Production of Hydroxycarboxylic Acid Receptor 3 (HCA3) Ligands by Bifidobacterium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacterial Strains
2.3. CSs
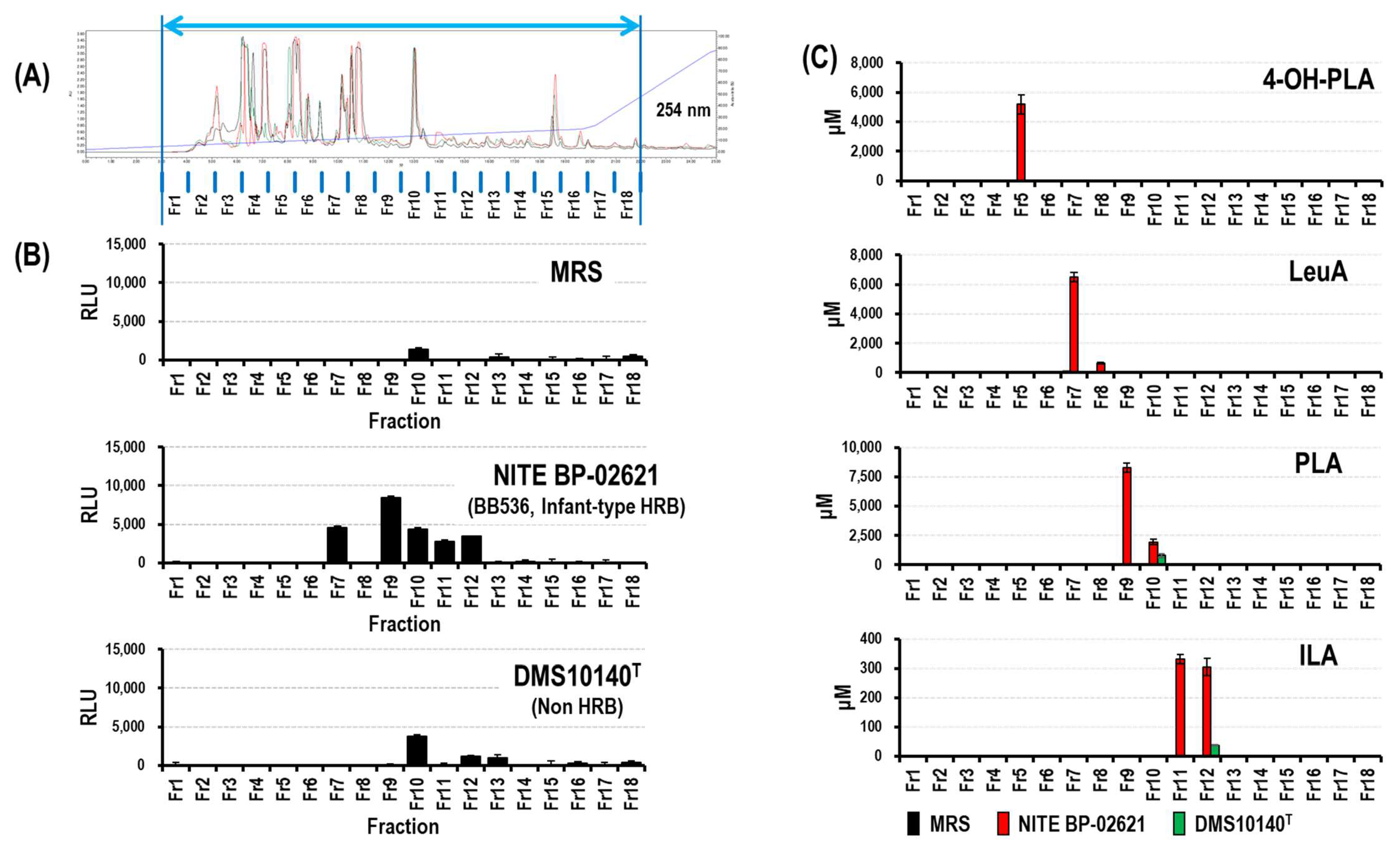
2.4. Quantification of Metabolite Concentrations
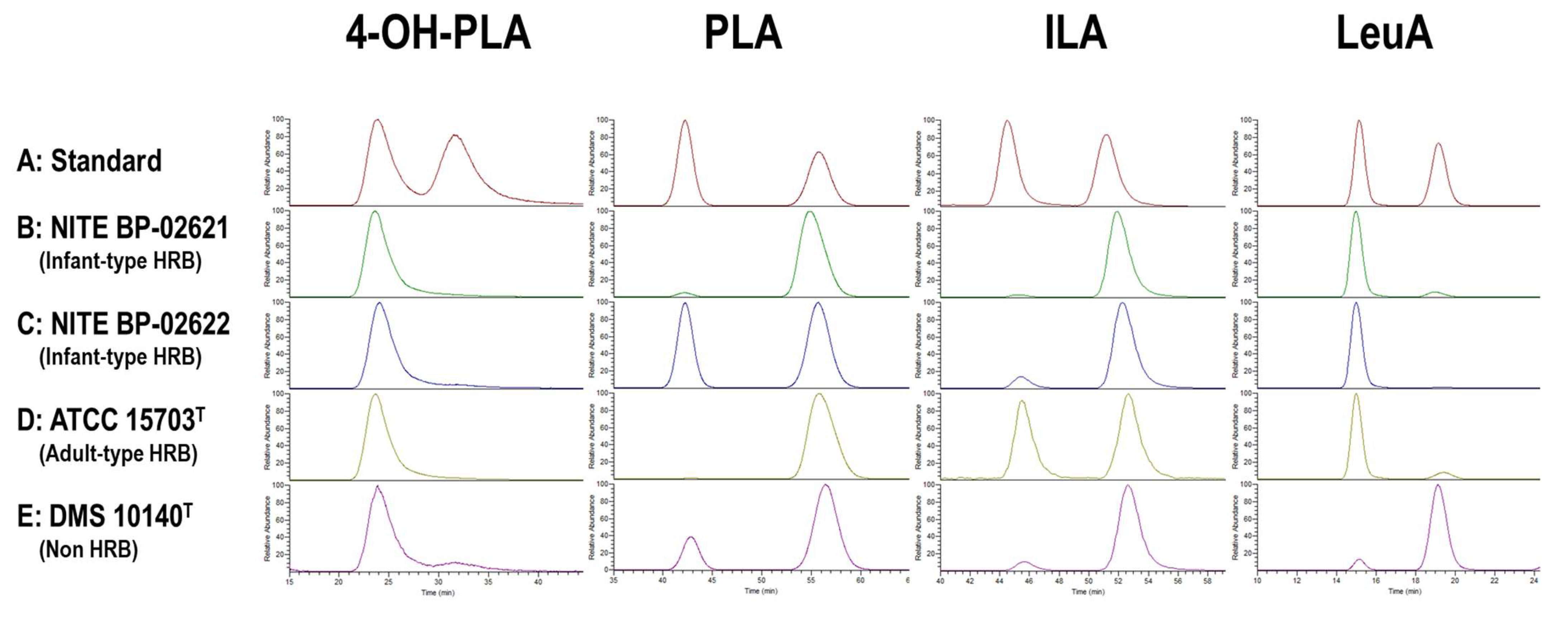
2.5. Analysis of the Optical Isomer of Aryl-LAs Using Chiral HPLC
2.6. Preparative LC of MRS and CSs
2.7. HCA3 Ligand Assay
2.8. Aryl Hydrocarbon Receptor (AhR) Ligand Assay
2.9. Construction of Insertional Mutant
3. Results
3.1. HCA3 Ligand Activity and Aryl-LAs Concentration of CSs of Bifidobacterium Strains
3.1.1. Identification of Novel HCA3 Ligand in CSs of Bifidobacterium Strains (Fraction 7)
3.1.2. Production of Aryl-LAs and LeuA by Bifidobacterium Strains
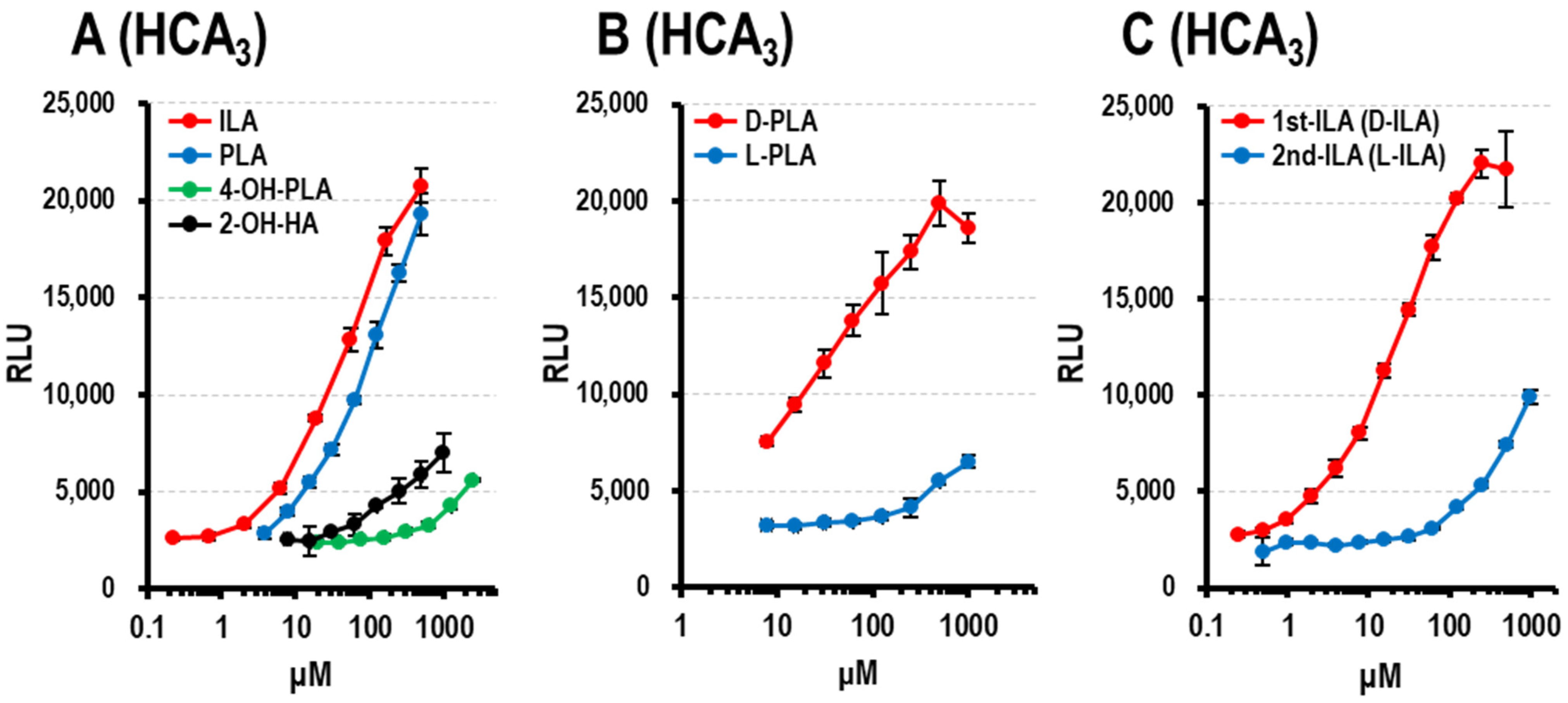
3.1.3. Optical Activity of Aryl-LAs and LeuA in CSs of Bifidobacterium Strains
3.1.4. HCA3 Ligand Activity of Aryl-LAs and Enantiomers of PLA, ILA, and LeuA
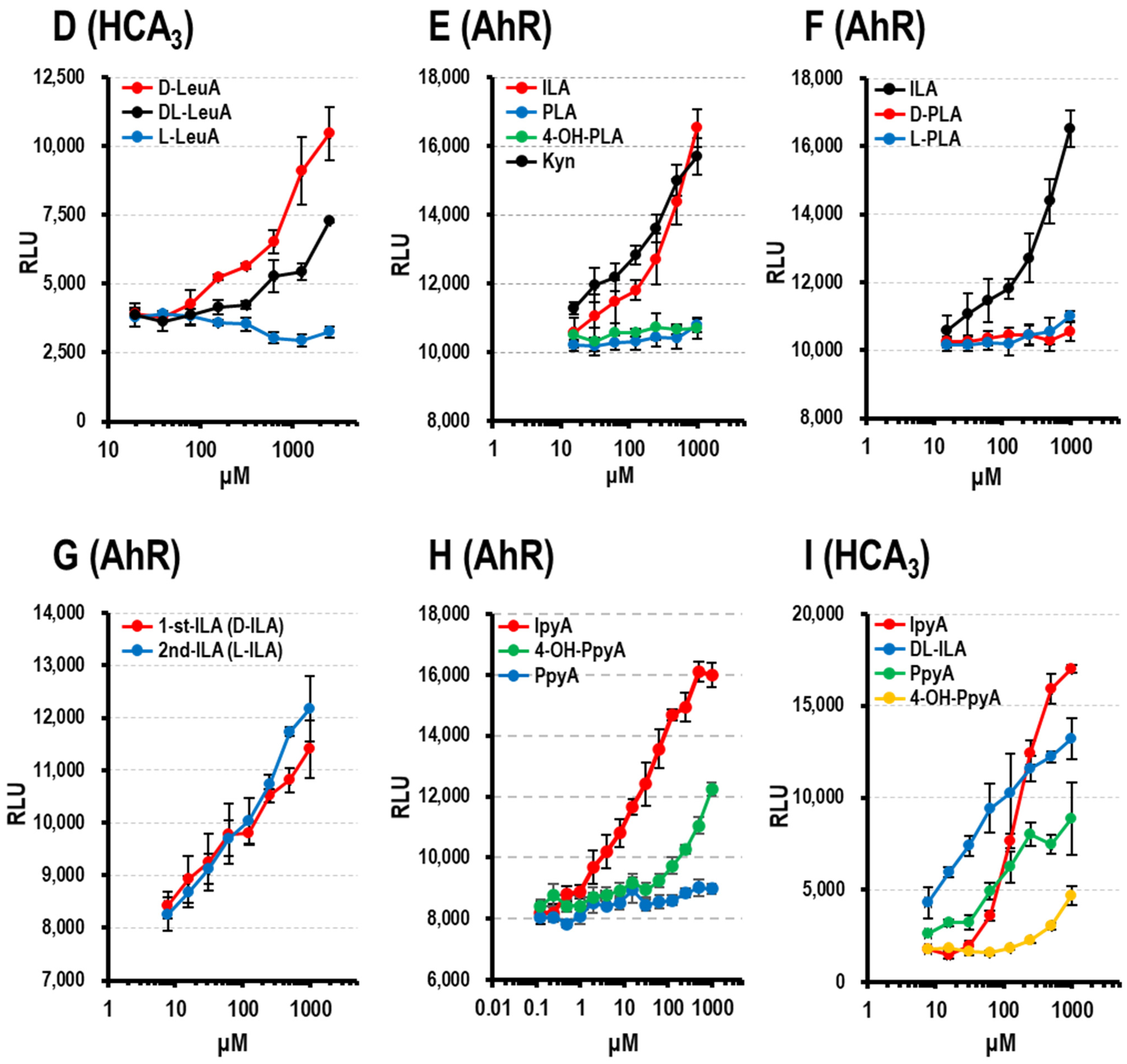
3.1.5. AhR Ligand Activity of Aryl-LAs
3.1.6. AhR and HCA3 Ligand Activity of Aryl-pyr-As
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, K. Biological roles and therapeutic potential of hydroxy-carboxylic Acid receptors. Front. Endocrinol. 2011, 2, 51. [Google Scholar] [CrossRef] [Green Version]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [Green Version]
- Peters, A.; Krumbholz, P.; Jäger, E.; Heintz-Buschart, A.; Çakir, M.V.; Rothemund, S.; Gaudl, A.; Ceglarek, U.; Schöneberg, T.; Stäubert, C. Metabolites of lactic acid bacteria present in fermented foods are highly potent agonists of human hydroxycarboxylic acid receptor 3. PLoS Genet. 2019, 15, e1008145. [Google Scholar] [CrossRef] [Green Version]
