From Data Mining of Chitinophaga sp. Genome to Enzyme Discovery of a Hyperthermophilic Metallocarboxypeptidase
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Sequencing of Chitinophaga sp. Genome
2.2. Selection of Potentially Interesting Proteases Via Data Mining
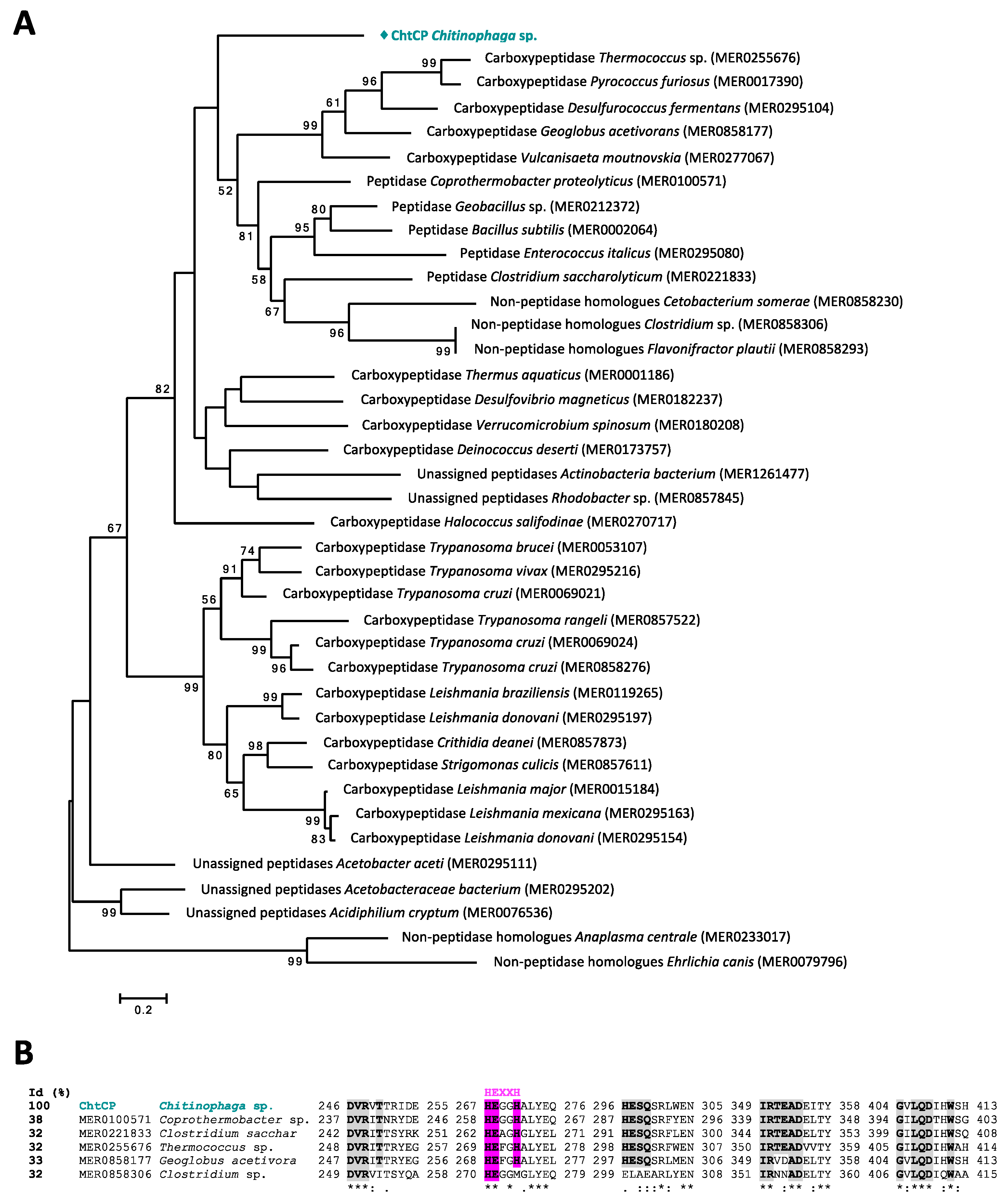
2.3. Sequence Analysis of Gene Chtcp
2.4. Cloning of Gene Chtcp
2.5. Expression, Extraction and Purification of Recombinant ChtCP
2.6. Enzyme Activity and Kinetic Parameters of ChtCP
2.7. The Effect of Temperature, pH and Cofactors on ChtCP Activity
2.8. Determination of Tm Via Differential Scanning Fluorometry (DSC)
2.9. Crystallisation of ChtCP
2.10. Data Collection and Structure Determination of ChtCP
3. Results
3.1. From Prediction of Peptidases in the Chitinophaga sp. Genome to Sequence Analysis of ChtCP
3.2. Expression and Evaluation of the ChtCP Quaternary Structure
3.3. ChtCP, An Enzyme with a Wide Range of pH and Temperature
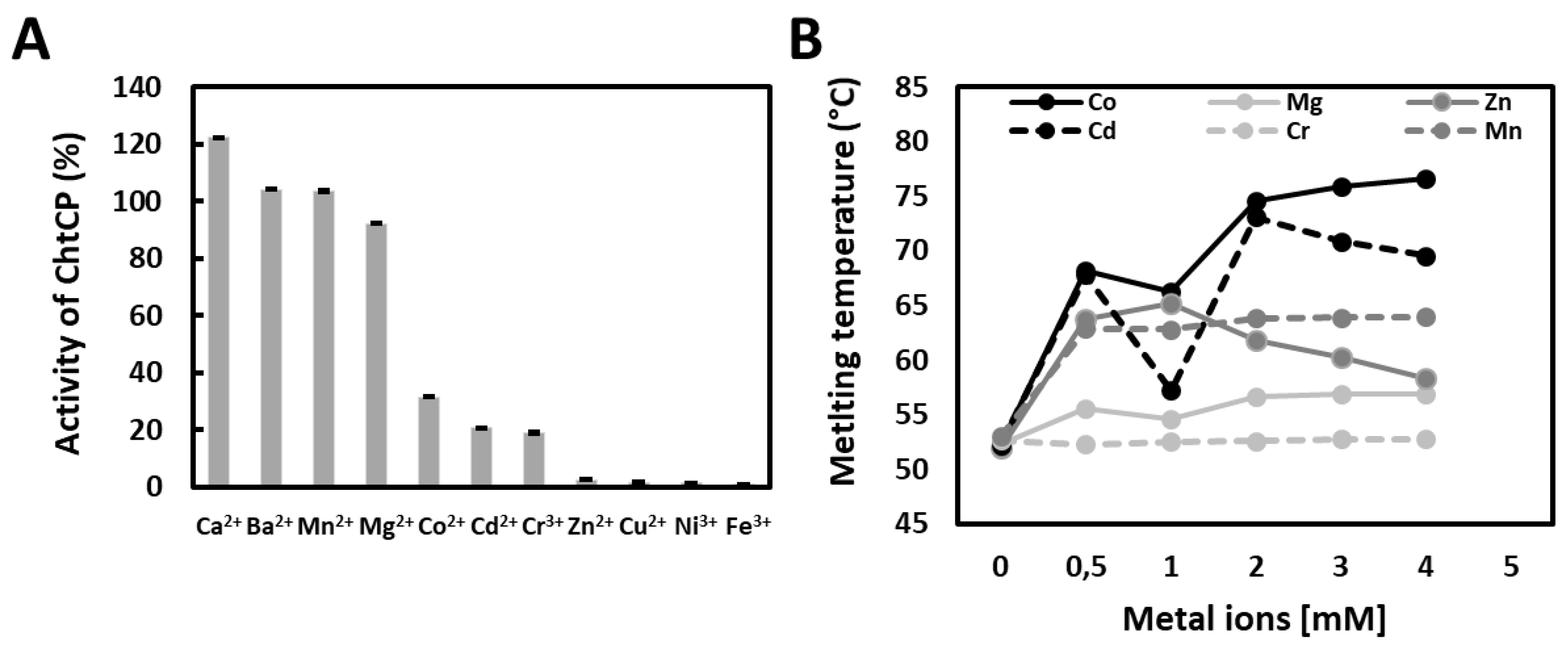
3.4. The Effect of Metal Ions on Enzymatic Activity and on Temperature of Denaturation
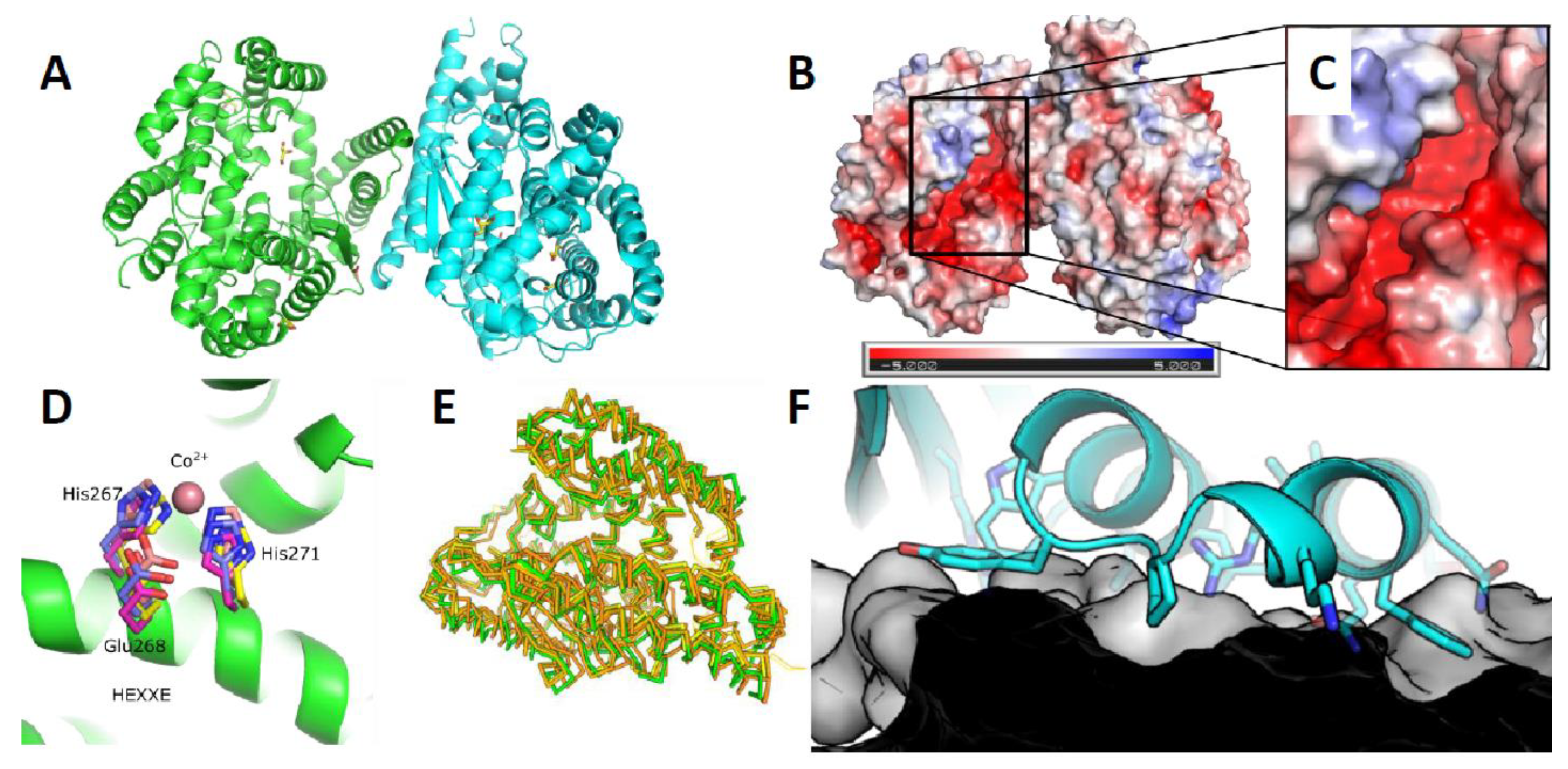
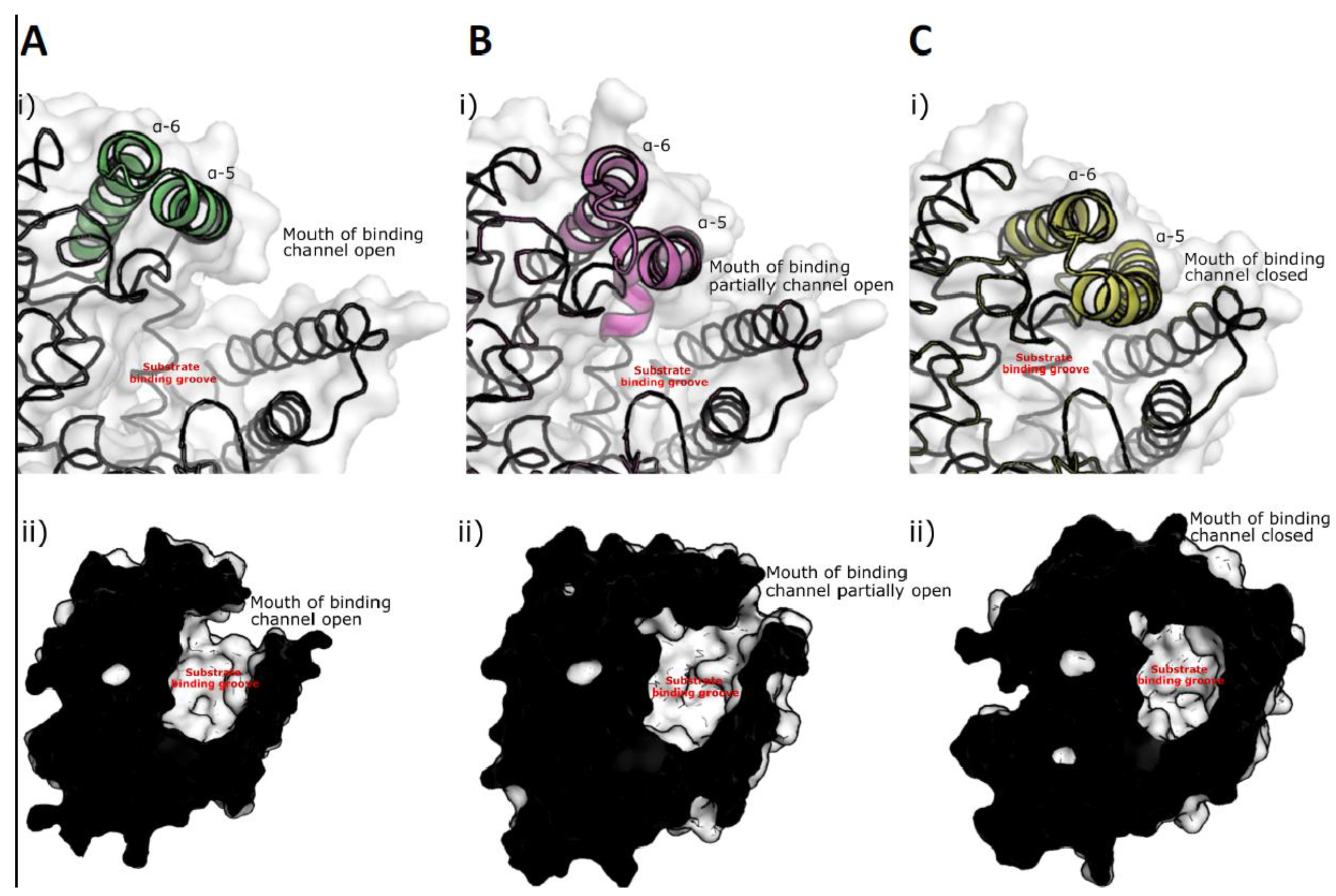
3.5. Crystal Scructure of ChtCP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dumorné, K.; Córdova, D.C.; Astorga-Eló, M.; Renganathan, P. Extremozymes: A potential source for industrial applications. J. Microbiol. Biotechnol. 2017, 27, 649–659. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S. Directed evolution: Tailoring biocatalysts for industrial applications. Crit. Rev. Biotechnol. 2013, 33, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Adrio, J.L.; Demain, A.L. Microbial Enzymes: Tools for Biotechnological Processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.E.; Choi, Y.J.; Lee, S.J.; Kang, K.-H.; Lee, H.; Oh, Y.H.; Lee, S.H.; Park, S.J.; Lee, S.Y. Metabolic engineering of Escherichia coli for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from glucose. Appl. Microbiol. Biotechnol. 2014, 98, 95–104. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, T.W.; Kim, M.K.; Lee, S.Y.; Lim, S.-C. Advanced bacterial polyhydroxyalkanoates: Towards a versatile and sustainable platform for unnatural tailor-made polyesters. Biotechnol. Adv. 2012, 30, 1196–1206. [Google Scholar] [CrossRef]
