Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Media, and Cell Growth
2.2. Generation of Gene Deletion Mutants in C. diphtheriae
2.3. Recombinant Plasmids
2.4. Protein Purification
2.5. RT-FeDEx Assay
2.6. RNA-Seq Analysis
2.7. Reverse Transcription Polymerase Chain Reactions
2.8. Detection of Diphtheria Toxin
2.9. Electron Microscopy
2.10. Chrome Azurol S (CAS) Assay
2.11. Caenorhabditis Elegans Killing Assay
2.12. Statistical Analysis
3. Results
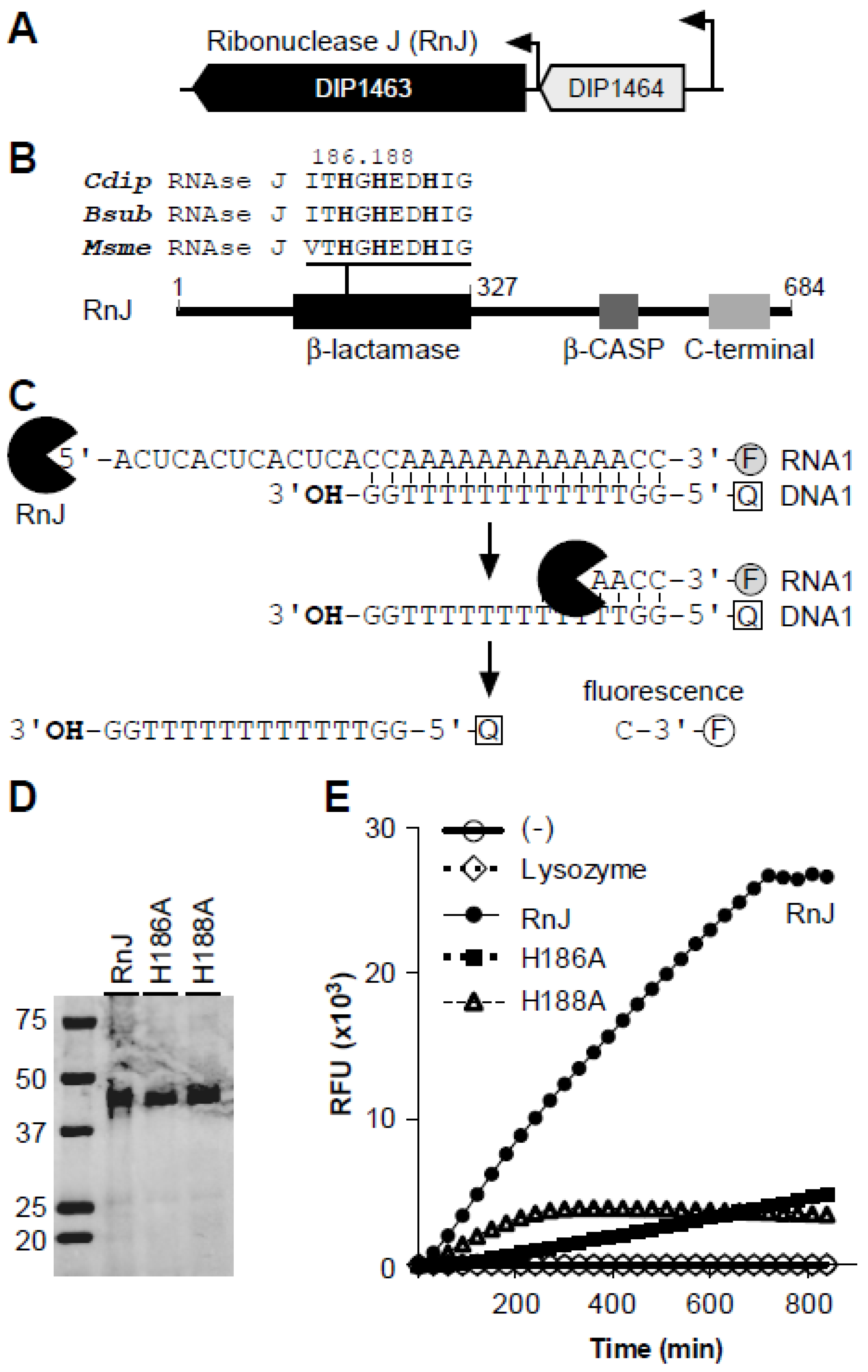
3.1. C. Diphtheriae Encodes Ribonuclease J
3.2. Genetic Disruption of rnj Alters Cell Growth and Morphology
3.3. Transcriptome Analysis of the rnj Mutant
3.4. Reduction of Secreted Diphtheria Toxin in the ∆rnJ Mutant
3.5. Virulence Attenuation of the ∆rnj Mutant in a Caenorhabditis Elegans Model of Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [Green Version]
- Hoe, C.-H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.-H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Repoila, F.; Majdalani, N.; Gottesman, S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: The RpoS paradigm. Mol. Microbiol. 2003, 48, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Bechhofer, D.H. Nucleotide specificity in bacterial mRNA recycling. Proc. Natl. Acad. Sci. USA 2013, 110, 8765–8766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, M.P.; Foley, P.L.; Belasco, J.G. Messenger RNA Degradation in Bacterial Cells. Annu. Rev. Genet. 2014, 48, 537–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apirion, D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics 1978, 90, 659–671. [Google Scholar]
- Grunberg-Manago, M. Messenger RNA Stability and Its Role in Control of Gene Expression in Bacteria and Phages. Annu. Rev. Genet. 1999, 33, 193–227. [Google Scholar] [CrossRef]
- Carpousis, A.J. The RNA Degradosome of Escherichia coli: An mRNA-Degrading Machine Assembled on RNase E. Annu. Rev. Microbiol. 2007, 61, 71–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condon, C.; Putzer, H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002, 30, 5339–5346. [Google Scholar] [CrossRef] [Green Version]
- Lehnik-Habrink, M.; Newman, J.; Rothe, F.M.; Solovyova, A.S.; Rodrigues, C.; Herzberg, C.; Commichau, F.M.; Lewis, R.J.; Stülke, J. RNase Y in Bacillus subtilis: A Natively Disordered Protein That is the Functional Equivalent of RNase E from Escherichia coli. J. Bacteriol. 2011, 193, 5431–5441. [Google Scholar] [CrossRef] [Green Version]
- Even, S. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005, 33, 2141–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condon, C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010, 7, 316–321. [Google Scholar] [CrossRef] [Green Version]