- Offermanns, S.; Colletti, S.L.; Lovenberg, T.W.; Semple, G.; Wise, A.; AP, I.J. International Union of Basic and Clinical Pharmacology. LXXXII: Nomenclature and Classification of Hydroxy-carboxylic Acid Receptors (GPR81, GPR109A, and GPR109B). Pharmacol. Rev. 2011, 63, 269–290. [Google Scholar] [CrossRef] [Green Version]
- Zellner, C.; Pullinger, C.R.; Aouizerat, B.E.; Frost, P.H.; Kwok, P.Y.; Malloy, M.J.; Kane, J.P. Variations in human HM74 (GPR109B) and HM74A (GPR109A) niacin receptors. Hum. Mutat. 2005, 25, 18–21. [Google Scholar] [CrossRef]
- Ahmed, K.; Tunaru, S.; Offermanns, S. GPR109A, GPR109B and GPR81, a family of hydroxy-carboxylic acid receptors. Trends Pharmacol. Sci. 2009, 30, 557–562. [Google Scholar] [CrossRef]
- Taggart, A.K.; Kero, J.; Gan, X.; Cai, T.Q.; Cheng, K.; Ippolito, M.; Ren, N.; Kaplan, R.; Wu, K.; Wu, T.J.; et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J. Biol. Chem. 2005, 280, 26649–26652. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, K.; Tunaru, S.; Langhans, C.D.; Hanson, J.; Michalski, C.W.; Kölker, S.; Jones, P.M.; Okun, J.G.; Offermanns, S. Deorphanization of GPR109B as a receptor for the beta-oxidation intermediate 3-OH-octanoic acid and its role in the regulation of lipolysis. J. Biol. Chem. 2009, 284, 21928–21933. [Google Scholar] [CrossRef] [Green Version]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Breastmilk-promoted bifidobacteria produce aromatic amino acids in the infant gut. Nature Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Wong, C.B.; Odamaki, T.; Xiao, J.Z. Insights into the reason of Human-Residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol. Rev. 2020, 44, 369–385. [Google Scholar] [CrossRef]