- Sun, J.; Alper, H.S. Metabolic engineering of strains: From industrial-scale to lab-scale chemical production. J. Ind. Microbiol. Biotechnol. 2015, 42, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Q.; Jiang, X.R. Next generation industrial biotechnology based on extremophilic bacteria. Curr. Opin. Biotechnol. 2018, 50, 94–100. [Google Scholar] [CrossRef]
- Belda, E.; van Heck, R.G.A.; Lopez-Sanchez, M.J.; Cruveiller, S.; Barbe, V.; Fraser, C.; Klenk, H.P.; Petersen, J.; Morgat, A.; Nikel, P.I.; et al. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 2016, 18, 3403–3424. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Van Lanen, S.G.; Baltz, R.H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado Sierra, E.; Pereira, M.R.; Maester, T.C.; Gomes-Pepe, E.S.; Mendoza, E.R.; de Macedo Lemos, E.G. Halotolerant aminopeptidase M29 from Mesorhizobium SEMIA 3007 with biotechnological potential and its impact on biofilm synthesis. Sci. Rep. 2017, 7, 10684. [Google Scholar] [CrossRef] [Green Version]
- Pereira, M.R.; Mercaldi, G.F.; Maester, T.C.; Balan, A.; de Macedo Lemos, E.G. Est16, a new esterase isolated from a metagenomic library of a microbial consortium specializing in diesel oil degradation. PLoS ONE 2015, 10, e0133723. [Google Scholar] [CrossRef] [PubMed]
- Maester, T.C.; Pereira, M.R.; Machado Sierra, E.G.; Balan, A.; de Macedo Lemos, E.G. Characterization of EST3: A metagenome-derived esterase with suitable properties for biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 5815–5827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes-Pepe, E.S.; Machado Sierra, E.G.; Pereira, M.R.; Castellane, T.C.L.; de Macedo Lemos, E.G. Bg10: A Novel Metagenomics Alcohol-Tolerant and Glucose-Stimulated GH1 ß-Glucosidase Suitable for Lactose-Free Milk Preparation. PLoS ONE 2016, 11, e0167932. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, T.G.; Abt, B.; Spring, S.; Lapidus, A.; Nolan, M.; Tice, H.; Copeland, A.; Cheng, J.F.; Chen, F.; Bruce, D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034 T). Stand. Genom. Sci. 2010, 2, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangkhobol, V.; Skerman, V.B.D. Chitinophaga, a New Genus of Chitinolytic Myxobacteria. Int. J. Syst. Bacteriol. 1981, 31, 285–293. [Google Scholar] [CrossRef]
- Kämpfer, P.; Young, C.C.; Sridhar, K.R.; Arun, A.B.; Lai, W.A.; Shen, F.T.; Rekha, P.D. Transfer of [Flexibacter] sancti, [Flexibacter] filiformis, [Flexibacter] japonensis and [Cytophaga] arvensicola to the genus Chitinophaga and description of Chitinophaga skermanii sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 2223–2228. [Google Scholar] [CrossRef] [Green Version]
- Weon, H.Y.; Yoo, S.H.; Kim, Y.J.; Son, J.A.; Kim, B.Y.; Kwon, S.W.; Koo, B.S.; Würdemann, D.; Tindall, B.J.; Pukall, R.; et al. Chitinophaga niabensis sp. nov. and Chitinophaga niastensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 1267–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishi, L.T.; Lopes, E.M.; Fernandes, C.C.; Fernandes, G.C.; Sacco, L.P.; Carareto Alves, L.M.; Lemos, E.G.M. Draft Genome Sequence of a Chitinophaga Strain Isolated from a Lignocellulose Biomass-Degrading Consortium. Genome Announc. 2017, 5, e01056-16. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Lv, Y.; Li, A.; Feng, G.; Bao, G.; Zhu, H.; Tan, Z. Chitinophaga silvisoli sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 909–913. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, X.; Song, H.; Liu, Y.; Cheng, Q. Chitinophaga alhagiae sp. nov., isolated from rhizosphere soil of Alhagi sparsifolia. Int. J. Syst. Evol. Microbiol. 2019, 69, 1179–1184. [Google Scholar] [CrossRef]
