Landscape Setting Drives the Microbial Eukaryotic Community Structure in Four Swedish Mountain Lakes over the Holocene
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Sediment Sampling
2.3. Dating and Chronology
2.4. Lake Water TOC and Sediment Chlorophyll
2.5. DNA Analysis
2.6. Bioinformatics
2.7. Data Analysis
3. Results
3.1. Past Diversity of the Microbial Eukaryotic Communities
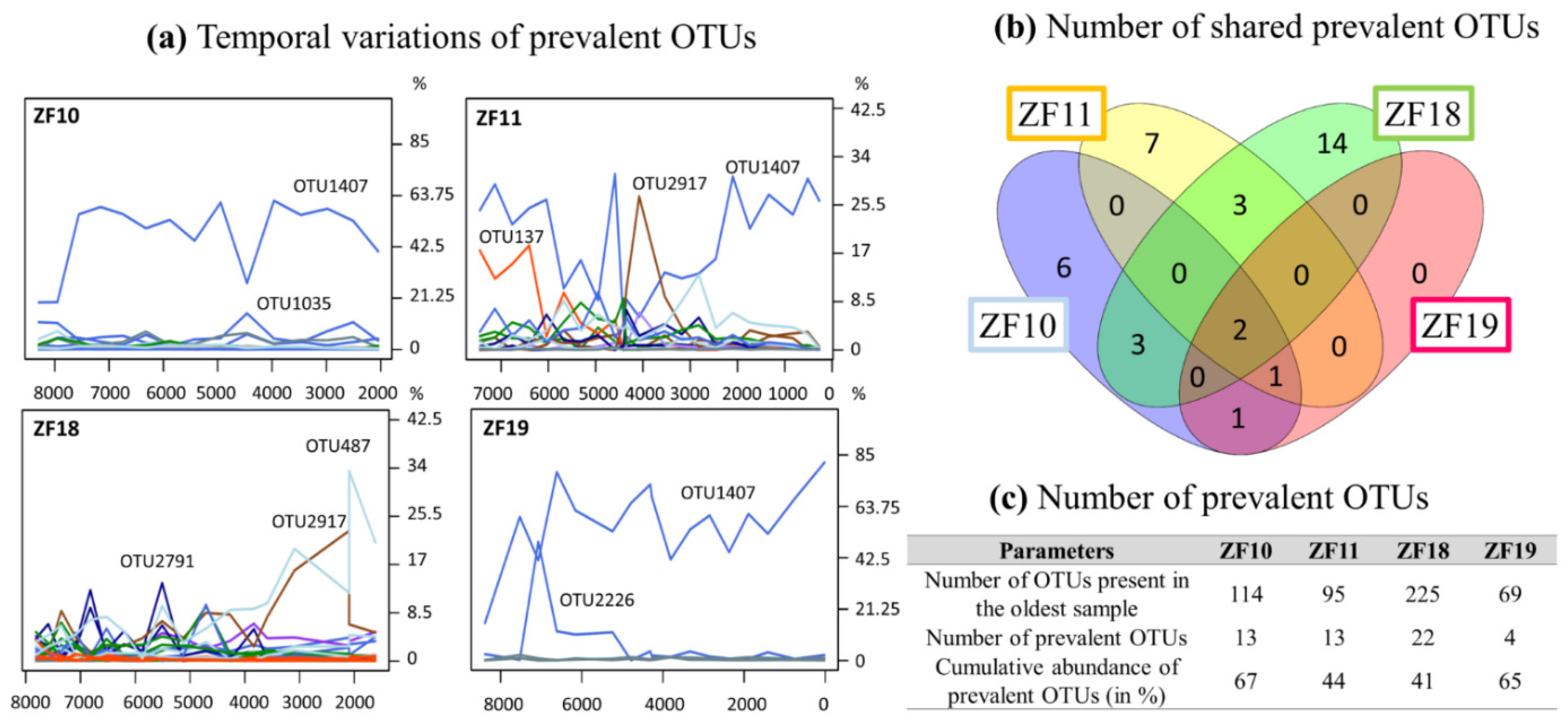
3.2. Long-Term Temporal Dynamics in Microbial Eukaryotic Communities
3.3. Landscape Setting and Microbial Eukaryotic Communities
4. Discussion
4.1. Landscape Setting, Not Climate Shapes Lake Microbial Eukaryotic Communities
4.2. The Importance of Early Community Structure for Shaping a Lake’s Microbial Eukaryotic Community at Later Dates
4.3. Reliability of the Sedimentary DNA Signal to Unravel Past Lake Microbial Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A.; Heinrich, F.; Bertilsson, S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012, 6, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Bunse, C.; Pinhassi, J. Marine Bacterioplankton Seasonal Succession Dynamics. Trends Microbiol. 2017, 25, 393–505. [Google Scholar] [CrossRef] [PubMed]
- Capo, E.; Rydberg, J.; Tolu, J.; Domaizon, I.; Debroas, D.; Bindler, R.; Bigler, C. How does environmental interannual fluctuations impact aquatic microbial communities? A 40 years record of protistan and cyanobacterial communities from a boreal lake (Nylandssjön, Sweden). Front. Ecol. Evol. 2019, 7, 1–13. [Google Scholar] [CrossRef]
- Adrian, R.; O’Reilly, C.M.; Zagarese, H.; Baines, S.B.; Hessen, D.O.; Keller, W.; Livingstone, D.M.; Sommaruga, R.; Straile, D.; Van Donk, E.; et al. Lakes as sentinels of climate change. Limnol. Ocean. 2009, 54, 2283–2297. [Google Scholar] [CrossRef]
- Williamson, C.E.; Saros, J.E.; Vincent, W.F.; Smol, J.P. Lakes and reservoirs as sentinels, integrators, and regulators of climate change. Limnol. Oceanogr. 2009, 54, 2273–2282. [Google Scholar] [CrossRef]
- Moser, K.A.; Baron, J.S.; Brahney, J.; Oleksy, I.A.; Saros, J.E.; Hundey, E.J.; Sadro, S.A.; Kopáček, J.; Sommaruga, R.; Kainz, M.J.; et al. Mountain lakes: Eyes on global environmental change. Glob. Planet. Change 2019, 178, 77–95. [Google Scholar] [CrossRef]
- Mao, D.; Cherkauer, K.A. Impacts of land-use change on hydrologic responses in the Great Lakes region. J. Hydrol. 2009, 374, 71–82. [Google Scholar] [CrossRef]
- Anderson, N.J.; Dietz, R.D.; Engstrom, D.R. Land-use change, not climate, controls organic carbon burial in lakes. Proc. R. Soc. B Biol. Sci. 2013, 280, 20131278. [Google Scholar] [CrossRef]
- Battarbee, R.W.; Anderson, N.J.; Higgitt, S.; Oldfield, F.; Appleby, P.G.; Jones, V.J.; Patrick, S.T.; Flower, R.J.; Kreiser, A.; Richardson, N.G.; et al. Lake Acidification in the United Kingdom 1800–1986; ENSIS Publishing: London, UK, 1988. [Google Scholar]
- Renberg, I.; Korsman, T.; Anderson, N. A temporal perspective of lake acidification in Sweden. Ambio 1993, 22, 264–271. [Google Scholar]