- Liu, S.; Fang, Z.; Wang, H.; Zhai, Q.; Hang, F.; Zhao, J.; Zhang, H.; Lu, W.; Chen, W. Gene-Phenotype Associations Involving Human-Residential Bifidobacteria (HRB) Reveal Significant Species- and Strain-Specificity in Carbohydrate Catabolism. Microorganisms 2021, 9, 883. [Google Scholar] [CrossRef]
- Odamaki, T.; Horigome, A.; Sugahara, H.; Hashikura, N.; Minami, J.; Xiao, J.Z.; Abe, F. Comparative Genomics Revealed Genetic Diversity and Species/Strain-Level Differences in Carbohydrate Metabolism of Three Probiotic Bifidobacterial Species. Int. J. Genom. 2015, 2015, 567809. [Google Scholar] [CrossRef] [Green Version]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef]
- Priyadarshi, A.; Lowe, G.; Saddi, V.; Trivedi, A.; Luig, M.; Tracy, M. Clinical Outcomes of Single vs. Two-Strain Probiotic Prophylaxis for Prevention of Necrotizing Enterocolitis in Preterm Infants. Front. Pediatr. 2021, 9, 729535. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.A.; Meng, D. Breast Milk and Microbiota in the Premature Gut: A Method of Preventing Necrotizing Enterocolitis. Nestle Nutr. Inst. Workshop Ser. 2020, 94, 103–112. [Google Scholar] [CrossRef]
- Meng, D.; Sommella, E.; Salviati, E.; Campiglia, P.; Ganguli, K.; Djebali, K.; Zhu, W.; Walker, W.A. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr. Res. 2020, 88, 209–217. [Google Scholar] [CrossRef]
- Ehrlich, A.M.; Pacheco, A.R.; Henrick, B.M.; Taft, D.; Xu, G.; Huda, M.N.; Mishchuk, D.; Goodson, M.L.; Slupsky, C.; Barile, D.; et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020, 20, 357. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 1415–1419. [Google Scholar] [CrossRef]