- Zong, Y.; Wu, M.; Liu, X.; Jin, Y.; Wang, G.; Li, M. Chitinophaga lutea sp. nov., isolated from arsenic-contaminated soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Chung, E.J.; Song, G.C.; Bibi, F.; Jeon, C.O.; Chung, Y.R. Chitinophaga eiseniae sp. nov., isolated from vermicompost. Int. J. Syst. Evol. Microbiol. 2011, 61, 2373–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsbrink, J.; Tuveng, T.R.; Pope, P.B.; Bulone, V.; Eijsink, V.G.H.; Brumer, H.; McKee, L.S. Proteomic insights into mannan degradation and protein secretion by the forest floor bacterium Chitinophaga pinensis. J. Proteom. 2017, 156, 63–74. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef]
- Nawrath, T.; Gerth, K.; Müller, R.; Schulz, S. The biosynthesis of the aroma volatile 2-methyltetrahydrothiophen-3-one in the bacterium Chitinophaga Fx7914. ChemBioChem 2010, 11, 1914–1919. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Gerth, K.; Steinmetz, H.; Reinecke, S.; Kessler, W.; Kirschning, A.; Müller, R. Elansolid A3, a unique p-quinone methide antibiotic from Chitinophaga sancti. Chem. A Eur. J. 2011, 17, 7739–7744. [Google Scholar] [CrossRef] [PubMed]
- Dickschat, J.S.; Rinkel, J.; Rabe, P.; Kashkooli, A.B.; Bouwmeester, H.J. 18-Hydroxydolabella-3,7-diene synthase—A diterpene synthase from Chitinophaga pinensis. Beilstein J. Org. Chem. 2017, 13, 1770–1780. [Google Scholar] [CrossRef]
- McKee, L.S.; Martínez-Abad, A.; Ruthes, A.C.; Vilaplana, F.; Brumer, H. Focused Metabolism of β-Glucans by the Soil Bacteroidetes Species Chitinophaga pinensis. Appl. Environ. Microbiol. 2019, 85, 85. [Google Scholar] [CrossRef] [Green Version]
- Lei, F.; Zhao, Q.; Sun-Waterhouse, D.; Zhao, M. Characterization of a salt-tolerant aminopeptidase from marine Bacillus licheniformis SWJS33 that improves hydrolysis and debittering efficiency for soy protein isolate. Food Chem. 2017, 214, 347–353. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.; Ren, J.; Zhao, M. Effects of Ultrasound Pretreatment on the Enzymatic Hydrolysis of Soy Protein Isolates and on the Emulsifying Properties of Hydrolysates. J. Agric. Food Chem. 2011, 59, 2600–2609. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Dhanasingh, I.; Ahn, J.-S.; Jin, H.-S.; Choi, J.M.; Lee, S.H.; Lee, D.-W. Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1. Biochem. Biophys. Res. Commun. 2015, 468, 927–933. [Google Scholar] [CrossRef]
