Wild Boars Carry Extended-Spectrum β-Lactamase- and AmpC-Producing Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
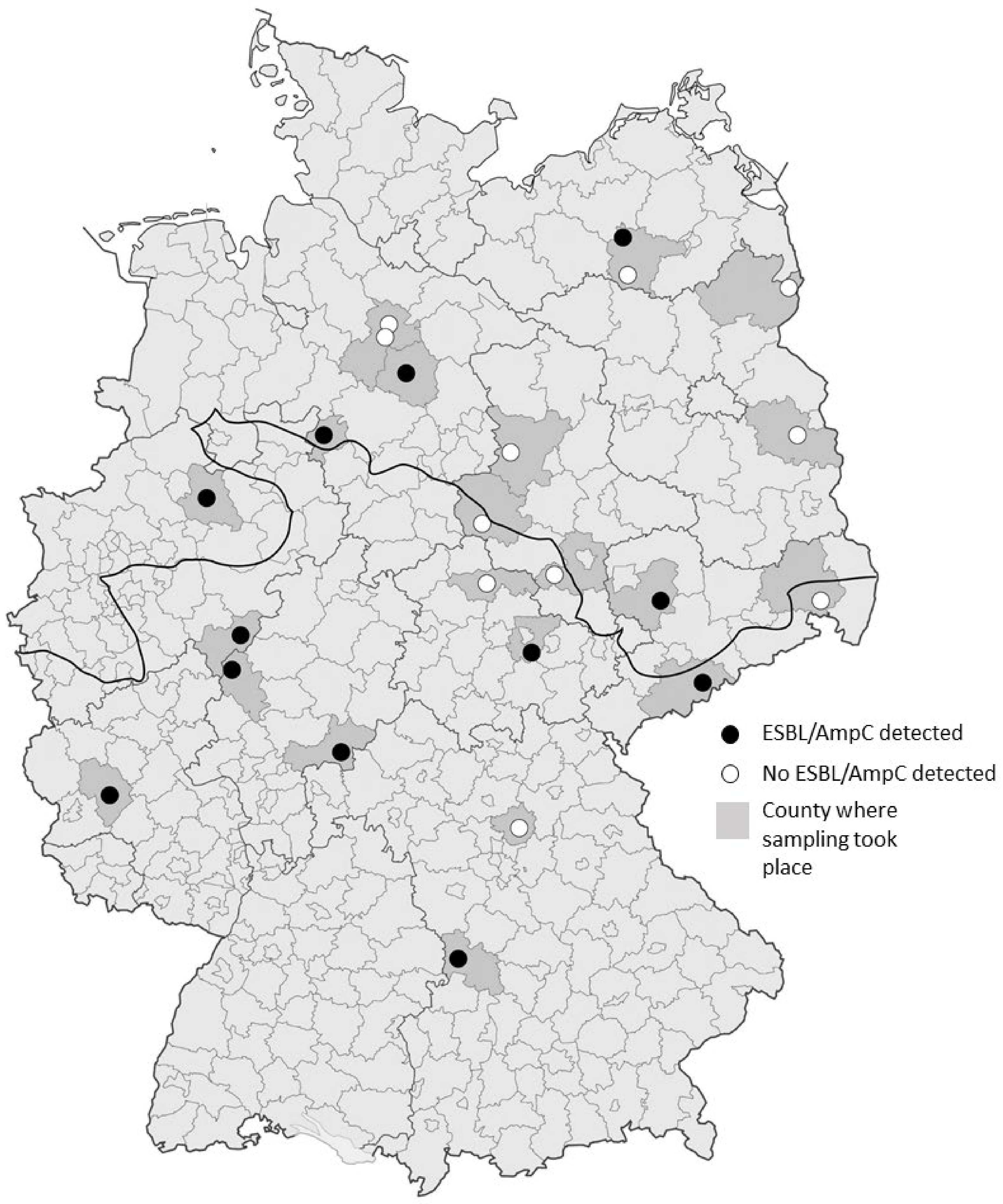
2.1. Sample Collection
2.2. MRSA Isolation
2.3. Isolation and Confirmation of ESBL/AmpC-Producing Escherichia coli
2.4. Antimicrobial Susceptibility Testing
2.5. DNA Isolation, Species Confirmation and PCR Analysis
2.6. Genotyping
2.7. Genome Sequencing
2.8. Statistical Analyses
3. Results
3.1. MRSA and ESBL/AmpC-E. coli Isolate Detection
3.2. Antimicrobial Susceptibility Testing and Detection of Resistance Genes
3.3. Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Becker, K.; Ballhausen, B.; Köck, R.; Kriegeskorte, A. Methicillin resistance in Staphylococcus isolates: The “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int. J. Med. Microbiol. 2014, 304, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant S. aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Fischer, J.; Hille, K.; Ruddat, I.; Mellmann, A.; Köck, R.; Kreienbrock, L. Simultaneous occurrence of MRSA and ESBL-producing Enterobacteriaceae on pig farms and in nasal and stool samples from farmers. Vet. Microbiol. 2017, 200, 107–113. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Goerge, T.; Lorenz, M.B.; van Alen, S.; Hübner, N.-O.; Becker, K.; Köck, R. MRSA colonization and infection among persons with occupational livestock exposure in Europe: Prevalence, preventive options and evidence. Vet. Microbiol. 2017, 200, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Meemken, D.; Blaha, T.; Tegeler, R.; Tenhagen, B.A.; Guerra, B.; Hammerl, J.A.; Hertwig, S.; Käsbohrer, A.; Appel, B.; Fetsch, A. Livestock associated methicillin-resistant S. aureus (LaMRSA) isolated from lesions of pigs at necropsy in northwest Germany between 2004 and 2007. Zoonoses Publ. Health 2010, 57, 143–148. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum beta-lactamase-producing and AmpC-producing E. coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef]
- Reich, F.; Atanassova, V.; Klein, G. Extended-spectrum beta-lactamase- and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 2013, 19, 1253–1259. [Google Scholar] [CrossRef]
- Carattoli, A. Review: Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef]
- Ferguson, D.D.; Smith, T.C.; Hanson, B.M.; Wardyn, S.E.; Donham, K.J. Detection of airborne methicillin-resistant S. aureus inside and downwind of a swine building, and in animal feed: Potential cccupational, animal health, and environmental implications. J. Agromed. 2016, 21, 149–153. [Google Scholar] [CrossRef]
- Friese, A.; Schulz, J.; Laube, H.; von Salviati, C.; Hartung, J.; Roesler, U. Faecal occurrence and emissions of livestock-associated methicillin-resistant S. aureus (laMRSA) and ESbl/AmpC-producing E. coli from animal farms in Germany. Berl. Munch. Tierarztl. Wochenschr. 2013, 126, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef]