- Mathy, N.; Hébert, A.; Mervelet, P.; Bénard, L.; Dorléans, A.; De La Sierra-Gallay, I.L.; Noirot, P.; Putzer, H.; Condon, C. Bacillus subtilisribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 2010, 75, 489–498. [Google Scholar] [CrossRef]
- Bugrysheva, J.V.; Scott, J.R. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol. Microbiol. 2009, 75, 731–743. [Google Scholar] [CrossRef]
- Gao, P.; Pinkston, K.L.; Nallapareddy, S.R.; Van Hoof, A.; Murray, B.E.; Harvey, B.R. Enterococcus faecalis rnjB is Required for Pilin Gene Expression and Biofilm Formation. J. Bacteriol. 2010, 192, 5489–5498. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Pinkston, K.L.; Bourgogne, A.; Murray, B.E.; Van Hoof, A.; Harvey, B.R. Functional studies of E. faecalis RNase J2 and its role in virulence and fitness. PLoS ONE 2017, 12, e0175212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Merritt, J.; Qi, F.; Khajotia, S.; Liu, N. RNases J1 and J2 are critical pleiotropic regulators in Streptococcus mutans. Microbiology 2015, 161, 797–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, R.; Shinde, P.; Zou, Z.; Kreth, J.; Merritt, J. Examining the Protein Interactome and Subcellular Localization of RNase J2 Complexes in Streptococcus mutans. Front. Microbiol. 2019, 10, 2150. [Google Scholar] [CrossRef] [PubMed]
- Laalami, S.; Zig, L.; Putzer, H. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2013, 71, 1799–1828. [Google Scholar] [CrossRef] [Green Version]
- Pei, X.-Y.; Bralley, P.; Jones, G.H.; Luisi, B.F. Linkage of catalysis and 5’ end recognition in ribonuclease RNase J. Nucleic Acids Res. 2015, 43, 8066–8076. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.E.; Leong, V.; Ortega, J.; Elliot, M.A. Development, Antibiotic Production, and Ribosome Assembly in Streptomyces venezuelae Are Impacted by RNase J and RNase III Deletion. J. Bacteriol. 2014, 196, 4253–4267. [Google Scholar] [CrossRef] [Green Version]
- Bralley, P.; Aseem, M.; Jones, G.H. SCO5745, a Bifunctional RNase J Ortholog, Affects Antibiotic Production in Streptomyces coelicolor. J. Bacteriol. 2014, 196, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Rogers, E.A.; Das, A.; Ton-That, H. Adhesion by Pathogenic Corynebacteria. Adv. Exp. Med. Biol. 2011, 715, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; Kandel, J. Structure and activity of diphtheria toxin. I. Thiol-dependent dissociation of a fraction of toxin into enzymically active and inactive fragments. J. Biol. Chem. 1971, 246, 24. [Google Scholar]
- Uchida, T.; Gill, D.M.; Pappenheimer, A.M. Mutation in the Structural Gene for Diphtheria Toxin carried by Temperate Phage β. Nat. New Biol. 1971, 233, 8–11. [Google Scholar] [CrossRef]
