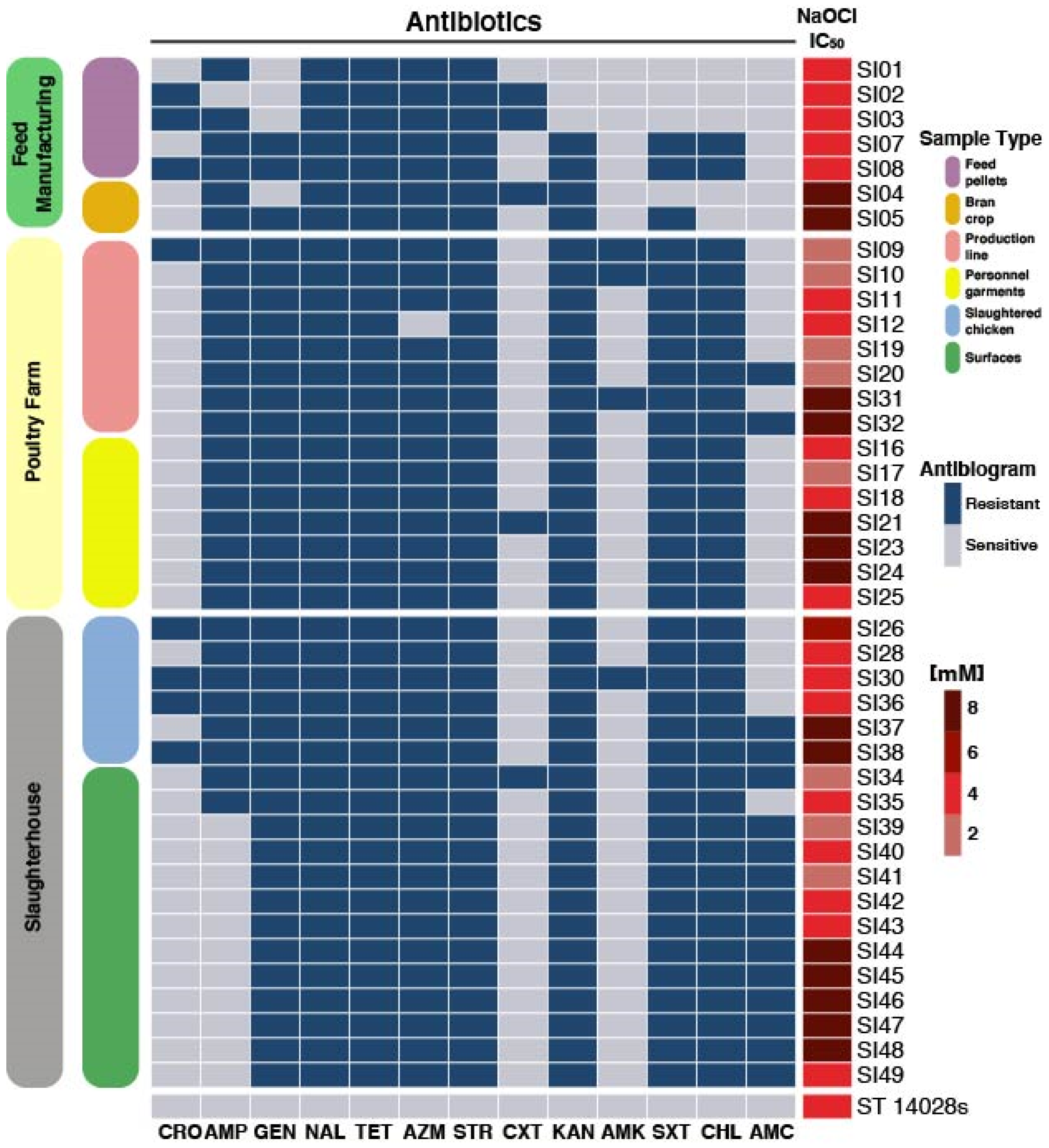
2.2. Antimicrobial Testing
2.3. NaOCl Resistance
2.4. Genome Sequencing
2.5. Genomic Analysis
3. Results
3.1. Serotyping and Antimicrobial Testing
3.2. Genomic Features of S. Infantis Strains
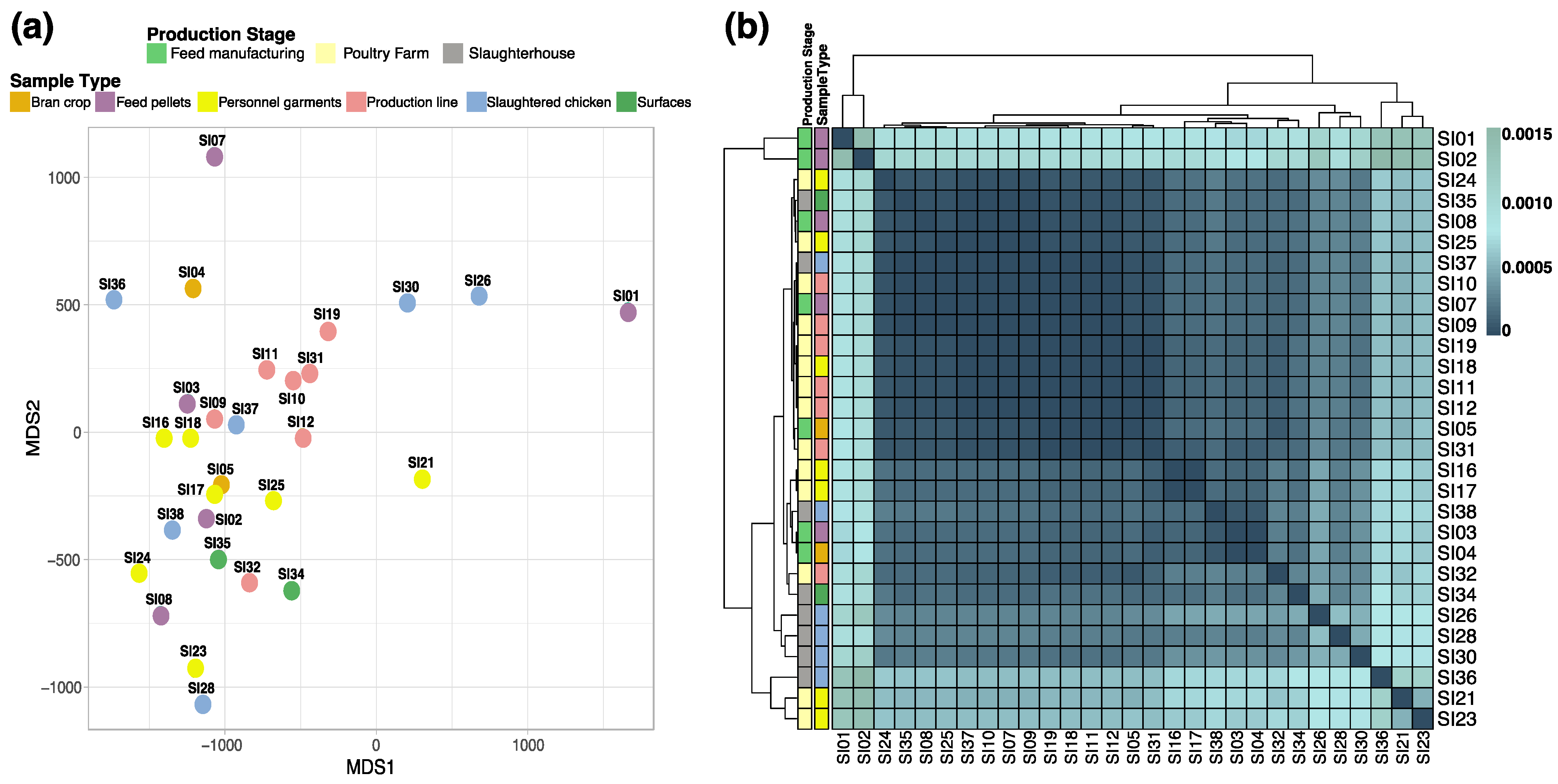
3.3. Genomic Similarity among the Strains
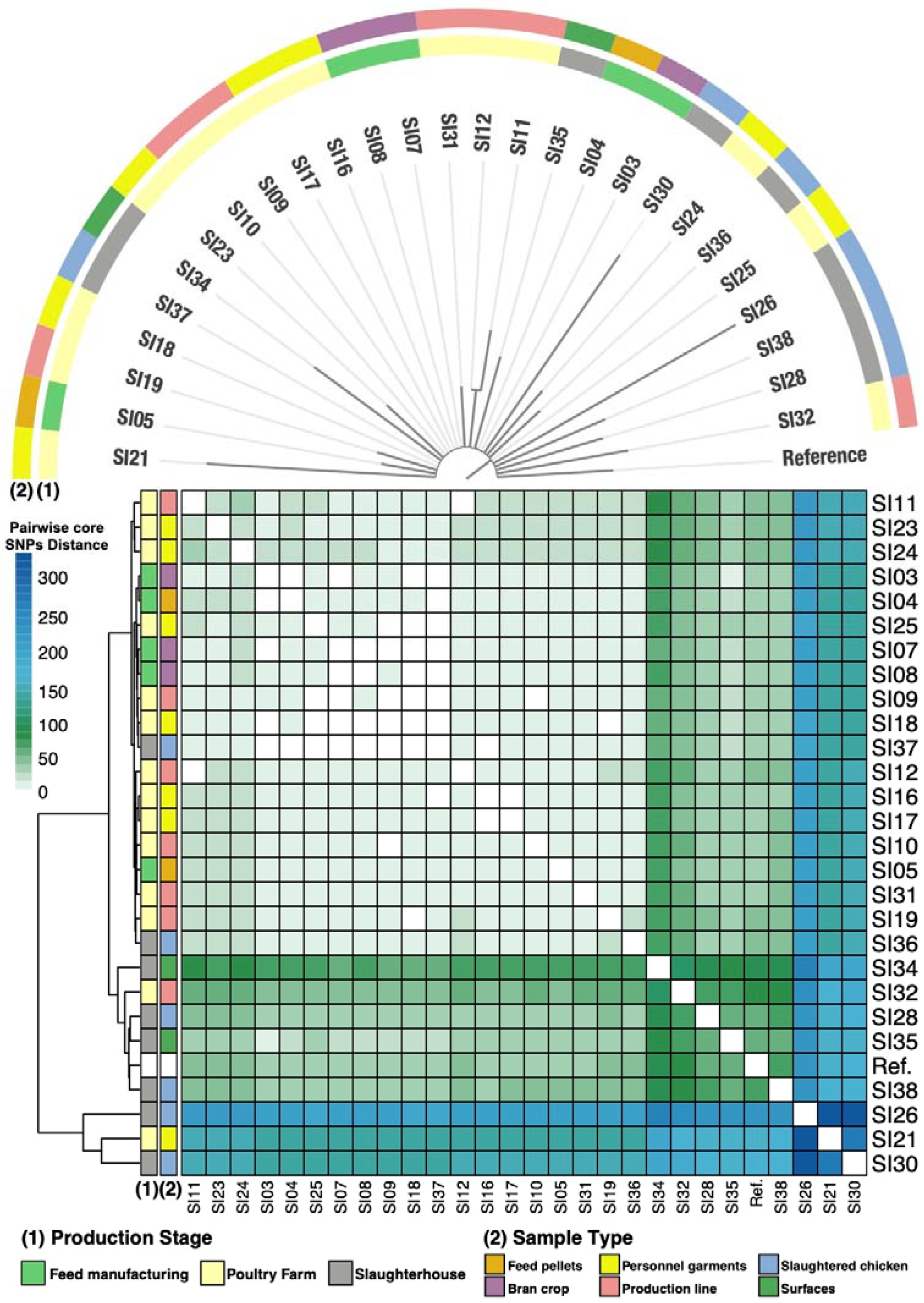
3.4. Phylogeny of the S. Infantis Isolates
3.5. Functional Profiles
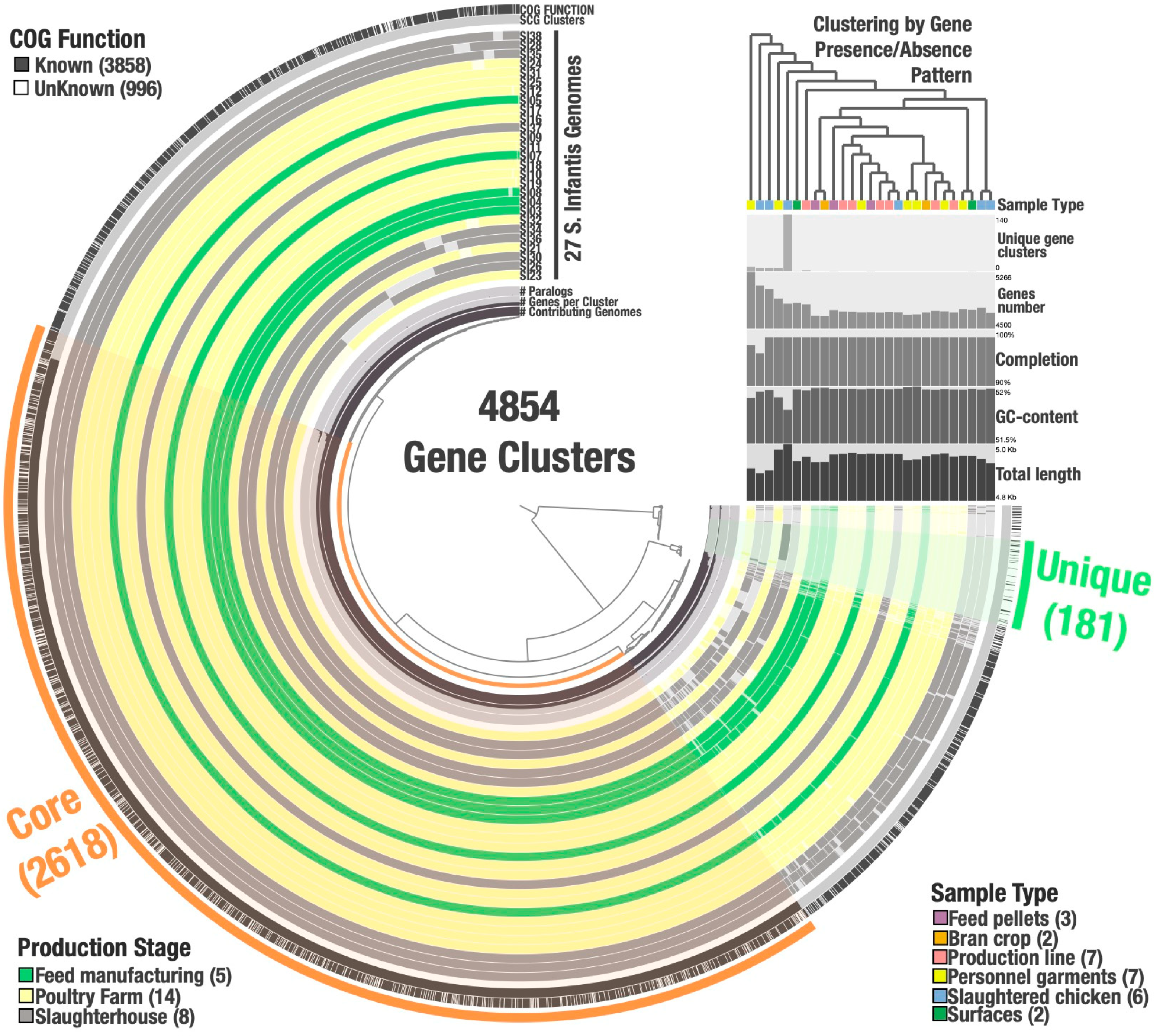
3.6. Salmonella Infantis Pangenome
3.7. In Silico Study of Genetic Determinants of Resistance and Virulence Genes in S. Infantis Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wotzka, S.Y.; Nguyen, B.D.; Hardt, W.D. Salmonella Typhimurium Diarrhea Reveals Basic Principles of Enteropathogen Infection and Disease-Promoted DNA Exchange. Cell Host Microbe. 2017, 21, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Yoon, Y. Etiological Agents Implicated in Foodborne Illness Worldwide. Food Sci. Anim. Resour. 2021, 41, 1. [Google Scholar] [CrossRef]
- Eng, S.K.; Pusparajah, P.; Ab Mutalib, N.S.; Ser, H.L.; Chan, K.G.; Lee, L.H. Salmonella: A Review on Pathogenesis, Epidemiology and Antibiotic Resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef] [Green Version]
- EFSA. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, E05926. [Google Scholar]
- Barrow, P.A.; Jones, M.A.; Smith, A.L.; Wigley, P. The Long View: Salmonella–the Last Forty Years. Avian Pathol. 2012, 41, 413–420. [Google Scholar] [CrossRef]
- Cosby, D.E.; Cox, N.A.; Harrison, M.A.; Wilson, J.L.; Buhr, R.J.; Fedorka-Cray, P.J. Salmonella and Antimicrobial Resistance in Broilers: A Review. J. Appl. Pult. Res. 2015, 24, 408–426. [Google Scholar] [CrossRef]
- Velasquez, C.; Macklin, K.; Kumar, S.; Bailey, M.; Ebner, P.; Oliver, H.; Martin-Gonzalez, F.; Singh, M. Prevalence and Antimicrobial Resistance Patterns of Salmonella Isolated from Poultry Farms in Southeastern United States. Poult. Sci. 2018, 97, 2144–2152. [Google Scholar] [CrossRef]
- Foley, S.L.; Nayak, R.; Hanning, I.B.; Johnson, T.J.; Han, J.; Ricke, S.C. Population Dynamics of Salmonella Enterica Serotypes in Commercial Egg and Poultry Production. Appl. Environ. Microbiol. 2011, 77, 4273–4279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendriksen, R.S.; Vieira, A.R.; Karlsmose, S.; Lo Fo Wong, D.M.; Jensen, A.B.; Wegener, H.C.; Aarestrup, F.M. Global Monitoring of Salmonella Serovar Distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: Results of Quality Assured Laboratories from 2001 to 2007. Foodborne Pathog. Dis. 2011, 8, 887–900. [Google Scholar] [CrossRef] [Green Version]