- Veldman, K.; van Tulden, P.; Kant, A.; Testerink, J.; Mevius, D. Characteristics of Cefotaxime-Resistant E. coli from wild birds in The Netherlands. Appl. Environ. Microbiol. 2013, 79, 7556–7561. [Google Scholar] [CrossRef] [PubMed]
- Furness, L.E.; Campbell, A.; Zhang, L.; Gaze, W.H.; McDonald, R.A. Wild small mammals as sentinels for the environmental transmission of antimicrobial resistance. Environ. Res. 2017, 154, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.T.; Fernandes, J.; Carvalho, J.; Cunha, M.V.; Caetano, T.; Mendo, S.; Serrano, E.; Fonseca, C. Wild boar as a reservoir of antimicrobial resistance. Sci. Total Environ. 2020, 717, 135001. [Google Scholar] [CrossRef]
- Kraushaar, B.; Fetsch, A. First description of PVL-positive methicillin-resistant S. aureus (MRSA) in wild boar meat. Int. J. Food Microbiol. 2014, 186, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Mateus-Vargas, R.H.; Atanassova, V.; Reich, F.; Klein, G. Antimicrobial susceptibility and genetic characterization of Escherichia coli recovered from frozen game meat. Food Microbiol. 2017, 63, 164–169. [Google Scholar] [CrossRef]
- de Boer, E.; Zwartkruis-Nahuis, J.T.M.; Wit, B.; Huijsdens, X.W.; de Neeling, A.J.; Bosch, T.; van Oosterom, R.A.A.; Vila, A.; Heuvelink, A.E. Prevalence of methicillin-resistant S. aureus in meat. Int. J. Food Microbiol. 2009, 134, 52–56. [Google Scholar] [CrossRef]
- Guenther, S.; Ewers, C.; Wieler, L.H. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef] [PubMed]
- Velhner, M.; Todorović, D.; Grego, E.; Jovčić, B.; Prunić, B.; Stojanov, I.; Kehrenberg, C. Fluoroquinolone-resistant and extended-spectrum beta-lactamase producing E. coli isolates from free-living wild animals. Vet. Microbiol. 2018, 223, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Silva, N.; Manageiro, V.; Ramos, S.; Coelho, A.; Gonçalves, D.; Caniça, M.; Torres, C.; Igrejas, G.; Poeta, P. First report on MRSA CC398 recovered from wild boars in the north of Portugal. Are we facing a problem? Sci. Total Environ. 2017, 596–597, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Poeta, P.; Radhouani, H.; Pinto, L.; Martinho, A.; Rego, V.; Rodrigues, R.; Goncalves, A.; Rodrigues, J.; Estepa, V.; Torres, C.; et al. Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 2009, 49, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Atanassova, V.; Apelt, J.; Reich, F.; Klein, G. Microbiological quality of freshly shot game in Germany. Meat Sci. 2008, 78, 414–419. [Google Scholar] [CrossRef]
- Marescotti, M.E.; Caputo, V.; Demartini, E.; Gaviglio, A. Discovering market segments for hunted wild game meat. Meat Sci. 2019, 149, 163–176. [Google Scholar] [CrossRef]
- Porrero, M.C.; Mentaberre, G.; Sánchez, S.; Fernández-Llario, P.; Gómez-Barrero, S.; Navarro-Gonzalez, N.; Serrano, E.; Casas-Díaz, E.; Marco, I.; Fernández-Garayzabal, J.F.; et al. Methicillin resistant S. aureus (MRSA) carriage in differentfree-living wild animal species in Spain. Vet. J. 2013, 198, 127–130. [Google Scholar] [CrossRef]