- Boyd, J.; Oza, M.N.; Murphy, J.R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 1990, 87, 5968–5972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourel, G.; Phalipon, A.; Kaczorek, M. Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtoxR, in Corynebacterium diphtheriae. Infect. Immun. 1989, 57, 3221–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, R.K. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 2000, 181, 156–167. [Google Scholar] [CrossRef] [Green Version]
- Kunkle, C.A.; Schmitt, M.P. Analysis of a DtxR-Regulated Iron Transport and Siderophore Biosynthesis Gene Cluster in Corynebacterium diphtheriae. J. Bacteriol. 2005, 187, 422–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trost, E.; Blom, J.; Soares, S.D.C.; Huang, I.-H.; Al-Dilaimi, A.; Schröder, J.; Jaenicke, S.; Dorella, F.A.; Rocha, F.S.; Miyoshi, A.; et al. Pangenomic Study of Corynebacterium diphtheriae That Provides Insights into the Genomic Diversity of Pathogenic Isolates from Cases of Classical Diphtheria, Endocarditis, and Pneumonia. J. Bacteriol. 2012, 194, 3199–3215. [Google Scholar] [CrossRef] [Green Version]
- Ton-That, H.; Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003, 50, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Swierczynski, A.; Ton-That, H. Type III Pilus of Corynebacteria: Pilus Length is Determined by the Level of Its Major Pilin Subunit. J. Bacteriol. 2006, 188, 6318–6325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, A.H.; Ton-That, H. Assembly of Distinct Pilus Structures on the Surface of Corynebacterium diphtheriae. J. Bacteriol. 2006, 188, 1526–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reardon-Robinson, M.E.; Osipiuk, J.; Jooya, N.; Chang, C.; Joachimiak, A.; Das, A.; Ton-That, H. A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriaeis essential for viability, pilus assembly, toxin production and virulence. Mol. Microbiol. 2015, 98, 1037–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broadway, M.M.; Rogers, E.A.; Chang, C.; Huang, I.-H.; Dwivedi, P.; Yildirim, S.; Schmitt, M.P.; Das, A.; Ton-That, H. Pilus Gene Pool Variation and the Virulence of Corynebacterium diphtheriae Clinical Isolates during Infection of a Nematode. J. Bacteriol. 2013, 195, 3774–3783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.; Nguyen, M.T.; Ton-That, H. Genetic Manipulation of Corynebacterium diphtheriae and Other Corynebacterium Species. Curr. Protoc. Microbiol. 2020, 58, 111. [Google Scholar] [CrossRef]
- Wittchen, M.; Busche, T.; Gaspar, A.H.; Lee, J.H.; Ton-That, H.; Kalinowski, J.; Tauch, A. Transcriptome sequencing of the human pathogen Corynebacterium diphtheriae NCTC 13129 provides detailed insights into its transcriptional landscape and into DtxR-mediated transcriptional regulation. BMC Genom. 2018, 19, 82. [Google Scholar] [CrossRef] [Green Version]
- Reyes, O.; Guyonvarch, A.; Bonainy, C.; Salti, V.; David, F.; LeBlon, G. ‘Integron’-bearing vectors: A method suitable for stable chromosomal integration in highly restrictive Corynebacteria. Gene 1991, 107, 61–68. [Google Scholar] [CrossRef]