- Capo, E.; Debroas, D.; Arnaud, F.; Perga, M.-E.; Chardon, C.; Domaizon, I. Tracking a century of changes in microbial eukaryotic diversity in lakes driven by nutrient enrichment and climate warming. Environ. Microbiol. 2017, 19, 2873–2892. [Google Scholar] [CrossRef] [PubMed]
- Monchamp, M.E.; Spaak, P.; Domaizon, I.; Dubois, N.; Bouffard, D.; Pomati, F. Homogenization of lake cyanobacterial communities over a century of climate change and eutrophication. Nat. Ecol. Evol. 2018, 2, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Smol, J. Pollution of Lakes and Rivers: A Paleoenvironmental Perspective, 2nd ed.; Blackwell Pub.: Hoboken, NJ, USA, 2009; ISBN 9781405159135. [Google Scholar]
- Battarbee, R.W.; Morley, D.; Bennion, H.; Simpson, G.L.; Hughes, M.; Bauere, V. A palaeolimnological meta-database for assessing the ecological status of lakes. J. Paleolimnol. 2011, 45, 405–414. [Google Scholar] [CrossRef]
- Bennion, H.; Battarbee, R.W.; Sayer, C.D.; Simpson, G.L.; Davidson, T.A. Defining reference conditions and restoration targets for lake ecosystems using palaeolimnology: A synthesis. J. Paleolimnol. 2011, 45, 533–544. [Google Scholar] [CrossRef]
- Birks, H.J.B.; Berglund, B.E. One hundred years of Quaternary pollen analysis 1916–2016. Veg. Hist. Archaeobot. 2018, 27, 271–309. [Google Scholar] [CrossRef]
- Coolen, M.J.L.; Gibson, J.A.E. Ancient DNA in lake sediment records. PAGES News 2009, 17, 104–106. [Google Scholar] [CrossRef]
- Domaizon, I.; Winegardner, A.; Capo, E.; Gauthier, J.; Gregory-Eaves, I. DNA-based methods in paleolimnology: New opportunities for investigating long-term dynamics of lacustrine biodiversity. J. Paleolimnol. 2017, 58, 1–21. [Google Scholar] [CrossRef]
- Li, G.; Dong, H.; Hou, W.; Wang, S.; Jiang, H.; Yang, J.; Wu, G. Temporal Succession of Ancient Phytoplankton Community in Qinghai Lake and Implication for Paleo-environmental Change. Sci. Rep. 2016, 6, 19769. [Google Scholar] [CrossRef][Green Version]
- Monchamp, M.E.; Walser, J.C.; Pomati, F.; Spaak, P. Sedimentary DNA reveals cyanobacterial community diversity over 200 years in two perialpine lakes. Appl. Environ. Microbiol. 2016, 82, 6472–6482. [Google Scholar] [CrossRef]
- Monchamp, M.-E.; Spaak, P.; Pomati, F. Long Term Diversity and Distribution of Non-photosynthetic Cyanobacteria in Peri-Alpine Lakes. Front. Microbiol. 2019, 9, 3344. [Google Scholar] [CrossRef]
- Keck, F.; Millet, L.; Debroas, D.; Etienne, D.; Galop, D.; Rius, D.; Domaizon, I. Assessing the response of micro-eukaryotic diversity to the Great Acceleration using lake sedimentary DNA. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; Porter, K.G.; Bennett, S.J.; DeBiase, A.E. Seasonal patterns of bacterivory by flagellates, ciliates, rotifers, and cladocerans in a freshwater planktonic community. Limnol. Oceanogr. 1989, 34, 673–687. [Google Scholar] [CrossRef]
- Gozlan, R.E.; St-Hilaire, S.; Feist, S.W.; Martin, P.; Kent, M.L. Disease threat to European fish. Nature 2005, 435, 1046. [Google Scholar] [CrossRef]