- Matschke, G.H. Aging European wild hogs by dentition. J. Wildl. Manage. 1967, 31, 109–113. [Google Scholar] [CrossRef]
- Straw, B.E.; Meuten, D.J. Physical examination. In Diseases of Swine, 7th ed.; Leman, A.D., Ed.; Iowa State University Press: Ames, IA, USA, 1992; p. 794. [Google Scholar]
- Meemken, D.; Blaha, T.; Hotzel, H.; Strommenger, B.; Klein, G.; Ehricht, R.; Monecke, S.; Kehrenberg, C. Genotypic and phenotypic characterization of S. aureus isolates from wild boars. Appl. Environ. Microbiol. 2013, 79, 1739–1742. [Google Scholar] [CrossRef]
- Seinige, D.; Von Altrock, A.; Kehrenberg, C. Genetic diversity and antibiotic susceptibility of S. aureus isolates from wild boars. Comp. Immunol. Microbiol. Infect. Dis. 2017, 54, 7–12. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority) Panel on Biological Hazards (BIOHAZ). Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 2011, 9, 2322. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 9th ed.; Approved Standard CLSI document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Prüller, S.; Rensch, U.; Meemken, D.; Kaspar, H.; Kopp, P.A.; Klein, G.; Kehrenberg, C. Antimicrobial susceptibility of Bordetella bronchiseptica isolates from swine and companion animals and detection of resistance genes. PLoS ONE 2015, 10, e0135703. [Google Scholar] [CrossRef] [PubMed]
- Saladin, M.; Cao, V.T.B.; Lambert, T.; Donay, J.-L.; Herrmann, J.-L.; Ould-Hocine, Z.; Verdet, C.; Delisle, F.; Philippon, A.; Arlet, G. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef]
- Chmelnitsky, I.; Carmeli, Y.; Leavitt, A.; Schwaber, M.J.; Navon-Venezia, S. CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple E. coli clones isolated in Tel Aviv, Israel. Antimicrob. Agents Chemother. 2005, 49, 4745–4750. [Google Scholar] [CrossRef]
- Chander, Y.; Oliveira, S.; Goyal, S.M. Characterisation of ceftiofur resistance in swine bacterial pathogens. Vet. J. 2011, 139. [Google Scholar] [CrossRef]
- Müller, A.; Jansen, W.; Grabowski, N.T.; Kehrenberg, C. Characterization of S. enterica serovars recovered from meat products legally and illegally imported into the EU reveals the presence of multiresistant and AmpC-producing isolates. Gut. Pathog. 2018, 10, 40. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- SAS® Institute Inc. Base SAS® 9.4 Procedures Guide: Statistical Procedures, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2013. [Google Scholar]
- Calvez, S.; Fournel, C.; Douet, D.G.; Daniel, P. Pulsed-field gel electrophoresis and multi locus sequence typing for characterizing genotype variability of Yersinia ruckeri isolated from farmed fish in France. Vet. Res. 2015, 46, 73. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, 278. [Google Scholar] [CrossRef]
- Aarestrup, F.M. The livestock reservoir for antimicrobial resistance: A personal view on changing patterns of risks, effects of interventions and the way forward. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140085. [Google Scholar] [CrossRef]