- Finazzi, G.; Bertasi, B.; Pavoni, E.; Filipello, V.; D’Incau, M.; Losio, M.N. Contamination of Poultry Meat with Salmonella Infantis Should Be Considered a Risk for Food Safety? Eur. J. Public Health 2019, 29, Ckz186-609. [Google Scholar] [CrossRef]
- Lapierre, L.; Cornejo, J.; Zavala, S.; Galarce, N.; Sánchez, F.; Benavides, M.B.; Guzmán, M.; Sáenz, L. Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile. Animals 2020, 10, 1049. [Google Scholar] [CrossRef]
- Lopez Romo, A.; Quirós, R. Appropriate Use of Antibiotics: An Unmet Need. TAU 2019, 11, 1756287219832174. [Google Scholar] [CrossRef] [Green Version]
- Carfora, V.; Alba, P.; Leekitcharoenphon, P.; Ballaro, D.; Cordaro, G.; Di Matteo, P.; Donati, V.; Ianzano, A.; Iurescia, M.; Stravino, F.; et al. Corrigendum: Colistin Resistance Mediated by Mcr-1 in ESBL- Producing, Multidrug Resistant Salmonella Infantis in Broiler Chicken Industry, Italy (2016-2017). Front. Microbiol. 2018, 9, 2395. [Google Scholar] [CrossRef] [Green Version]
- Dionisi, A.M.; Lucarelli, C.; Benedetti, I.; Owczarek, S.; Luzzi, I. Molecular Characterisation of Multidrug-Resistant Salmonella Enterica Serotype Infantis from Humans, Animals and the Environment in Italy. Int. J. Antimicrob. Agents 2011, 38, 384–389. [Google Scholar] [CrossRef]
- Nógrády, N.; Király, M.; Davies, R.; Nagy, B. Multidrug Resistant Clones of Salmonella Infantis of Broiler Origin in Europe. Int. J. Food Microbiol. 2012, 157, 108–112. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The Role of Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Oladeinde, A.; Abdo, Z.; Press, M.O.; Cook, K.; Cox, N.A.; Zwirzitz, B.; Woyda, R.; Lakin, S.; Thomas, J.; Looft, T.; et al. Horizontal Gene Transfer Is the Main Driver of Antimicrobial Resistance in Broiler Chicks Infected with Salmonella Enterica Serovar Heidelberg. Msystems 2021, 6, E00729-21. [Google Scholar] [CrossRef]
- Cohen, E.; Rahav, G.; Gal-Mor, O. Genome Sequence of an Emerging Salmonella Enterica Serovar Infantis and Genomic Comparison with Other S. Infantis Strains. Genome Biol. Evol. 2020, 12, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Lei, C.W.; Zhang, Y.; Kang, Z.Z.; Kong, L.H.; Tang, Y.Z.; Zhang, A.Y.; Yang, X.; Wang, H.N. Vertical Transmission of Salmonella Enteritidis with Heterogeneous Antimicrobial Resistance from Breeding Chickens to Commercial Chickens in China. Vet. Microbiol. 2020, 240, 108538. [Google Scholar] [CrossRef]