- Tayyab, M.; Rashid, N.; Angkawidjaja, C.; Kanaya, S.; Akhtar, M. Highly active metallocarboxypeptidase from newly isolated Geobacillus strain SBS-4S: Cloning and characterization. J. Biosci. Bioeng. 2011, 111, 259–265. [Google Scholar] [CrossRef]
- Okai, M.; Yamamura, A.; Hayakawa, K.; Tsutsui, S.; Miyazono, K.; Lee, W.C.; Nagata, K.; Inoue, Y.; Tanokura, M. Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase. Biochem. Biophys. Res. Commun. 2017, 484, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Minagawa, E.; Taguchi, H.; Matsuzawa, H.; Ohta, T.; Kaminogawa, S.; Yamauchi, K. Purification and Characterization of a Thermostable Carboxypeptidase (Carboxypeptidase Taq) from Thermus aquaticus YT-1. Biosci. Biotechnol. Biochem. 1992, 56, 1839–1844. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Kim, Y.J.; Bae, S.S.; Jeon, J.H.; Lim, J.K.; Kang, S.G.; Lee, J.-H. Overexpression and Characterization of a Carboxypeptidase from the Hyperthermophilic Archaeon Thermococcus sp. NA1. Biosci. Biotechnol. Biochem. 2006, 70, 1140–1147. [Google Scholar] [CrossRef] [Green Version]
- Cheng, T.C.; Ramakrishnan, V.; Chan, S.I. Purification and characterization of a cobalt-activated carboxypeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. Protein Sci. 2008, 8, 2474–2486. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.M.; Isaza, C.E.; White, J.D.; Chen, R.P.-Y.; Liang, G.F.-C.; He, H.T.-F.; Chan, S.I.; Chan, M.K. Insight into the substrate length restriction of M32 carboxypeptidases: Characterization of two distinct subfamilies. Proteins 2009, 77, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Anandharaj, M.; Sivasankari, B.; Siddharthan, N.; Rani, R.P.; Sivakumar, S. Production, Purification, and Biochemical Characterization of Thermostable Metallo-Protease from Novel Bacillus alkalitelluris TWI3 Isolated from Tannery Waste. Appl. Biochem. Biotechnol. 2016, 178, 1666–1686. [Google Scholar] [CrossRef]