- Kagami, M.; De Bruin, A.; Ibelings, B.W.; Van Donk, E. Parasitic chytrids: Their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 2007, 578, 113–129. [Google Scholar] [CrossRef]
- Kyle, M.; Haande, S.; Ostermaier, V.; Rohrlack, T. The Red Queen Race between Parasitic Chytrids and Their Host, Planktothrix: A Test Using a Time Series Reconstructed from Sediment DNA. PLoS ONE 2015, 10, e0118738. [Google Scholar] [CrossRef]
- Capo, E.; Debroas, D.; Arnaud, F.; Guillemot, T.; Bichet, V.; Millet, L.; Gauthier, E.; Massa, C.; Develle, A.-L.; Pignol, C.; et al. Long-term dynamics in microbial eukaryotes communities: A palaeolimnological view based on sedimentary DNA. Mol. Ecol. 2016, 25, 5925–5943. [Google Scholar] [CrossRef]
- Coolen, M.J.L.; Orsi, W.D.; Balkema, C.; Quince, C.; Harris, K.; Sylva, S.P.; Filipova-Marinova, M.; Giosan, L. Evolution of the plankton paleome in the Black Sea from the Deglacial to Anthropocene. Proc. Natl. Acad. Sci. USA 2013, 110, 8609–8614. [Google Scholar] [CrossRef] [PubMed]
- Kisand, V.; Talas, L.; Kisand, A.; Stivrins, N.; Reitalu, T.; Alliksaar, T.; Vassiljev, J.; Liiv, M.; Heinsalu, A.; Seppä, H.; et al. From microbial eukaryotes to metazoan vertebrates: Wide spectrum paleo-diversity in sedimentary ancient DNA over the last ~14,500 years. Geobiology 2018, 16, 628–639. [Google Scholar] [CrossRef] [PubMed]
- Capo, E.; Spong, G.; Norman, S.; Königsson, H.; Bartels, P.; Byström, P. Droplet digital PCR assays for the quantification of brown trout (Salmo trutta) and Arctic char (Salvelinus alpinus) from environmental DNA collected in the water of mountain lakes. PLoS ONE 2019, 14, e0226638. [Google Scholar] [CrossRef]
- Renberg, I.; Bindler, R.; Brännvall, M.-L. Using the historical atmospheric lead-deposition record as a chronological marker in sediment deposits in Europe. The Holocene 2001, 11, 511–516. [Google Scholar] [CrossRef]
- Rydberg, J. Wavelength dispersive X-ray fluorescence spectroscopy as a fast, non-destructive and cost-effective analytical method for determining the geochemical composition of small loose-powder sediment samples. J. Paleolimnol. 2014, 52, 265–276. [Google Scholar] [CrossRef]
- Reimer, P.J.; Bard, E.; Bayliss, A.; Beck, J.W.; Blackwell, P.G.; Ramsey, C.B.; Buck, C.E.; Cheng, H.; Edwards, R.L.; Friedrich, M.; et al. IntCal13 and Marine13 Radiocarbon Age Calibration Curves 0–50,000 Years cal BP. Radiocarbon 2013, 55, 1869–1887. [Google Scholar] [CrossRef]
- Blaauw, M. Methods and code for ´classical´ age-modelling of radiocarbon sequences. Quat. Geochronol. 2010, 5, 512–518. [Google Scholar] [CrossRef]
- Meyer-Jacob, C.; Michelutti, N.; Paterson, A.M.; Monteith, D.; Yang, H.; Weckström, J.; Smol, J.P.; Bindler, R. Inferring Past Trends in Lake Water Organic Carbon Concentrations in Northern Lakes Using Sediment Spectroscopy. Environ. Sci. Technol. 2017, 51, 13248–13255. [Google Scholar] [CrossRef]