- Quino, W.; Hurtado, C.V.; Escalante-Maldonado, O.; Flores-León, D.; Mestanza, O.; Vences-Rosales, F.; Zamudio, M.L.; Gavilán, R.G. Multidrogorresistencia de Salmonella Infantis En Perú: Un Estudio Mediante Secuenciamiento de Nueva Generación. Rev. Peru. Med. Exp. Salud Publica 2019, 36, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Nagy, T.; Szmolka, A.; Wilk, T.; Kiss, J.; Szabó, M.; Pászti, J.; Nagy, B.; Olasz, F. Comparative Genome Analysis of Hungarian and Global Strains of Salmonella Infantis. Front. Microbiol. 2020, 11, 539. [Google Scholar] [CrossRef] [Green Version]
- Tyson, G.H.; Li, C.; Harrison, L.B.; Martin, G.; Hsu, C.H.; Tate, H.; Tran, T.; Strain, E.; Zhao, S. A Multidrug-Resistant Salmonella Infantis Clone Is Spreading and Recombining in the United States. Microb. Drug Res. 2021, 27, 792–799. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2016. EFSA J. 2018, 16, 270. [Google Scholar]
- Aviv, G.; Tsyba, K.; Steck, N.; Salmon-Divon, M.; Cornelius, A.; Rahav, G.; Grassl, G.A.; Gal-Mor, O. A Unique Megaplasmid Contributes to Stress Tolerance and Pathogenicity of an Emergent Salmonella Enterica Serovar Infantis Strain. Environ. Microbiol. 2014, 16, 977–994. [Google Scholar] [CrossRef]
- Olasz, F.; Nagy, T.; Szabó, M.; Kiss, J.; Szmolka, A.; Barta, E.; van Tonder, A.; Thomson, N.; Barrow, P.; Nagy, B. Genome Sequences of Three Salmonella Enterica Subsp. Enterica Serovar Infantis Strains from Healthy Broiler Chicks in Hungary and in the United Kingdom. Genome Announc. 2015, 3, E01468-14. [Google Scholar] [CrossRef] [Green Version]
- Acar, S.; Bulut, E.; Stasiewicz, M.J.; Soyer, Y. Genome Analysis of Antimicrobial Resistance, Virulence, and Plasmid Presence in Turkish Salmonella Serovar Infantis Isolates. Int. J. Food Microbiol. 2019, 307, 108275. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, E0144802. [Google Scholar]
- Nógrády, N.; Toth, A.; Kostyak, A.; Paszti, J.; Nagy, B. Emergence of Multidrug- Resistant Clones of Salmonella Infantis in Broiler Chickens and Humans in Hungary. J. Antimicrob. Chemother. 2007, 60, 645–648. [Google Scholar] [CrossRef] [Green Version]
- Bogomazova, A.N.; Gordeeva, V.D.; Krylova, E.V.; Soltynskaya, I.V.; Davydova, E.E.; Ivanova, O.E.; Komarov, A.A. Mega-Plasmid Found Worldwide Confers Multiple Antimicrobial Resistance in Salmonella Infantis of Broiler Origin in Russia. Int. J. Food Microbiol. 2020, 319, 108497. [Google Scholar] [CrossRef]
- Marín, M.; Holani, R.; Blyth, G.A.; Drouin, D.; Odeón, A.; Cobo, E.R. Human Cathelicidin Improves Colonic Epithelial Defenses against Salmonella Typhimurium by Modulating Bacterial Invasion, TLR4 and pro-Inflammatory Cytokines. Cell Tissue Res. 2019, 376, 433–442. [Google Scholar] [CrossRef]