- Traore, T.; Junping, Z.; Zhiming, R.; Genchu, H.; Taowei, Y.; Xian, Z.; Meijuan, X. Cloning, over-expression, and characterization of a new carboxypeptidase A gene of Bacillus pumilus ML413 in Bacillus subtilis 168. Afr. J. Biotechnol. 2016, 15, 684–695. [Google Scholar] [CrossRef]
- Van Albertus, A.D.; Baukje, F.; Petrus Jacobus, T.D. Carboxypeptidase for Cheese Ripening. Patent No. WO2005074695A1, 18 August 2004. [Google Scholar]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Schomburg, I.; Placzek, S.; Jeske, L.; Ulbrich, M.; Xiao, M.; Sensen, C.W.; Schomburg, D. BRENDA in 2015: Exciting developments in its 25th year of existence. Nucleic Acids Res. 2015, 43, D439–D446. [Google Scholar] [CrossRef] [Green Version]
- Consortium, T.U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Ku, T.; Lu, P.; Chan, C.; Wang, T.; Lai, S.; Lyu, P.; Hsiao, N. Predicting melting temperature directly from protein sequences. Comput. Biol. Chem. 2009, 33, 445–450. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Peränen, J.; Rikkonen, M.; Hyvönen, M.; Kääriäinen, L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996, 236, 371–373. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Doi, E.; Shibata, D.; Matoba, T. Modified colorimetric ninhydrin methods for peptidase assay. Anal. Biochem. 1981, 118, 173–184. [Google Scholar] [CrossRef]
- Kazuhiko, I.; Hiroyasu, I.; Ikuo, M.; Yutaka, K.; Hisasi, K. Novel Bifunctional Hyperthermostable Carboxypeptidase/Aminoacylase from Pyrococcus horikoshii OT3. Appl. Environ. Microbiol. 2001, 67, 673–679. [Google Scholar] [CrossRef] [Green Version]