- Sakurai, T.; Odamaki, T.; Xiao, J.Z. Production of Indole-3-Lactic Acid by Bifidobacterium Strains Isolated from Human Infants. Microorganisms 2019, 7, 340. [Google Scholar] [CrossRef] [Green Version]
- Matelska, D.; Shabalin, I.G.; Jabłońska, J.; Domagalski, M.J.; Kutner, J.; Ginalski, K.; Minor, W. Classification, substrate specificity and structural features of D-2-hydroxyacid dehydrogenases: 2HADH knowledgebase. BMC Evol. Biol. 2018, 18, 199. [Google Scholar] [CrossRef]
- Sakanaka, M.; Hansen, M.E.; Gotoh, A.; Katoh, T.; Yoshida, K.; Odamaki, T.; Yachi, H.; Sugiyama, Y.; Kurihara, S.; Hirose, J.; et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv. 2019, 5, eaaw7696. [Google Scholar] [CrossRef] [Green Version]
- Yasui, K.; Kano, Y.; Tanaka, K.; Watanabe, K.; Shimizu-Kadota, M.; Yoshikawa, H.; Suzuki, T. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 2009, 37, e3. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Di Luccia, B.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8αα(+) T cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Lu, C.; Fan, D.; Lu, X.; Xia, Y.; Zhao, H.; Xu, H.; Zhu, Y.; Li, J.; Liu, H.; et al. Human Umbilical Mesenchymal Stem Cells Display Therapeutic Potential in Rheumatoid Arthritis by Regulating Interactions Between Immunity and Gut Microbiota via the Aryl Hydrocarbon Receptor. Front. Cell Dev. Biol. 2020, 8, 131. [Google Scholar] [CrossRef]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Sadik, A.; Somarribas Patterson, L.F.; Öztürk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfänder, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gan, M.; Li, J.; Li, H.; Su, M.; Tan, D.; Wang, S.; Jia, M.; Zhang, L.; Chen, G. Endogenous Indole Pyruvate Pathway for Tryptophan Metabolism Mediated by IL4I1. J. Agric. Food Chem. 2020, 68, 10678–10684. [Google Scholar] [CrossRef]