- Michelutti, N.; Blais, J.M.; Cumming, B.F.; Paterson, A.M.; Rühland, K.; Wolfe, A.P.; Smol, J.P. Do spectrally inferred determinations of chlorophyll a reflect trends in lake trophic status? J. Paleolimnol. 2010, 43, 205–217. [Google Scholar] [CrossRef]
- Gast, R.J.; Dennett, M.R.; Caron, D.A. Characterization of Protistan Assemblages in the Ross Sea, Antarctica, by Denaturing Gradient Gel Electrophoresis. Appl. Environ. Microbiol. 2004, 70, 2028–2037. [Google Scholar] [CrossRef]
- Van de Peer, Y.; De Rijk, P.; Wuyts, J.; Winkelmans, T.; De Wachter, R. The European Small Subunit Ribosomal RNA database. Nucleic Acids Res. 2000, 28, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Esling, P.; Lejzerowicz, F.; Pawlowski, J. Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res. 2015, 43, 2513–2524. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Mangot, J.-F.; Domaizon, I.; Taib, N.; Marouni, N.; Duffaud, E.; Bronner, G.; Debroas, D. Short-term dynamics of diversity patterns: Evidence of continual reassembly within lacustrine small eukaryotes. Environ. Microbiol. 2013, 15, 1745–1758. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Lefranc, M.; Thénot, A.; Lepère, C.; Debroas, D. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 2005, 71, 5935–5942. [Google Scholar] [CrossRef] [PubMed]
- Lepère, C.; Boucher, D.; Jardillier, L.; Domaizon, I.; Debroas, D. Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin). Appl. Environ. Microbiol. 2006, 72, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Lepère, C.; Domaizon, I.; Debroas, D. Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl. Environ. Microbiol. 2008, 74, 2940–2949. [Google Scholar] [CrossRef]
- Oksanen, A.J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. The Vegan Package. Community Ecology Package. 2015. Available online: http://CRAN.R-project.org/package=vegan (accessed on 12 May 2019).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Oliveros, J.C. VENNY: An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 3 December 2020).
- Triadó-Margarit, X.; Casamayor, E.O. Genetic diversity of planktonic eukaryotes in high mountain lakes (Central Pyrenees, Spain). Environ. Microbiol. 2012, 14, 2445–2456. [Google Scholar] [CrossRef]
- Simon, M.; López-García, P.; Deschamps, P.; Moreira, D.; Restoux, G.; Bertolino, P.; Jardillier, L. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J. 2015, 9, 1941–1953. [Google Scholar] [CrossRef]
- Kammerlander, B.; Breiner, H.-W.; Filker, S.; Sommaruga, R.; Sonntag, B.; Stoeck, T. High diversity of protistan plankton communities in remote high mountain lakes in the European Alps and the Himalayan mountains. FEMS Microbiol. Ecol. 2015, 91, 1–10. [Google Scholar] [CrossRef][Green Version]