- Corcoran, M.; Morris, D.; De Lappe, N.; O’connor, J.; Lalor, P.; Dockery, P.; Cormican, M. Commonly Used Disinfectants Fail to Eradicate Salmonella Enterica Biofilms from Food Contact Surface Materials. Appl. Environ. Microbiol. 2014, 80, 1507–1514. [Google Scholar] [CrossRef] [Green Version]
- Hassan, A.R.H.; Salam, H.S.; Abdel-Latef, G.K. Serological Identification and Antimicrobial Resistance of Salmonella Isolates from Broiler Carcasses and Human Stools in Beni-Suef, Egypt. BJBAS 2016, 5, 202–207. [Google Scholar] [CrossRef] [Green Version]
- Jajere, S.M. A Review of Salmonella Enterica with Particular Focus on the Pathogenicity and Virulence Factors, Host Specificity and Antimicrobial Resistance Including Multidrug Resistance. Vet. World 2019, 12, 504. [Google Scholar] [CrossRef] [Green Version]
- Mourão, J.; Rebelo, A.; Ribeiro, S.; Peixe, L.; Novais, C.; Antunes, P. Tolerance to Arsenic Contaminant among Multidrug-resistant and Copper-tolerant Salmonella Successful Clones Is Associated with Diverse Ars Operons and Genetic Contexts. Environ. Microbiol. 2020, 22, 2829–2842. [Google Scholar] [CrossRef]
- Mahnert, A.; Vaishampayan, P.; Probst, A.J.; Auerbach, A.; Moissl-Eichinger, C.; Venkateswaran, K.; Berg, G. Cleanroom Maintenance Significantly Reduces Abundance but Not Diversity of Indoor Microbiomes. PLoS ONE 2015, 10, E0134848. [Google Scholar] [CrossRef]
- Capita, R.; Fernández-Pérez, S.; Buzón-Durán, L.; Alonso-Calleja, C. Effect of Sodium Hypochlorite and Benzalkonium Chloride on the Structural Parameters of the Biofilms Formed by Ten Salmonella Enterica Serotypes. Pathogens 2019, 8, 154. [Google Scholar] [CrossRef] [Green Version]
- Mejía, L.; Medina, J.L.; Bayas, R.; Salazar, C.S.; Villavicencio, F.; Zapata, S.; Vinueza-Burgos, C. Genomic Epidemiology of Salmonella Infantis in Ecuador: From Poultry Farms to Human Infections. Front. Vet. Sci 2020, 7, 691. [Google Scholar] [CrossRef]
- Roy, P.K.; Ha, A.J.W.; Mizan, M.F.R.; Hossain, M.I.; Ashrafudoulla, M.; Toushik, S.H.; Nahar, S.; Kim, Y.K.; Ha, S.D. Effects of Environmental Conditions (Temperature, PH, and Glucose) on Biofilm Formation of Salmonella Enterica Serotype Kentucky and Virulence Gene Expression. Poult. Sci. 2021, 100, 101209. [Google Scholar] [CrossRef]
- Zeng, X.; Lv, S.; Qu, C.; Lan, L.; Tan, D.; Li, X.; Bai, L. Serotypes, Antibiotic Resistance, and Molecular Characterization of Non-Typhoidal Salmonella Isolated from Diarrheic Patients in Guangxi Zhuang Autonomous Region, China, 2014–2017. Food Control. 2021, 120, 107478. [Google Scholar] [CrossRef]
- Instituto de Salud Pública de Chile (ISP). Boletín de Vigilancia de Laboratorio. 2019. Available online: http://Www.Ispch.Cl/Boletines (accessed on 1 October 2020).