- Smit, B.A.; Engels, W.J.; Wouters, J.T.; Smit, G. Diversity of L-leucine catabolism in various microorganisms involved in dairy fermentations, and identification of the rate-controlling step in the formation of the potent flavour component 3-methylbutanal. Appl. Microbiol. Biotechnol. 2004, 64, 396–402. [Google Scholar] [CrossRef]
- Sakko, M.; Moore, C.; Novak-Frazer, L.; Rautemaa, V.; Sorsa, T.; Hietala, P.; Järvinen, A.; Bowyer, P.; Tjäderhane, L.; Rautemaa, R. 2-hydroxyisocaproic acid is fungicidal for Candida and Aspergillus species. Mycoses 2014, 57, 214–221. [Google Scholar] [CrossRef]
- Ohshima, M.; Sugahara, K.; Kasahara, K.; Katakura, A. Metabolomic analysis of the saliva of Japanese patients with oral squamous cell carcinoma. Oncol. Rep. 2017, 37, 2727–2734. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, S.; Mitsuyama, E.; Yoshida, K.; Odamaki, T.; Xiao, J.Z. Enriched metabolites that potentially promote age-associated diseases in subjects with an elderly-type gut microbiota. Gut Microbes 2021, 13, 1865705. [Google Scholar] [CrossRef] [PubMed]
- Bonner, C.A.; Disz, T.; Hwang, K.; Song, J.; Vonstein, V.; Overbeek, R.; Jensen, R.A. Cohesion group approach for evolutionary analysis of TyrA, a protein family with wide-ranging substrate specificities. Microbiol. Mol. Biol. Rev. 2008, 72, 13–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Concentration (μM) | |||||||
|---|---|---|---|---|---|---|---|
| Species | Isolation Source | Strain | OD600 | 4-OH-PLA | PLA | ILA | LeuA |
| Infant-type HRB | |||||||
| B. bifidum | Infant feces | ATCC29521T | 0.78 ± 0.03 | 14.5 ± 1.3 | 492.9 ± 127.5 | 19.0 ± 5.1 | 131.6 ± 8.2 |