- Kirsch, P.D.; Ekerdt, J.G. KaleidaGraph: Graphing and Data Analysis. Version 3.5 for Windows Synergy Software, 2457 Perkiomen Ave., Reading, PA 19606-2049. www.Synergy.com. $155.00. J. Am. Chem. Soc. 2000, 122, 11755. [Google Scholar] [CrossRef]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Marion, A.; Groll, M.; Scharf, D.H.; Scherlach, K.; Glaser, M.; Sievers, H.; Schuster, M.; Hertweck, C.; Brakhage, A.A.; Antes, I.; et al. Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ. ACS Chem. Biol. 2017, 12, 1874–1882. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Number 4 Collaborative Computational Project. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.; Jamdar, S.N.; Ghosh, B.; Yadav, P.; Kumar, A.; Kundu, S.; Goyal, V.D.; Makde, R.D. Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Arndt, J.W.; Hao, B.; Ramakrishnan, V.; Cheng, T.; Chan, S.I.; Chan, M.K. Crystal structure of a novel carboxypeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. Structure 2002, 10, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Niemirowicz, G.; Fernández, D.; Solà, M.; Cazzulo, J.J.; Avilés, F.X.; Gomis-Rüth, F.X. The molecular analysis of Trypanosoma cruzi metallocarboxypeptidase 1 provides insight into fold and substrate specificity. Mol. Microbiol. 2008, 70, 853–866. [Google Scholar] [CrossRef]
- Colombo, S.; D’auria, S.; Fusi, P.; Zecca, L.; Raia, C.A.; Tortora, P. Purification and characterization of a thermostable carboxypeptidase from the extreme thermophilic archaebacterium Sulfolobus solfataricus. Eur. J. Biochem. 1992, 206, 349–357. [Google Scholar] [CrossRef]
- Lee, S.H.; Taguchi, H.; Yoshimura, E.; Minagawa, E.; Kaminogawa, S.; Ohta, T.; Matsuzawa, H. Carboxypeptidase Taq, a thermostable zinc enzyme, from Thermus aquaticus YT-1: Molecular cloning, sequencing, and expression of the encoding gene in Escherichia coli. Biosci. Biotechnol. Biochem. 1994, 58, 1490–1495. [Google Scholar] [CrossRef] [Green Version]
- Petrik, I.D.; Liu, J.; Lu, Y. Metalloenzyme design and engineering through strategic modifications of native protein scaffolds. Curr. Opin. Chem. Biol. 2014, 19, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Brandts, J.F.; Hu, C.Q.; Lin, L.N.; Mas, M.T. A simple model for proteins with interacting domains. Applications to scanning calorimetry data. Biochemistry 1989, 28, 8588–8596. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Using Dali for Protein Structure Comparison. In Structural Bioinformatics; Humana Press: New York, NY, USA, 2020; pp. 29–42. [Google Scholar]