- Cadena, M.; Kelman, T.; Marco, M.L.; Pitesky, M. Understanding Antimicrobial Resistance (AMR) Profiles of Salmonella Biofilm and Planktonic Bacteria Challenged with Disinfectants Commonly Used during Poultry Processing. Foods 2019, 8, 275. [Google Scholar] [CrossRef] [Green Version]
- Aviv, G.; Rahav, G.; Gal-Mor, O. Horizontal Transfer of the Salmonella Enterica Serovar Infantis Resistance and Virulence Plasmid PESI to the Gut Microbiota of Warm-Blooded Hosts. MBio 2016, 7, E01395-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kürekci, C.; Sahin, S.; Iwan, E.; Kwit, R.; Bomba, A.; Wasyl, D. Whole-Genome Sequence Analysis of Salmonella Infantis Isolated from Raw Chicken Meat Samples and Insights into PESI-like Megaplasmid. Int. J. Food Microbiol. 2021, 337, 108956. [Google Scholar] [CrossRef]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas Immune System: Biology, Mechanisms and Applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef]
- Alba, P.; Leekitcharoenphon, P.; Carfora, V.; Amoruso, R.; Cordaro, G.; Di Matteo, P.; Engage-Eurl-Ar Network Study Group. Molecular Epidemiology of Salmonella Infantis in Europe: Insights into the Success of the Bacterial Host and Its Parasitic PESI-like Megaplasmid. Microb. Genom. 2020, 6, e000365. [Google Scholar] [CrossRef]
- Andrews, S. FastQC a Quality-Control Tool for High-Throughput Sequence Data. 2010. Available online: http://www.bioinformaticsbabraham.ac.uk/projects/fastqc (accessed on 28 July 2021).
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by EggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.; Letunic, I.; Rattei, T.; Jense, L.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Zdobnov, E. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Zdobnov, E.M.; Tegenfeldt, F.; Kuznetsov, D.; Waterhouse, R.M.; Simao, F.A.; Ioannidis, P.; Seppey, M.; Loetscher, A.; Kriventseva, E.V. OrthoDB v9. 1: Cataloging Evolutionary and Functional Annotations for Animal, Fungal, Plant, Archaeal, Bacterial and Viral Orthologs. Nucleic Acids Res. 2017, 45, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Kaiser, B.L.D.; Dinsmore, B.A.; Deng, X. Salmonella Serotype Determination Utilizing High-Throughput Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Mlst Github. Available online: https://Github.Com/Tseemann/Mlst (accessed on 20 October 2021).
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable Analysis of Bacterial Genome Variation at the Population Level. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 6 July 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis 2016; Springer: New York, NY, USA, 2016. [Google Scholar]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Broder, A.Z. On the Resemblance and Containment of Documents. In Proceedings of the Compression and Complexity of SEQUENCES 1997 (Cat. No. 97TB100171), Salerno, Italy, 13 June 1997. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.8 2015. Available online: https://cran.r-project.org/package=pheatmap (accessed on 10 August 2019).
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads. 2015. Available online: Https://Github.Com/Tseemann/Snippy (accessed on 22 October 2021).
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-Sites: Rapid Efficient Extraction of SNPs from Multi-FASTA Alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Leaché, A.D.; Banbury, B.L.; Felsenstein, J.; De Oca, A.N.M.; Stamatakis, A. Short Tree, Long Tree, Right Tree, Wrong Tree: New Acquisition Bias Corrections for Inferring SNP Phylogenies. Syst. Biol. 2015, 64, 1032–1047. [Google Scholar] [CrossRef] [Green Version]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-Led, Integrated, Reproducible Multi-Omics with Anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.O.; Eren, A.M. Linking Pangenomes and Metagenomes: The Prochlorococcus Metapangenome. PeerJ 2018, 6, E4320. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA Homology Search with Profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankavaram, U.T.; Rao, B.S.; Kiryuntin, B.; Galperin, M.Y.; Fedorova, N.D.; Koonin, E.V. The COG Database: New Developments in Phylogenetic Classification of Proteins from Complete Genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Benedict, M.N.; Henriksen, J.R.; Metcalf, W.W.; Whitaker, R.J.; Price, N.D. ITEP: An Integrated Toolkit for Exploration of Microbial Pan-Genomes. BMC Genom. 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dongen, S.; Abreu-Goodger, C. Using MCL to Extract Clusters from Networks. In Bacterial Molecular Networks; Springer: New York, NY, USA, 2012; pp. 281–295. [Google Scholar]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antibiotic Resistance Genes. 2020. Available online: https://Github.Com/Tseemann/Abricate (accessed on 19 July 2021).