| Infant feces | NITE BP-02429 | 0.84 ± 0.05 | 6.8 ± 0.6 | 184.5 ± 25.5 | 7.4 ± 1.9 | 26.9 ± 6.2 | |
| Infant feces | NITE BP-02431 | 1.10 ± 0.02 | 17.6 ± 1.8 | 248.3 ± 57.5 | 13.8 ± 3.6 | 60.8 ± 12.1 | |
| B. breve | Intestine of infant | ATCC15700T | 1.24 ± 0.03 | 4.2 ± 1.3 | 23.0 ± 3.6 | 10.0 ± 2.1 | 99.3 ± 6.6 |
| Infant feces | FERM BP-11175 (MCC 1274) | 1.22 ± 0.02 | 12.6 ± 2.5 | 26.1 ± 4.2 | 9.5 ± 2.3 | 48.8 ± 2.4 | |
| Infant feces | NITE BP-02622 (M-16V) | 1.33 ± 0.01 | 8.1 ± 2.0 | 33.3 ± 7.3 | 10.6 ± 2.7 | 122.6 ± 4.7 | |
| B. longum subsp. infantis | Intestine of infant | ATCC15697T | 1.35 ± 0.02 | 16.2 ± 3.7 | 63.3 ± 11.3 | 6.9 ± 2.0 | 44.1 ± 1.1 |
| Intestine of infant | NITE BP-02623 (M-63) | 1.34 ± 0.02 | 12.9 ± 2.7 | 72.3 ± 13.6 | 6.6 ± 1.9 | 63.2 ± 2.3 | |
| B. longum subsp. longum | Intestine of adult | ATCC15707T | 1.14 ± 0.01 | 8.9 ± 2.4 | 123.0 ± 20.8 | 23.6 ± 5.0 | 181.8 ± 18.1 |
| Infant feces | NITE BP-02621 (BB536) | 1.24 ± 0.03 | 23.6 ± 2.9 | 152.2 ± 11.4 | 10.2 ± 1.9 | 230.1 ± 38.7 | |
| Average of infant-type HRB | 1.16 ± 0.2 | 12.5 ± 5.7 | 141.9 ± 144.2 | 11.7 ± 5.6 | 78.2 ± 65.9 | ||
| Adult-type HRB | |||||||
| B. adolescentis | Intestine of adult | ATCC15703T | 1.20 ± 0.01 | 2.2 ± 1.2 | 18.2 ± 3.9 | 0.2 ± 0.1 | 46.1 ± 3.2 |
| B. angulatum | Feces, human | ATCC27535T | 1.18 ± 0.03 | 10.7 ± 1.0 | 72.4 ± 10.3 | 4.6 ± 1.7 | 255.6 ± 34.2 |
| B. dentium | Dental caries | DSM20436T | 1.14 ± 0.05 | 0.7 ± 0.7 | 9.9 ± 2.0 | 0.9 ± 0.3 | 112.7 ± 12.2 |
| B. pseudocatenulatum | Feces, human | ATCC27919T | 1.19 ± 0.04 | 0.6 ± 0.9 | 14.8 ± 4.2 | 0.8 ± 0.2 | 136.4 ± 14.9 |
| Average of adult-type HRB | 1.18 ± 0.02 | 3.6 ± 4.8 * | 28.8 ± 29.2 | 1.6 ± 2.0 ** | 137.7 ± 87.4 | ||