- Filker, S.; Sommaruga, R.; Vila, I.; Stoeck, T. Microbial eukaryote plankton communities of high-mountain lakes from three continents exhibit strong biogeographic patterns. Mol. Ecol. 2016, 25, 2286–2301. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, L.; Jensen, M.; Pandey, R.V.; Jost, S.; Bass, D.; Psenner, R.; Boenigk, J. Molecular investigation of protistan diversity along an elevation transect of alpine lakes. Aquat. Microb. Ecol. 2016, 78, 25–37. [Google Scholar] [CrossRef]
- Grossmann, L.; Jensen, M.; Heider, D.; Jost, S.; Glücksman, E.; Hartikainen, H.; Mahamdallie, S.S.; Gardner, M.; Hoffmann, D.; Bass, D.; et al. Protistan community analysis: Key findings of a large-scale molecular sampling. ISME J. 2016, 10, 2269–2279. [Google Scholar] [CrossRef] [PubMed]
- Debroas, D.; Domaizon, I.; Humbert, J.-F.; Jardillier, L.; Lepère, C.; Oudart, A.; Taïb, N. Overview of freshwater microbial eukaryotes diversity: A first analysis of publicly available metabarcoding data. FEMS Microbiol. Ecol. 2017, 93, 1–14. [Google Scholar] [CrossRef]
- Jeppesen, E.; Søndergaard, M.; Jensen, J.P.; Havens, K.E.; Anneville, O.; Carvalho, L.; Coveney, M.F.; Deneke, R.; Dokulil, M.T.; Foy, B.; et al. Lake responses to reduced nutrient loading—An analysis of contemporary long-term data from 35 case studies. Freshw. Biol. 2005, 50, 1747–1771. [Google Scholar] [CrossRef]
- Ibrahim, A.; Capo, E.; Wessels, M.; Martin, I.; Meyer, A.; Schleheck, D.; Epp, L.S. Anthropogenic impact on the historical phytoplankton community of Lake Constance reconstructed by multimarker analysis of sediment-core environmental DNA. Mol. Ecol. 2020, 1–17. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef]
- Herzschuh, U.; Zhang, C.; Mischke, S.; Herzschuh, R.; Mohammadi, F.; Mingram, B.; Kürschner, H.; Riedel, F. A late Quaternary lake record from the Qilian Mountains (NW China): Evolution of the primary production and the water depth reconstructed from macrofossil, pollen, biomarker, and isotope data. Glob. Planet. Change 2005, 46, 361–379. [Google Scholar] [CrossRef]
- Fritz, S.C.; Anderson, N.J. The relative influences of climate and catchment processes on Holocene lake development in glaciated regions. J. Paleolimnol. 2013, 49, 349–362. [Google Scholar] [CrossRef]
- Hou, W.; Dong, H.; Li, G.; Yang, J.; Coolen, M.J.L.; Liu, X.; Wang, S.; Jiang, H.; Wu, X.; Xiao, H.; et al. Identification of Photosynthetic Plankton Communities Using Sedimentary Ancient DNA and Their Response to late-Holocene Climate Change on the Tibetan Plateau. Sci. Rep. 2014, 4, 6648. [Google Scholar] [CrossRef] [PubMed]
- Seppä, H.; Bjune, A.E.; Telford, R.J.; Birks, H.J.B.; Veski, S. Last nine-thousand years of temperature variability in Northern Europe. Clim. Past 2009, 5, 523–535. [Google Scholar] [CrossRef]
- De Meester, L.; Vanoverbeke, J.; Kilsdonk, L.J.; Urban, M.C. Evolving perspectives on monopolization and priority effects. Trends Ecol. Evol. 2016, 31, 136–146. [Google Scholar] [CrossRef]
- Vass, M.; Langenheder, S. The legacy of the past: Effects of historical processes on microbial metacommunities. Aquat. Microb. Ecol. 2017, 79, 13–19. [Google Scholar] [CrossRef]