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. AAC 2014, 58, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33 (Suppl. 1), D325–D328. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Hasman, H. PlasmidFinder and PMLST: In Silico Detection and Typing of Plasmids. ACC 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lynne, A.M.; David, D.E.; Tang, H.; Xu, J.; Nayak, R.; Kaldhone, P.; Logue, C.; Foley, S.L. DNA Sequence Analysis of Plasmids from Multidrug Resistant Salmonella Enterica Serotype Heidelberg Isolates. PLoS ONE 2012, 7, E51160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khajanchi, B.K.; Hasan, N.A.; Choi, S.Y.; Han, J.; Zhao, S.; Colwell, R.R.; Cerniglia, C.; Foley, S.L. Comparative Genomic Analysis and Characterization of Incompatibility Group FIB Plasmid Encoded Virulence Factors of Salmonella Enterica Isolated from Food Sources. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srikanth, C.V.; Mercado-Lubo, R.; Hallstrom, K.; McCormick, B.A. Salmonella Effector Proteins and Host-Cell Responses. Cell Mol. Life Sci. 2011, 68, 3687–3697. [Google Scholar] [CrossRef]
- Hao, L.Y.; Willis, D.K.; Andrews-Polymenis, H.; McClelland, M.; Barak, J.D. Requirement of Siderophore Biosynthesis for Plant Colonization by Salmonella Enterica. Appl. Environ. Microbiol. 2012, 78, 4561–4570. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Fujishiro, T.; Huang, G.; Koch, J.; Takabayashi, A.; Yokono, M.; Tanaka, A.; Xu, T.; Hu, X.; Ermler, U.; et al. Towards Artificial Methanogenesis: Biosynthesis of the [Fe]-Hydrogenase Cofactor and Characterization of the Semi-Synthetic Hydrogenase. Faraday Discuss. 2017, 198, 37–58. [Google Scholar] [CrossRef]
- Pate, M.; Mičunovič, J.; Golob, M.; Vestby, L.K.; Ocepek, M. Salmonella Infantis in Broiler Flocks in Slovenia: The Prevalence of Multidrug Resistant Strains with High Genetic Homogeneity and Low Biofilm-Forming Ability. BioMed Res. Int. 2019, 2019, 4981463. [Google Scholar] [CrossRef] [Green Version]
- Kingsbury, J.M.; Thom, K.; Erskine, H.; Olsen, L.; Soboleva, T. Prevalence and Genetic Analysis of Salmonella Enterica from a Cross-Sectional Survey of the New Zealand Egg Production Environment. J. Food Prot. 2019, 82, 2201–2214. [Google Scholar] [CrossRef]
- Gymoese, P.; Kiil, K.; Torpdahl, M.; Østerlund, M.T.; Sørensen, G.; Olsen, J.E.; Nielsen, E. WGS Based Study of the Population Structure of Salmonella Enterica Serovar Infantis. BMC Genom. 2019, 20, 1–11. [Google Scholar] [CrossRef]
- Abbasoglu, D.; Akcelık, M. Phenotypic and Genetic Characterization of Multidrug-Resistant Salmonella Infantis Strains Isolated from Broiler Chicken Meats in Turkey. Biologia 2011, 66, 406–410. [Google Scholar] [CrossRef]
- Rahmani, M.; Peighambari, S.M.; Svendsen, C.A.; Cavaco, L.M.; Agersø, Y.; Hendriksen, R.S. Molecular Clonality and Antimicrobial Resistance in Salmonella Enterica Serovars Enteritidis and Infantis from Broilers in Three Northern Regions of Iran. BMC Vet. Res. 2013, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Asai, T.; Ishihara, K.; Harada, K.; Kojima, A.; Tamura, Y.; Sato, S.; Takahashi, T. Long-term Prevalence of Antimicrobial-resistant Salmonella Enterica Subspecies Enterica Serovar Infantis in the Broiler Chicken Industry in Japan. Microbiol. Immunol. 2007, 51, 111–115. [Google Scholar] [CrossRef]
- Medeiros, M.A.N.; de Oliveira, D.C.N.; Rodrigues, D.D.P.; de Freitas, D.R.C. Prevalence and Antimicrobial Resistance of Salmonella in Chicken Carcasses at Retail in 15 Brazilian Cities. Rev. Panam. Salud Publica 2011, 30, 555–560. [Google Scholar] [CrossRef]
- Vallejos-Sánchez, K.; Tataje-Lavanda, L.; Villanueva-Pérez, D.; Bendezú, J.; Montalván, Á.; Zimic-Peralta, M.; Fernández-Sáchez, M.; Fernández-Díaz, M. Whole-Genome Sequencing of a Salmonella Enterica Subsp. Enterica Serovar Infantis Strain Isolated from Broiler Chicken in Peru. Microbiol. Resour. Announc. 2019, 8, E00826-19. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abebe, W.; Adesiyun, A.A. Characterization of Salmonella Isolates Recovered from Stages of the Processing Lines at Four Broiler Processing Plants in Trinidad and Tobago. Microorganisms 2021, 9, 1048. [Google Scholar] [CrossRef]
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic Resistance in Bacteria Associated with Food Animals: A United States Perspective of Livestock Production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef] [Green Version]