| Non-HRB | |||||||
| B. animalis subsp.lactis | yoghurt | DSM10140T | 1.03 ± 0.00 | 2.6 ± 1.0 | 16.6 ± 3.2 | 0.9 ± 0.1 | 60.1 ± 12.9 |
| B. animalis subsp. animalis | Rat feces | ATCC25527T | 1.05 ± 0.02 | 0.7 ± 0.5 | 18.6 ± 4.2 | 0.7 ± 0.2 | 103.0 ± 10.4 |
| B. pseudolongum subsp. globosum | Rumen | JCM5820T | 0.85 ± 0.02 | 1.5 ± 0.7 | 8.9 ± 2.8 | 1.0 ± 0.8 | 46.8 ± 2.3 |
| B. pseudolongum subsp. pseudolongum | Swine feces | ATCC25526T | 0.95 ± 0.01 | 4.6 ± 0.9 | 35.4 ± 3.9 | 1.3 ± 0.2 | 216.9 ± 36.8 |
| B. thermophilum | Swine feces | ATCC 25525T | 1.25 ± 0.03 | 0.6 ± 0.3 | 23.3 ± 3.9 | 1.3 ± 0.3 | 170.9 ± 29.3 |
| Average of non-HRB | 1.03 ± 0.15 | 1.98 ± 1.69 ** | 20.53 ± 9.78 | 1.04 ± 0.27 *** | 119.55 ± 72.85 | ||
| 1st % | |||||
|---|---|---|---|---|---|
| Species | Strain | 4-OH-PLA | PLA | ILA | LeuA |
| B. bifidum | ATCC29521T | 96.9 ± 0.0 | 0.7 ± 0.1 | 0.3 ± 0.2 | 41.5 ± 2.4 |
| NITE BP-02429 | 96.0 ± 1.5 | 0.2 ± 0.0 | 0.3 ± 0.1 | 32.4 ± 2.0 | |
| NITE BP-02431 | 96.8 ± 0.6 | 0.3 ± 0.0 | 0.3 ± 0.0 | 31.8 ± 2.1 | |
| B. breve | ATCC15700T | 95.9 ± 2.4 | 44.3 ± 2.3 | 8.4 ± 0.9 | 97.8 ± 0.2 |
| FERM BP-11175 (MCC 1274) | 96.8 ± 0.1 | 13.8 ± 0.6 | 3.4 ± 0.2 | 94.5 ± 0.2 | |
| NITE BP-02622 (M-16V) | 95.6 ± 1.5 | 39.2 ± 3.5 | 10.8 ± 1.0 | 98.1 ± 0.4 | |
| B. longum subsp. infantis | ATCC15697T | 96.4 ± 0.3 | 1.9 ± 0.1 | 0.7 ± 0.0 | 84.7 ± 1.3 |
| NITE BP-02623 (M-63) | 97.7 ± 1.7 | 2.9 ± 0.1 | 0.8 ± 0.0 | 88.1 ± 1.2 | |
| B. longum subsp. longum | ATCC15707T | 98.3 ± 2.3 | 2.9 ± 0.1 | 2.1 ± 0.1 | 92.8 ± 0.2 |
| NITE BP-02621 (BB536) | 95.8 ± 0.1 | 2.8 ± 0.1 | 2.7 ± 0.2 | 92.2 ± 0.7 | |
| B. adolescentis | ATCC15703T | 96.2 ± 4.0 | 4.3 ± 0.3 | 87.6 ± 13.8 | 89.6 ± 0.9 |