- Capo, E.; Debroas, D.; Arnaud, F.; Domaizon, I. Is Planktonic Diversity Well Recorded in Sedimentary DNA? Toward the Reconstruction of Past Protistan Diversity. Microb. Ecol. 2015, 70. [Google Scholar] [CrossRef]
- Capo, E.; Domaizon, I.; Maier, D.; Debroas, D.; Bigler, C. To what extent is the DNA of microbial eukaryotes modified during burying into lake sediments? A repeat-coring approach on annually laminated sediments. J. Paleolimnol. 2017, 58, 479–495. [Google Scholar] [CrossRef]
- Capo, E.; Giguet-Covex, C.; Rouillard, A.; Nota, K.; Heintzman, P.D.; Vuillemin, A.; Ariztegui, D.; Arnaud, F.; Belle, S.; Bertilsson, S.; et al. Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: Overview and recommendations. Quaternary 2020, in press. [Google Scholar]
- Monchamp, M.E.; Enache, I.; Turko, P.; Pomati, F.; Rîşnoveanu, G.; Spaak, P. Sedimentary and egg-bank DNA from 3 European lakes reveal concurrent changes in the composition and diversity of cyanobacterial and Daphnia communities. Hydrobiologia 2017, 800, 155–172. [Google Scholar] [CrossRef]
- Giguet-Covex, C.; Ficetola, G.F.; Walsh, K.; Poulenard, J.; Bajard, M.; Fouinat, L.; Sabatier, P.; Gielly, L.; Messager, E.; Develle, A.L.; et al. New insights on lake sediment DNA from the catchment: Importance of taphonomic and analytical issues on the record quality. Sci. Rep. 2019, 9, 1–21. [Google Scholar] [CrossRef]
- Crump, B.C.; Amaral-zettler, L.A.; Kling, G.W. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J. 2012, 6, 1629–1639. [Google Scholar] [CrossRef]
- Ruiz-González, C.; Niño-García, J.P.; del Giorgio, P.A. Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol. Lett. 2015, 18, 1198–1206. [Google Scholar] [CrossRef]
- Mangot, J.F.; Debroas, D.; Domaizon, I. Perkinsozoa, a well-known marine protozoan flagellate parasite group, newly identified in lacustrine systems: A review. Hydrobiologia 2011, 659, 37–48. [Google Scholar] [CrossRef]
- Annenkova, N.V.; Giner, C.R.; Logares, R. Tracing the origin of planktonic protists in an ancient lake. Microorganisms 2020, 8, 543. [Google Scholar] [CrossRef]
- Delmont, T.O.; Kiefl, E.; Kilinc, O.; Esen, O.C.; Uysal, I.; Rappé, M.S.; Giovannoni, S.; Eren, A.M. Single-amino acid variants reveal evolutionary processes that shape the biogeography of a global SAR11 subclade. Elife 2019, 8, 1–26. [Google Scholar] [CrossRef]
- Pedersen, M.W.; Ruter, A.; Schweger, C.; Friebe, H.; Staff, R.A.; Kjeldsen, K.K.; Mendoza, M.L.Z.; Beaudoin, A.B.; Zutter, C.; Larsen, N.K.; et al. Postglacial viability and colonization in North America’s ice-free corridor. Nature 2016, 537, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Parducci, L.; Unneberg, P.; Ågren, R.; Schenk, F.; Rattray, J.E.; Han, L.; Muschitiello, F.; Pedersen, M.W.; Smittenberg, R.H.; et al. Archaeal community changes in Lateglacial lake sediments: Evidence from ancient DNA. Quat. Sci. Rev. 2018, 181, 19–29. [Google Scholar] [CrossRef]