- Gyles, C.L. Antimicrobial Resistance in Selected Bacteria from Poultry. Anim. Health Res. Rev. 2008, 9, 149–158. [Google Scholar] [CrossRef]
- McDermott, P.F.; Zhao, S.; Tate, H. Antimicrobial Resistance in Nontyphoidal Salmonella. Microbiol. Spectr. 2018, 6, 780–790. [Google Scholar] [CrossRef] [Green Version]
- Douarre, P.E.; Mallet, L.; Radomski, N.; Felten, A.; Mistou, M.Y. Analysis of COMPASS, a New Comprehensive Plasmid Database Revealed Prevalence of Multi- Replicon and Extensive Diversity of IncF Plasmids. Front. Microbiol. 2020, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kondo, K.; Kawano, M.; Sugai, M. Distribution of Antimicrobial Resistance and Virulence Genes within the Prophage- Associated Regions in Nosocomial Pathogens. MSphere 2021, 6, E00452-21. [Google Scholar] [CrossRef]
- Mansour, M.N.; Yaghi, J.; El Khoury, A.; Felten, A.; Mistou, M.Y.; Atoui, A.; Radomski, N. Prediction of Salmonella Serovars Isolated from Clinical and Food Matrices in Lebanon and Genomic-Based Investigation Focusing on Enteritidis Serovar. Int. J. Food Microbiol. 2020, 333, 108831. [Google Scholar] [CrossRef]
- Smorawinska, M.; Szuplewska, M.; Zaleski, P.; Wawrzyniak, P.; Maj, A.; Plucienniczak, A.; Bartosik, D. Mobilizable Narrow Host Range Plasmids as Natural Suicide Vectors Enabling Horizontal Gene Transfer among Distantly Related Bacterial Species. FEMS Microbiol. Lett. 2012, 326, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Leyer, G.; Johnson, A. Acid Adaptation Sensitizes Salmonella Typhimurium to Hypochlorous Acid. AEM 1997, 63, 461–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urano, H.; Fukuzaki, S. The Mode of Action of Sodium Hypochlorite in the Cleaning Process. Biocontrol. Sci. 2005, 10, 21–29. [Google Scholar] [CrossRef]
- Ramirez-Hernandez, A.; Carrascal-Camacho, A.K.; Varón-García, A.; Brashears, M.M.; Sanchez-Plata, M.X. Genotypic Characterization of Antimicrobial Resistant Salmonella Spp. Strains from Three Poultry Processing Plants in Colombia. Foods 2021, 10, 491. [Google Scholar] [CrossRef]
- Borges, K.A.; Furian, T.Q.; Borsoi, A.; Moraes, H.L.; Salle, C.T.; Nascimento, V.P. Detection of Virulence-Associated Genes in Salmonella Enteritidis Isolates from Chicken in South of Brazil. Pesqui. Vet. Bras. 2013, 33, 1416–1422. [Google Scholar] [CrossRef]
- Almeida, F.; Pitondo-Silva, A.; Oliveira, M.A.; Falcão, J.P. Molecular Epidemiology and Virulence Markers of Salmonella Infantis Isolated over 25 Years in São Paulo State, Brazil. Infect. Genet. Evol. 2013, 19, 145–151. [Google Scholar]
- Sever, N.K.; Akan, M. Molecular Analysis of Virulence Genes of Salmonella Infantis Isolated from Chickens and Turkeys. Microb. Pathog. 2019, 126, 199–204. [Google Scholar] [CrossRef]
- Joseph, B.; Otta, S.K.; Karunasagar, I.; Karunasagar, I. Biofilm Formation by Salmonella Spp. on Food Contact Surfaces and Their Sensitivity to Sanitizers. Int. J. Food Microbiol. 2001, 64, 367–372. [Google Scholar] [CrossRef]
- Ashrafudoulla, M.; Na, K.W.; Byun, K.H.; Kim, D.H.; Yoon, J.W.; Mizan, M.F.R.; Kang, I. Isolation and Characterization of Salmonella Spp. from Food and Food Contact Surfaces in a Chicken Processing Factory. Poult. Sci. J. 2021, 100, 101234. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Hafeez, M.A.; Ahmad, R.; Mahmood, S. CRISPR-Cas System in Regulation of Immunity and Virulence of Bacterial Pathogens. Gene Rep. 2018, 13, 151–157. [Google Scholar] [CrossRef]
- Nussenzweig, P.M.; Marraffini, L.A. Molecular Mechanisms of CRISPR-Cas Immunity in Bacteria. Annu. Rev. Genet. 2020, 54, 93–120. [Google Scholar] [CrossRef] [PubMed]

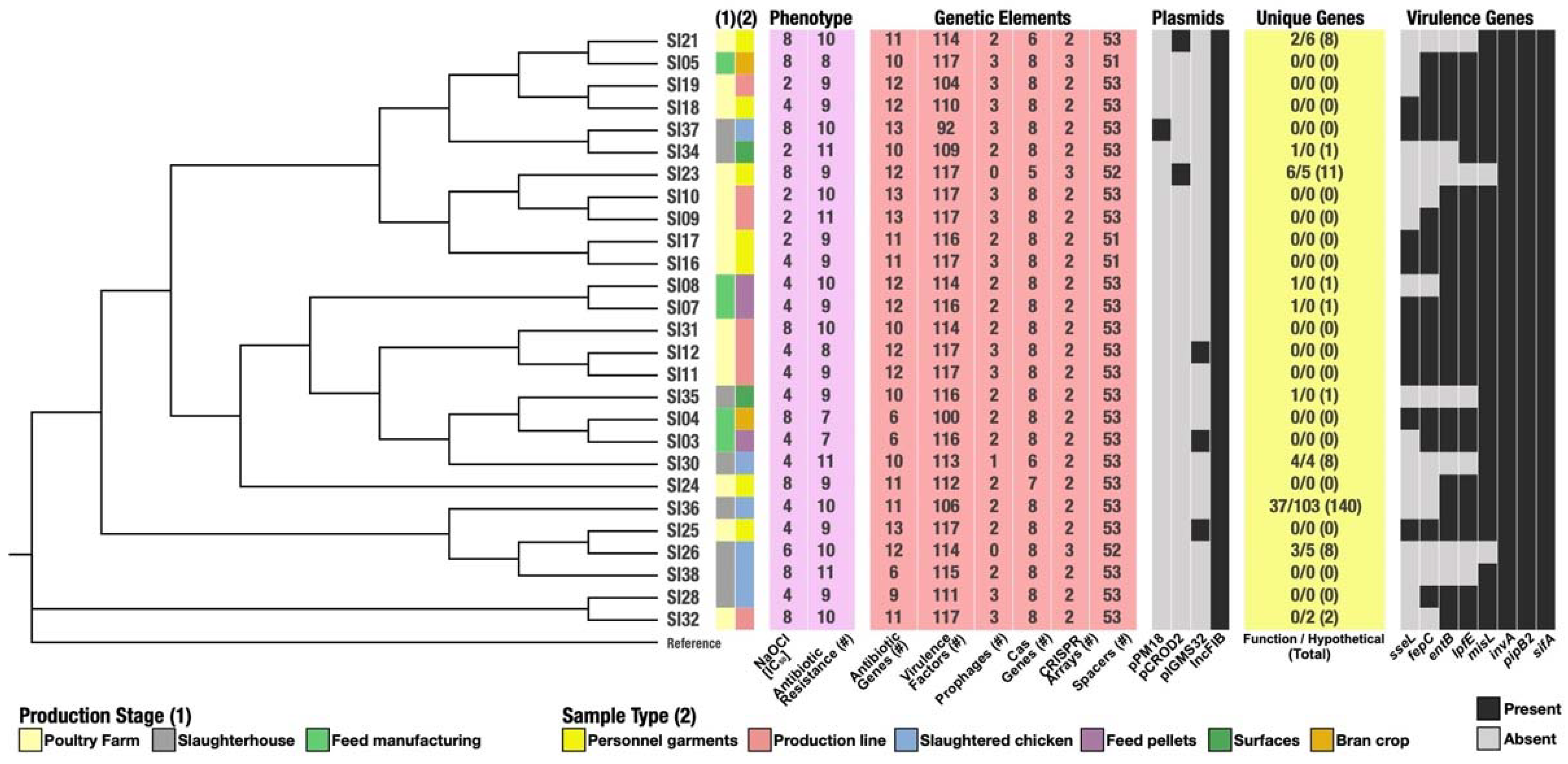
| Strain | Production Stage | Sample Type |
|---|---|---|
| SI01 | Feed manufacturing | Feed pellets |
| SI02 | Feed manufacturing | Feed pellets |
| SI03 | Feed manufacturing | Feed pellets |
| SI07 | Feed manufacturing | Feed pellets |
| SI08 | Feed manufacturing | Feed pellets |
| SI04 | Feed manufacturing | Bran crop |
| SI05 | Feed manufacturing | Bran crop |
| SI09 | Poultry Farm | Production line swab |
| SI10 | Poultry Farm | Production line swab |
| SI11 | Poultry Farm | Production line washing |
| SI12 | Poultry Farm | Production line washing |
| SI19 | Poultry Farm | Production line swab |
| SI20 | Poultry Farm | Production line washing |