- Deutscher Jagdverband (DJV). DJV-Handbuch Jagd [German Hunting Association Handbook]; DJV-Service & Marketing, GmbH: Bonn, Germany, 2016. [Google Scholar]
- Schulz, J.; Friese, A.; Klees, S.; Tenhagen, B.A.; Fetsch, A.; Rösler, U.; Hartung, J. Longitudinal study of the contamination of air and of soil surfaces in the vicinity of pig barns by livestock-associated methicillin-resistant S. aureus. Appl. Environ. Microbiol. 2012, 78, 5666–5671. [Google Scholar] [CrossRef]
- von Salviati, C.; Laube, H.; Guerra, B.; Roesler, U.; Friese, A. Emission of ESBL/AmpC-producing E. coli from pig fattening farms to surrounding areas. Vet. Microbiol. 2015, 175, 77–84. [Google Scholar] [CrossRef]
- Lombardini, M.; Meriggi, A.; Fozzi, A. Factors influencing wild boar damage to agricultural crops in Sardinia (Italy). Curr. Zool. 2017, 63, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Konstantin, B.; Stillfreid, M. Berlin-Hauptstadt der Wildschweine. In Wildforschung Baden-Württemberg, Proceedings of the Schwarzwildtagung, Vortragsveranstaltung zu aktuellen Themen, Hofgartensaal Aulendorf, Germany, 12 October 2016; Landwirtschaftliches Zentrum für Rinderhaltung, Grünlandwirtschaft, Milchwirtschaft, Wild und Fischerei Baden-Württemberg (LAZBW)—Wildforschungsstelle Aulendorf: Aulendorf, Germany, 2017; Volume 12, pp. 17–22. Available online: https://www.schwarzwild-hainich.de/docs/Tagungsband_Aulendorf.pdf (accessed on 5 February 2021).
- Sauter-Louis, C.J.; Forth, H.; Probst, C.; Staubach, C.; Hlinak, A.; Rudovsky, A.; Holland, D.; Schlieben, P.; Göldner, M.; Schatz, J.; et al. Joining the club: First detection of African swine fever in wild boar in Germany. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Wasyl, D.; Zając, M.; Lalak, A.; Skarżyńska, M.; Samcik, I.; Kwit, R.; Jabłoński, A.; Bocian, L.; Woźniakowski, G.; Hoszowski, A.; et al. Antimicrobial resistance in E. coli isolated from wild animals in Poland. Microb. Drug Resist. 2018, 24, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Literak, I.; Dolejska, M.; Radimersky, T.; Klimes, J.; Friedman, M.; Aarestrup, F.M.; Hasman, H.; Cizek, A. Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: Multiresistant E. coli producing extended-spectrum beta-lactamases in wild boars. J. Appl. Microbiol. 2010, 108, 1702–1711. [Google Scholar] [CrossRef]
- BVL (Bundesamt für Verbraucherschutz und Lebensmittelsicherheit), 2017. BVL-Report 12.2, Berichte zur Lebensmittelsicherheit—Zoonosen-Monitoring 2016. Berlin, Germany. Available online: https://www.bvl.bund.de/ZoonosenMonitoring (accessed on 20 March 2020).
- BVL (Bundesamt für Verbraucherschutz und Lebensmittelsicherheit), 2018. BVL-Report 13.2, Berichte zur Lebensmittelsicherheit—Zoonosen-Monitoring 2017. Berlin, Germany. Available online: https://www.bvl.bund.de/ZoonosenMonitoring (accessed on 7 April 2020).
- Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eur. Surveill. 2008, 13, 19044. [Google Scholar]
- Day, M.J.; Rodríguez, I.; van Essen-Zandbergen, A.; Dierikx, C.; Kadlec, K.; Schink, A.-K.; Wu, G.; Chattaway, M.A.; DoNascimento, V.; Wain, J.; et al. Diversity of STs, plasmids and ESBL genes among E. coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 2016, 71, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- García-Cobos, S.; Köck, R.; Mellmann, A.; Frenzel, J.; Friedrich, A.W.; Rossen, J.W.A. Molecular typing of enterobacteriaceae from pig holdings in North-Western Germany reveals extended- Spectrum and AmpC β-lactamases producing but no carbapenem resistant ones. PLoS ONE 2015, 10, e0134533. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Stamm, I.; Grobbel, M.; Kopp, P.A.; Guerra, B.; Stubbe, M.; Doi, Y.; Zong, Z.; Kola, A.; et al. CTX-M-15-D-ST648 E. coli from companion animals and horses: Another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 2014, 69, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, N.; Casas-Díaz, E.; Porrero, C.M.; Mateos, A.; Domínguez, L.; Lavín, S.; Serrano, E. Food-borne zoonotic pathogens and antimicrobial resistance of indicator bacteria in urban wild boars in Barcelona, Spain. Vet. Microbiol. 2013, 167, 686–689. [Google Scholar] [CrossRef] [PubMed]

| ESBL/AmpC-Positive | ESBL/AmpC-Negative | |
|---|---|---|
| Qualitative data | ||
| Sex | ||
| Male (n = 187) | 8.0% (4.5–12.9%, n = 15) | 92.0% (87.1–95.4%, n = 172) |
| Female (n = 186) | 3.8% (1.5–7.6%, n = 7) | 96.2% (92.4–98.5%, n = 179) |
| Age group (years) | ||
| 0–1 (n = 197) | 6.1% (3.2–10.4%, n = 12) | 93.9% (89.6–96.8%, n = 185) |
| 1–2 (n = 99) | 5.1% (1.7–11.4%, n = 5) | 94.9% (88.6–98.3%, n = 94) |
| >2 (n = 78) | 6.4% (2.1–14.3%, n = 5) | 93.6% (85.7–97.9%, n = 73) |
| Nutritional status | ||
| Normal (n = 334) | 5.7% (3.5–8.7%, n = 19) | 94.3% (91.3–96.5%, n = 315) |
| Low (n = 41) | 7.3% (1.5–19.9%, n = 3) | 92.7% (80.1–98.5%, n = 38) |
| Sampling Region | ||
| Northern German Lowlands (n = 181) | 5.5% (2.7–9.9%, n = 10) | 94.5% (90.1–97.3%, n = 171) |
| Middle & Southern Germany (n = 194) | 6.2% (3.2–10.6%, n = 12) | 93.8% (89.4–96.7%, n = 182) |
| Quantitative data | ||
| Average weight (kg) | 40.95 (28.86–53.04) | 39.03 (36.68–41.38) |
| Average population density of county (people/km2) | 192.64 (158.67–226.6) | 149.26 (140.05–158.47) |
| Effect | Odds Ratio | 95% CI | p-Value |
|---|---|---|---|
| Sex: male (vs. female) | 2.23 | 0.89–5.60 | 0.088 |
| Age: | |||
| <1 year (vs. 1–2 years) | 1.22 | 0.42–3.56 | 0.717 |
| >2 years (vs. <1 year) | 1.06 | 0.36–3.10 | 0.921 |
| 1–2 years (vs. >2 years) | 0.78 | 0.22–2.78 | 0.698 |
| Nutritional status: low (vs. normal) | 1.31 | 0.37–4.63 | 0.676 |
| Region: north (vs. mid/south) | 1.13 | 0.48–2.68 | 0.786 |
| Population density 1 | 8.67 | 1.45–51.89 | 0.018 |
| Weight | 1.00 | 0.98–1.02 | 0.708 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holtmann, A.R.; Meemken, D.; Müller, A.; Seinige, D.; Büttner, K.; Failing, K.; Kehrenberg, C. Wild Boars Carry Extended-Spectrum β-Lactamase- and AmpC-Producing Escherichia coli. Microorganisms 2021, 9, 367. https://doi.org/10.3390/microorganisms9020367
Holtmann AR, Meemken D, Müller A, Seinige D, Büttner K, Failing K, Kehrenberg C. Wild Boars Carry Extended-Spectrum β-Lactamase- and AmpC-Producing Escherichia coli. Microorganisms. 2021; 9(2):367. https://doi.org/10.3390/microorganisms9020367
Chicago/Turabian StyleHoltmann, Anna R., Diana Meemken, Anja Müller, Diana Seinige, Kathrin Büttner, Klaus Failing, and Corinna Kehrenberg. 2021. "Wild Boars Carry Extended-Spectrum β-Lactamase- and AmpC-Producing Escherichia coli" Microorganisms 9, no. 2: 367. https://doi.org/10.3390/microorganisms9020367
APA StyleHoltmann, A. R., Meemken, D., Müller, A., Seinige, D., Büttner, K., Failing, K., & Kehrenberg, C. (2021). Wild Boars Carry Extended-Spectrum β-Lactamase- and AmpC-Producing Escherichia coli. Microorganisms, 9(2), 367. https://doi.org/10.3390/microorganisms9020367