- Garner, R.E.; Gregory-Eaves, I.; Walsh, D.A. Sediment Metagenomes as Time Capsules of Lake Microbiomes. mSphere 2020, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lammers, Y.; Heintzman, P.D.; Alsos, I.G. Environmental palaeogenomic reconstruction of an Ice Age algal population. bioRxiv 2020. [Google Scholar] [CrossRef]
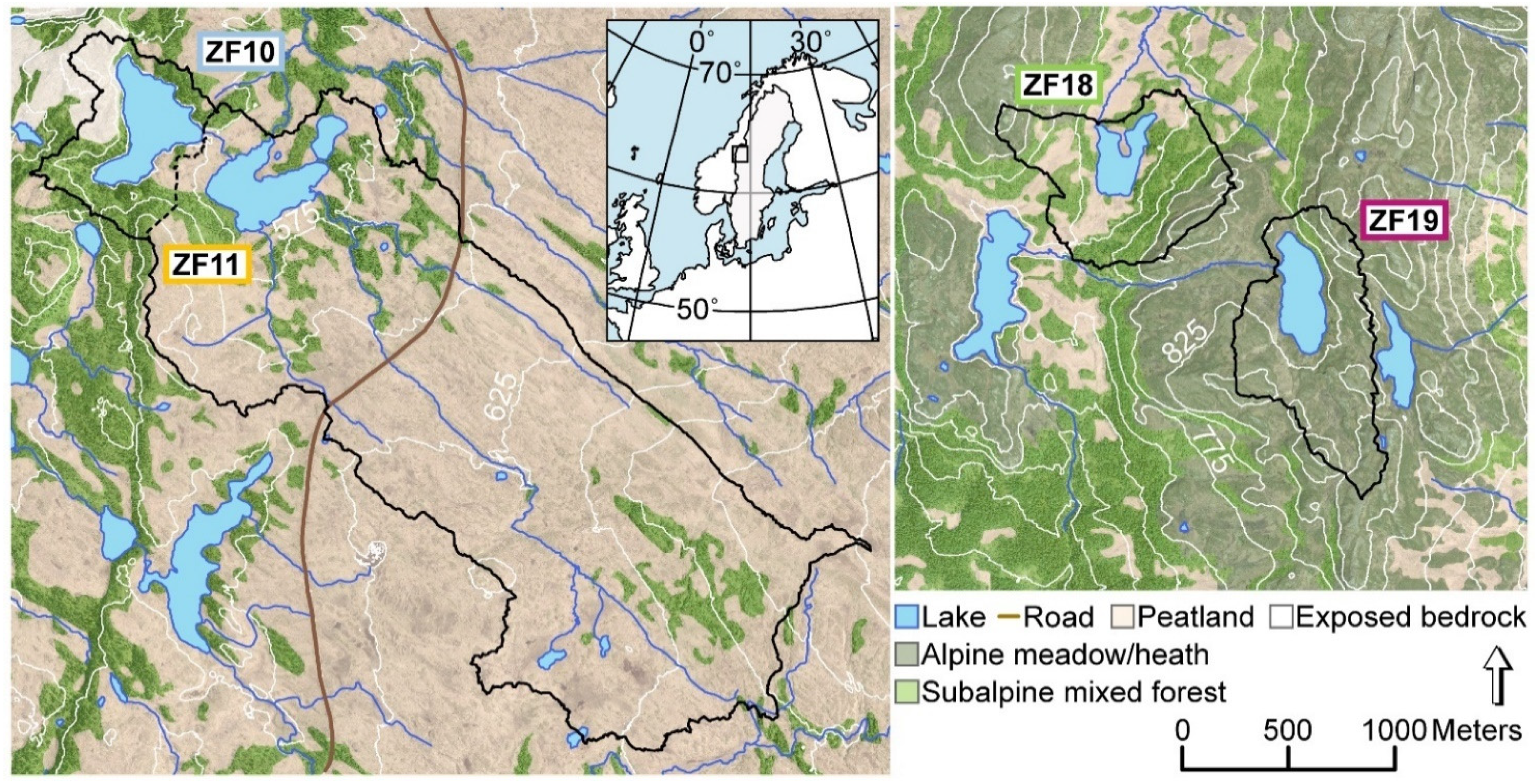



| Lake | Coordinates | Elevation (m) | Lake Area (ha) | Catchment Area (ha) | Max. Lake Depth (m) | Avg. Catchment Slope (°) |
|---|---|---|---|---|---|---|
| ZF10 (Bergtjärnen) | 63°37’27” N | 573 | 14 | 31 | 341 | 16.5 |
| 12°15’35” E | ||||||
| ZF11 (Vargtjärnen) | 63°37’17” N | 564 | 12 | 454 | 14 | 6.6 |
| 12°16’12” E | ||||||
| ZF18 (Hästskotjärnen) | 63°18’52” N | 735 | 6 | 51 | 8 | 12.6 |
| 12°54’11” E | ||||||
| ZF19 (Högdalstjärnen) | 63°18’3” N | 812 | 9 | 51 | 14 | 12.3 |
| 12°55’14” E |
| Microbial Eukaryotic Groups | All lakes | Lake ZF10 | Lake ZF11 | Lake ZF18 | Lake ZF19 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #DNA-Reads | #OTUs | #DNA-Reads | #OTUs | #DNA-Reads | #OTUs | #DNA-Reads | #OTUs | #DNA-Reads | #OTUs | ||
| Number of Molecular Inventories | 72 | 15 | 22 | 17 | 17 | ||||||
| Alveolata | Apicomplexa | 114,977 | 86 | 10,822 | 22 | 41,631 | 63 | 54974 | 47 | 7550 | 24 |
| Ciliophora | 100,614 | 196 | 5918 | 46 | 40,639 | 145 | 47,189 | 127 | 6868 | 48 | |
| Dinophyceae | 82,078 | 135 | 24,297 | 51 | 26,267 | 106 | 20,734 | 89 | 10,780 | 49 | |
| Perkinsea | 60,465 | 69 | 28,327 | 28 | 7885 | 54 | 15,889 | 42 | 8364 | 30 | |
| Voromonas | 1944 | 8 | 202 | 5 | 1346 | 4 | 233 | 5 | 163 | 3 | |
| Other_Alveolata | 643,595 | 105 | 220,235 | 45 | 136,264 | 93 | 26,718 | 63 | 260,378 | 47 | |
| Amoebozoa | Centramoebida | 68 | 3 | 42 | 1 | 24 | 2 | 2 | 1 | 0 | 0 |
| Tubulinea | 1362 | 19 | 28 | 1 | 1219 | 18 | 101 | 6 | 14 | 3 | |
| Cryptophyta | Cryptomonadales | 1571 | 4 | 473 | 1 | 248 | 3 | 310 | 2 | 540 | 2 |
| Cryptophyta_2 | 608 | 1 | 243 | 1 | 39 | 1 | 325 | 1 | 1 | 1 | |
| Cryptophyta_3 | 4 | 1 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | |
| Cryptophyta_4 | 289 | 3 | 2 | 1 | 174 | 1 | 111 | 3 | 2 | 1 | |
| Pyrenomonadales | 1705 | 7 | 329 | 2 | 1062 | 5 | 313 | 6 | 1 | 1 | |
| Other_Cryptophyta | 313 | 7 | 0 | 0 | 46 | 7 | 266 | 2 | 1 | 1 | |
| Haptophyta | Coccolithales | 6 | 2 | 0 | 0 | 0 | 0 | 6 | 2 | 0 | 0 |
| Pavlovales | 113 | 1 | 0 | 0 | 0 | 0 | 113 | 1 | 0 | 0 | |
| Phaeocystales | 974 | 3 | 15 | 1 | 341 | 3 | 618 | 1 | 0 | 0 | |
| Prymnesiales | 1820 | 7 | 458 | 2 | 1008 | 7 | 350 | 4 | 4 | 1 | |
| Syracosphaerales | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Other_Haptophyta | 17436 | 17 | 6604 | 12 | 5557 | 13 | 1490 | 12 | 3785 | 10 | |
| Opisthokonta | Choanoflagellida | 806 | 8 | 375 | 1 | 254 | 5 | 177 | 4 | 0 | 0 |
| Chytrids | 183,194 | 233 | 7126 | 58 | 58,565 | 191 | 107,699 | 145 | 9804 | 84 | |
| Opisthokonta_incertae_sedis | 954 | 1 | 1 | 1 | 261 | 1 | 690 | 1 | 2 | 1 | |
| Rhizaria | Cercozoa | 56,498 | 244 | 10,334 | 75 | 23,998 | 194 | 10,745 | 131 | 11,421 | 61 |