| B. angulatum | ATCC27535T | 96.9 ± 0.2 | 7.1 ± 0.5 | 1.1 ± 0.2 | 95.2 ± 0.5 |
| B. dentium | DSM20436T | 39.1 ± 7.2 | 77.5 ± 3.5 | 99.9 ± 0.0 | 96.2 ± 0.2 |
| B. pseudocatenulatum | ATCC27919T | 92.8 ± 8.6 | 21.3 ± 2.2 | 63.4 ± 2.5 | 49.6 ± 3.5 |
| B. animalis subsp. lactis | DSM10140T | 98.2 ± 3.4 | 4.3 ± 0.3 | 8.7 ± 1.3 | 14.1 ± 2.5 |
| B. animalis subsp. animalis | ATCC25527T | 98.2 ± 2.8 | 5.1 ± 0.6 | 13.7 ± 1.1 | 7.7 ± 0.3 |
| B. pseudolongum subsp. globosum | JCM5820T | 95.0 ± 0.4 | 26.2 ± 3.0 | 68.2 ± 5.4 | 73.3 ± 1.5 |
| B. pseudolongum subsp. pseudolongum | ATCC25526T | 98.0 ± 3.2 | 1.3 ± 0.1 | 8.5 ± 0.2 | 12.5 ± 0.2 |
| B. thermophilum | ATCC 25525T | 87.8 ± 4.1 | 28.6 ± 3.0 | 81.0 ± 9.2 | 96.9 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakurai, T.; Horigome, A.; Odamaki, T.; Shimizu, T.; Xiao, J.-Z. Production of Hydroxycarboxylic Acid Receptor 3 (HCA3) Ligands by Bifidobacterium. Microorganisms 2021, 9, 2397. https://doi.org/10.3390/microorganisms9112397
Sakurai T, Horigome A, Odamaki T, Shimizu T, Xiao J-Z. Production of Hydroxycarboxylic Acid Receptor 3 (HCA3) Ligands by Bifidobacterium. Microorganisms. 2021; 9(11):2397. https://doi.org/10.3390/microorganisms9112397
Chicago/Turabian StyleSakurai, Takuma, Ayako Horigome, Toshitaka Odamaki, Takashi Shimizu, and Jin-Zhong Xiao. 2021. "Production of Hydroxycarboxylic Acid Receptor 3 (HCA3) Ligands by Bifidobacterium" Microorganisms 9, no. 11: 2397. https://doi.org/10.3390/microorganisms9112397
APA StyleSakurai, T., Horigome, A., Odamaki, T., Shimizu, T., & Xiao, J.-Z. (2021). Production of Hydroxycarboxylic Acid Receptor 3 (HCA3) Ligands by Bifidobacterium. Microorganisms, 9(11), 2397. https://doi.org/10.3390/microorganisms9112397