- Luong, T.T.; Reardon-Robinson, M.E.; Siegel, S.D.; Ton-That, H. Reoxidation of the Thiol-Disulfide Oxidoreductase MdbA by a Bacterial Vitamin K Epoxide Reductase in the Biofilm-Forming Actinobacterium Actinomyces oris. J. Bacteriol. 2017, 199, e00817-16. [Google Scholar] [CrossRef] [Green Version]
- Siegel, S.D.; Amer, B.R.; Wu, C.; Sawaya, M.R.; Gosschalk, J.E.; Clubb, R.T.; Ton-That, H.; Otto, M.; Wu, H. Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein. mBio 2019, 10, e01580-18. [Google Scholar] [CrossRef] [Green Version]
- Sinturel, F.; Pellegrini, O.; Xiang, S.; Tong, L.; Condon, C.; Bénard, L. Real-time fluorescence detection of exoribonucleases. RNA 2009, 15, 2057–2062. [Google Scholar] [CrossRef] [Green Version]
- Louden, B.C.; Lynne, A.M.; Haarmann, D. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ton-That, H. Corynebacterium diphtheriae Virulence Analyses Using a Caenorhabditis elegans Model. Curr. Protoc. Microbiol. 2020, 58, e109. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A.; Sifri, C.D.; Mylonakis, E.; Qin, X.; Singh, K.V.; Murray, B.E.; Calderwood, S.B.; Ausubel, F.M. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 2001, 98, 10892–10897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, D.; De Almeida, D.F. Isolation and characterization of a new temperature-sensitive cell division mutant of Escherichia coli K-12. J. Bacteriol. 1975, 124, 1502–1507. [Google Scholar] [CrossRef] [Green Version]
- Zellmeier, S.; Zuber, U.; Schumann, W.; Wiegert, T. The absence of FtsH metalloprotease activity causes overexpression of the sigmaW-controlled pbpE gene, resulting in filamentous growth of Bacillus subtilis. J. Bacteriol. 2003, 185, 973–982. [Google Scholar] [CrossRef] [Green Version]
- Tao, X.; Schiering, N.; Zeng, H.-Y.; Ringe, D.; Murphy, J.R. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 1994, 14, 191–197. [Google Scholar] [CrossRef]
- Zajdowicz, S.; Haller, J.C.; Krafft, A.E.; Hunsucker, S.W.; Mant, C.T.; Duncan, M.W.; Hodges, R.S.; Jones, D.N.M.; Holmes, R.K. Purification and Structural Characterization of Siderophore (Corynebactin) from Corynebacterium diphtheriae. PLoS ONE 2012, 7, e34591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, L.M.; Cryz, S.J.; Holmes, R.K. Genetic and biochemical evidence for a siderophore-dependent iron transport system in Corynebacterium diphtheriae. Infect. Immun. 1984, 45, 143–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, C.E.; Burgos, J.M.; Schmitt, M.P. Analysis of Novel Iron-Regulated, Surface-Anchored Hemin-Binding Proteins in Corynebacterium diphtheriae. J. Bacteriol. 2013, 195, 2852–2863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Lu, M.; Zhang, H.; Hu, J.; Zhou, C.; Xu, Q.; Shah, A.M.U.H.; Xu, H.; Wang, L.; Hua, Y. Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J. Nucleic Acids Res. 2015, 43, 5550–5559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deikus, G.; Condon, C.; Bechhofer, D.H. Role of Bacillus subtilis RNase J1 Endonuclease and 5′-Exonuclease Activities in trp Leader RNA Turnover. J. Biol. Chem. 2008, 283, 17158–17167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langklotz, S.; Baumann, U.; Narberhaus, F. Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta Bioenerg. 2012, 1823, 40–48. [Google Scholar] [CrossRef]
- Yanofsky, C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA 2007, 13, 1141–1154. [Google Scholar] [CrossRef] [Green Version]




| Strains | Width (µm) | Length (µm) |
|---|---|---|
| WT | 0.64 ± 0.06 | 1.26 ± 0.40 |
| ∆rnj | 0.99 ± 0.13 | 1.39 ± 0.36 |
| WT/pFtsH | 0.90 ± 0.07 | 1.32 ± 0.33 |
| Gene Name | Function | Log2-Fold Change |
|---|---|---|
| iutD | Putative ABC-type iron protein | 1.03 |
| ciuC | Iron transport system membrane protein | 1.18 |
| ciuE | Corynebactin biosynthetic gene | 1.24 |
| sufB | Fe–S cluster assembly protein | 1.33 |
| sufR | Iron–sulfur cluster biosynthesis transcriptional regulator | 1.56 |
| piuB | Iron-uptake factor | 1.83 |
| irp4 | DtxR-dependent, iron-regulated promoter/operator | 2.65 |
| irp6A | Ferrisiderophore receptor (putative ABC transporter) | −1.30 |
| irp5 | DtxR-dependent, iron-regulated promoter/operator | −1.33 |
| Strains | +Fe3+ | −Fe3+ | ||
|---|---|---|---|---|
| OD600 | Siderophore Production (%) | OD600 | Siderophore Production (%) | |
| WT | 3.38 ± 0.08 | 19.30 ± 2.99 | 2.62 ± 0.29 | 67.77 ± 2.90 |
| ∆rnj | 2.92 ± 0.31 | 20.42 ± 1.97 | 2.38 ± 0.23 | 44.49 ± 3.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luong, T.T.; Nguyen, M.T.; Chen, Y.-W.; Chang, C.; Lee, J.H.; Wittchen, M.; Ton-That, H.; Cruz, M.; Garsin, D.A.; Das, A.; et al. Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae. Microorganisms 2021, 9, 389. https://doi.org/10.3390/microorganisms9020389
Luong TT, Nguyen MT, Chen Y-W, Chang C, Lee JH, Wittchen M, Ton-That H, Cruz M, Garsin DA, Das A, et al. Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae. Microorganisms. 2021; 9(2):389. https://doi.org/10.3390/microorganisms9020389
Chicago/Turabian StyleLuong, Truc Thanh, Minh Tan Nguyen, Yi-Wei Chen, Chungyu Chang, Ju Huck Lee, Manuel Wittchen, HyLam Ton-That, Melissa Cruz, Danielle A. Garsin, Asis Das, and et al. 2021. "Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae" Microorganisms 9, no. 2: 389. https://doi.org/10.3390/microorganisms9020389
APA StyleLuong, T. T., Nguyen, M. T., Chen, Y.-W., Chang, C., Lee, J. H., Wittchen, M., Ton-That, H., Cruz, M., Garsin, D. A., Das, A., Tauch, A., & Ton-That, H. (2021). Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae. Microorganisms, 9(2), 389. https://doi.org/10.3390/microorganisms9020389