| SI31 | Poultry Farm | Production line washing |
| SI32 | Poultry Farm | Production line washing |
| SI16 | Poultry Farm | Personnel garments swab |
| SI17 | Poultry Farm | Personnel garments swab |
| SI18 | Poultry Farm | Personnel garments swab |
| SI21 | Poultry Farm | Personnel garments swab |
| SI23 | Poultry Farm | Personnel garments swab |
| SI24 | Poultry Farm | Personnel garments swab |
| SI25 | Poultry Farm | Personnel garments swab |
| SI26 | Slaughterhouse | Slaughtered chicken cecum |
| SI28 | Slaughterhouse | Slaughtered chicken cecum |
| SI30 | Slaughterhouse | Slaughtered chicken cecum |
| SI36 | Slaughterhouse | Slaughtered chicken entrails |
| SI37 | Slaughterhouse | Slaughtered chicken entrails |
| SI38 | Slaughterhouse | Slaughtered chicken breast |
| SI34 | Slaughterhouse | Surfaces washing |
| SI35 | Slaughterhouse | Surfaces washing |
| SI39 | Slaughterhouse | Surfaces washing |
| SI40 | Slaughterhouse | Surfaces washing |
| SI41 | Slaughterhouse | Surfaces washing |
| SI42 | Slaughterhouse | Surfaces washing |
| SI43 | Slaughterhouse | Surfaces washing |
| SI44 | Slaughterhouse | Surfaces washing |
| SI45 | Slaughterhouse | Surfaces washing |
| SI46 | Slaughterhouse | Surfaces washing |
| SI47 | Slaughterhouse | Surfaces washing |
| SI48 | Slaughterhouse | Surfaces washing |
| SI49 | Slaughterhouse | Surfaces washing |
| Sample | Strain | Size (mb) | GC (%) | # Contigs | N50 | % Completion | Genome Cov. | |
|---|---|---|---|---|---|---|---|---|
| Feed manufacturing | Feed pellets | SI01 | 4.96 | 52.15 | 48 | 333,150 | 88 | 48X |
| SI02 | 4.95 | 52.14 | 153 | 56,434 | 77.4 | 37X | ||
| SI03 | 4.95 | 52.15 | 65 | 194,600 | 98.4 | 34X | ||
| SI07 | 4.98 | 52.14 | 70 | 148,284 | 99.2 | 32X | ||
| SI08 | 4.99 | 52.14 | 49 | 310,053 | 100 | 44X | ||
| Bran crop | SI04 | 4.95 | 52.15 | 55 | 204,015 | 99.2 | 63X | |
| SI05 | 4.98 | 52.14 | 54 | 333,144 | 99.2 | 63X | ||
| Poultry Farm | Production line | SI09 | 4.98 | 52.13 | 65 | 194,600 | 99.2 | 30X |
| SI10 | 4.99 | 52.14 | 47 | 245,770 | 98.4 | 44X | ||
| SI11 | 4.99 | 52.14 | 46 | 245,770 | 98.4 | 44X | ||
| SI12 | 4.98 | 52.14 | 55 | 245,776 | 99.2 | 67X | ||
| SI19 | 4.99 | 52.14 | 49 | 333,144 | 98.4 | 41X | ||
| SI31 | 4.97 | 52.14 | 77 | 161,152 | 100 | 39X | ||
| SI32 | 4.99 | 52.14 | 52 | 333,144 | 98.4 | 58X | ||
| Personnelgarments swab | SI16 | 4.96 | 52.16 | 64 | 204,015 | 98.4 | 40X | |
| SI17 | 4.97 | 52.17 | 55 | 333,144 | 99.2 | 105X | ||
| SI18 | 4.99 | 52.14 | 53 | 333,574 | 100 | 82X | ||
| SI21 | 5.02 | 52.07 | 51 | 310,053 | 95.2 | 69X | ||
| SI23 | 5.02 | 52.06 | 75 | 181,952 | 92.7 | 66X | ||
| SI24 | 4.99 | 52.14 | 44 | 333,144 | 100 | 51X | ||
| SI25 | 4.99 | 52.14 | 58 | 333,574 | 99.2 | 89X | ||
| Slaughterhouse | Slaughtered chicken | SI26 | 4.99 | 52.14 | 49 | 333,145 | 92.7 | 52X |
| SI28 | 4.98 | 52.15 | 53 | 217,253 | 98.4 | 60X | ||
| SI30 | 4.99 | 52.14 | 46 | 333,443 | 92.7 | 55X | ||
| SI36 | 5.03 | 51.9 | 50 | 333,145 | 100 | 73X | ||
| SI37 | 4.99 | 52.14 | 68 | 201,754 | 98.4 | 28X | ||
| SI38 | 4.96 | 52.14 | 47 | 333,144 | 100 | 43X | ||
| Surfaces | SI34 | 4.98 | 52.15 | 44 | 333,144 | 98.4 | 39X | |
| SI35 | 4.98 | 52.14 | 45 | 333,144 | 100 | 39X | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardo-Esté, C.; Lorca, D.; Castro-Severyn, J.; Krüger, G.; Alvarez-Thon, L.; Zepeda, P.; Sulbaran-Bracho, Y.; Hidalgo, A.; Tello, M.; Molina, F.; et al. Genetic Characterization of Salmonella Infantis with Multiple Drug Resistance Profiles Isolated from a Poultry-Farm in Chile. Microorganisms 2021, 9, 2370. https://doi.org/10.3390/microorganisms9112370
Pardo-Esté C, Lorca D, Castro-Severyn J, Krüger G, Alvarez-Thon L, Zepeda P, Sulbaran-Bracho Y, Hidalgo A, Tello M, Molina F, et al. Genetic Characterization of Salmonella Infantis with Multiple Drug Resistance Profiles Isolated from a Poultry-Farm in Chile. Microorganisms. 2021; 9(11):2370. https://doi.org/10.3390/microorganisms9112370
Chicago/Turabian StylePardo-Esté, Coral, Diego Lorca, Juan Castro-Severyn, Gabriel Krüger, Luis Alvarez-Thon, Phillippi Zepeda, Yoelvis Sulbaran-Bracho, Alejandro Hidalgo, Mario Tello, Franck Molina, and et al. 2021. "Genetic Characterization of Salmonella Infantis with Multiple Drug Resistance Profiles Isolated from a Poultry-Farm in Chile" Microorganisms 9, no. 11: 2370. https://doi.org/10.3390/microorganisms9112370