| Other_Rhizaria | 60,868 | 78 | 4897 | 18 | 24,895 | 64 | 17,820 | 38 | 13,256 | 31 | |
| Rhodophyta | Bangiophyceae | 202 | 3 | 0 | 0 | 202 | 3 | 0 | 0 | 0 | 0 |
| Florideophyceae | 873 | 7 | 581 | 1 | 235 | 5 | 57 | 2 | 0 | 0 | |
| Stramenopiles | Bacillariophyta | 26,914 | 32 | 2231 | 14 | 6479 | 25 | 9207 | 20 | 8997 | 16 |
| Bicosoecida | 21,556 | 49 | 1145 | 12 | 6678 | 39 | 4434 | 26 | 9299 | 15 | |
| Chrysophyceae | 39,585 | 44 | 1532 | 13 | 34283 | 39 | 1953 | 28 | 1817 | 19 | |
| Dictyochophyceae | 63 | 4 | 0 | 0 | 8 | 3 | 53 | 2 | 2 | 1 | |
| Eustigmatophyceae | 6828 | 9 | 1632 | 3 | 489 | 5 | 3530 | 7 | 1177 | 2 | |
| Labyrinthulida | 940 | 8 | 1 | 1 | 135 | 5 | 730 | 6 | 74 | 2 | |
| MAST (1, 2, 3, 4, 7) | 696 | 15 | 1 | 1 | 528 | 14 | 145 | 7 | 22 | 3 | |
| Oomycetes | 12,365 | 38 | 2346 | 14 | 4694 | 28 | 3137 | 24 | 2188 | 18 | |
| PX_clade | 4751 | 13 | 394 | 7 | 1544 | 10 | 2772 | 10 | 41 | 3 | |
| Synurophyceae | 3521 | 15 | 4 | 3 | 894 | 9 | 926 | 15 | 1697 | 5 | |
| Other_Stramenopiles | 65,265 | 55 | 1301 | 15 | 33,985 | 50 | 7677 | 30 | 22,302 | 15 | |
| Viridiplantae | Chlorophyta | 147,386 | 147 | 20,410 | 43 | 52,483 | 126 | 56,888 | 94 | 17605 | 47 |
| Streptophyta | 5240 | 40 | 171 | 11 | 2619 | 30 | 1105 | 23 | 1345 | 14 | |
| Other_Viridiplantae | 51 | 3 | 21 | 1 | 17 | 3 | 13 | 1 | 0 | 0 | |
| Total | 1,668,500 | 1721 | 352,500 | 512 | 517,000 | 1375 | 399,500 | 1028 | 399,500 | 558 | |
| Parameters | Lake ZF10 | Lake ZF11 | Lake ZF18 | Lake ZF19 | ||||
|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | |
| chlorophyll | 0.34 | 0.08 | 0.64 | 0.035 | 0.56 | 0.004 | 0.02 | 0.891 |
| LW-TOC | 0.31 | 0.1 | 0.47 | 0.031 | 0.4 | 0.033 | 0.38 | 0.029 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capo, E.; Ninnes, S.; Domaizon, I.; Bertilsson, S.; Bigler, C.; Wang, X.-R.; Bindler, R.; Rydberg, J. Landscape Setting Drives the Microbial Eukaryotic Community Structure in Four Swedish Mountain Lakes over the Holocene. Microorganisms 2021, 9, 355. https://doi.org/10.3390/microorganisms9020355
Capo E, Ninnes S, Domaizon I, Bertilsson S, Bigler C, Wang X-R, Bindler R, Rydberg J. Landscape Setting Drives the Microbial Eukaryotic Community Structure in Four Swedish Mountain Lakes over the Holocene. Microorganisms. 2021; 9(2):355. https://doi.org/10.3390/microorganisms9020355
Chicago/Turabian StyleCapo, Eric, Sofia Ninnes, Isabelle Domaizon, Stefan Bertilsson, Christian Bigler, Xiao-Ru Wang, Richard Bindler, and Johan Rydberg. 2021. "Landscape Setting Drives the Microbial Eukaryotic Community Structure in Four Swedish Mountain Lakes over the Holocene" Microorganisms 9, no. 2: 355. https://doi.org/10.3390/microorganisms9020355
APA StyleCapo, E., Ninnes, S., Domaizon, I., Bertilsson, S., Bigler, C., Wang, X.-R., Bindler, R., & Rydberg, J. (2021). Landscape Setting Drives the Microbial Eukaryotic Community Structure in Four Swedish Mountain Lakes over the Holocene. Microorganisms, 9(2), 355. https://doi.org/10.3390/microorganisms9020355