- Merheb-Dini, C.; Cabral, H.; Leite, R.S.R.; Zanphorlin, L.M.; Okamoto, D.N.; Rodriguez, G.O.B.; Juliano, L.; Arantes, E.C.; Gomes, E.; Da Silva, R. Biochemical and functional characterization of a metalloprotease from the thermophilic fungus thermoascus aurantiacus. J. Agric. Food Chem. 2009, 57, 9210–9217. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.C.; Inampudi, K.K.; Bean, D.P.; Wilson, C.J. Understanding Thermal Adaptation of Enzymes through the Multistate Rational Design and Stability Prediction of 100 Adenylate Kinases. Structure 2014, 22, 218–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-masó, J.A.; Bordanaba-ruiseco, L.; Sanz, M.; Menéndez, M.; Del Solar, G. Metal-Induced Stabilization and Activation of Plasmid Replication Initiator RepB. Front. Mol. Biosci. 2016, 3, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
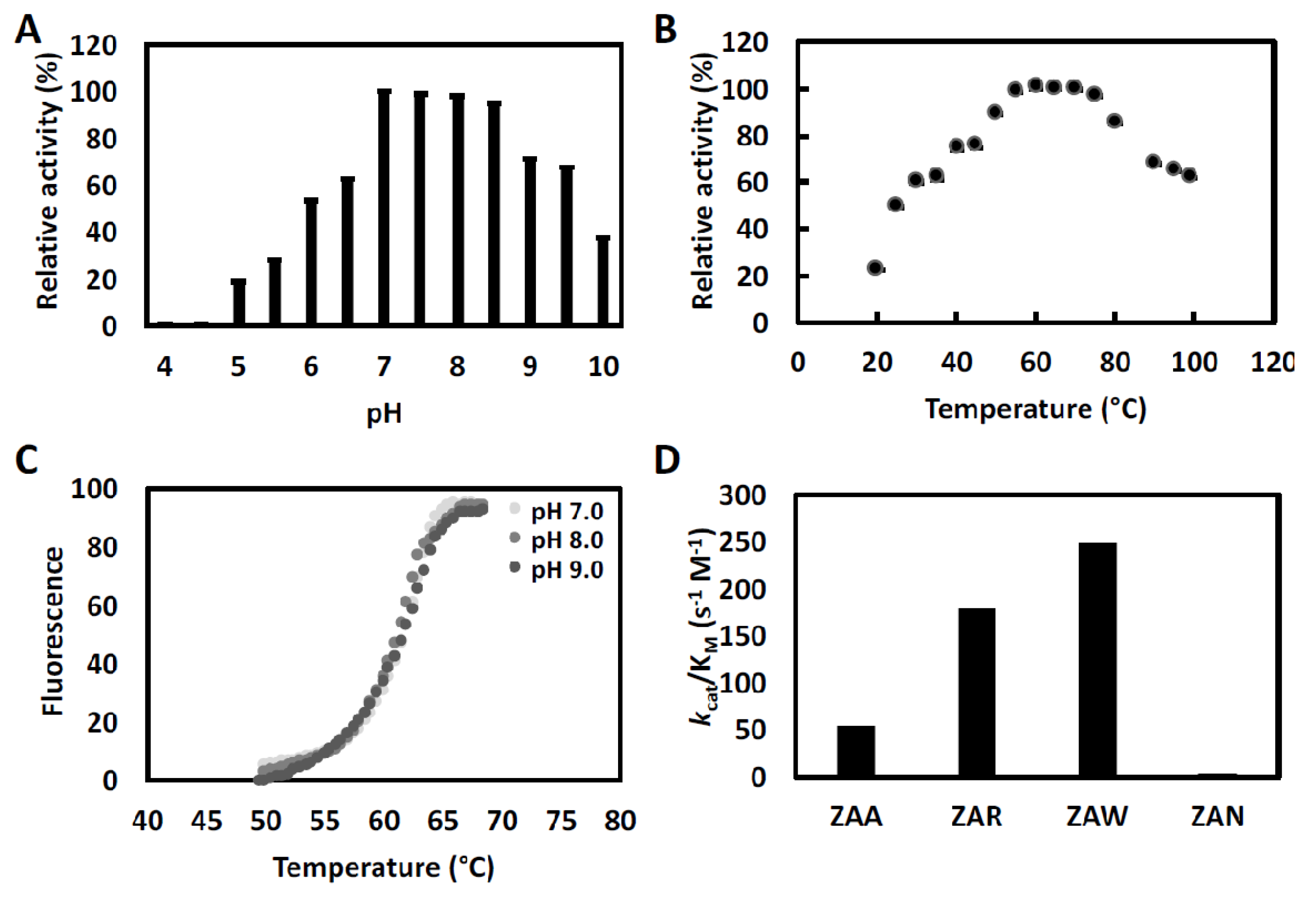
| Substrate | Abbreviation | kcat (s−1) | kM (M) | kcat/kM (s−1·M−1) |
|---|---|---|---|---|
| Z-Ala-Ala-OH | ZAA | 1.55 | 2.81 × 10−2 | 5.53 × 101 |
| Z-Ala-Arg-OH | ZAR | 6.28 × 10−1 | 3.51 × 10−3 | 1.79 × 102 |
| Z-Ala-Trp-OH | ZAW | 2.72 × 10−1 | 1.10 × 10−3 | 2.48 × 102 |
| Z-Ala-Asn-OH | ZAN | 1.48 × 10−-1 | 3.41 × 10−2 | 4.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, G.C.; Sierra, E.G.M.; Brear, P.; Pereira, M.R.; Lemos, E.G.M. From Data Mining of Chitinophaga sp. Genome to Enzyme Discovery of a Hyperthermophilic Metallocarboxypeptidase. Microorganisms 2021, 9, 393. https://doi.org/10.3390/microorganisms9020393
Fernandes GC, Sierra EGM, Brear P, Pereira MR, Lemos EGM. From Data Mining of Chitinophaga sp. Genome to Enzyme Discovery of a Hyperthermophilic Metallocarboxypeptidase. Microorganisms. 2021; 9(2):393. https://doi.org/10.3390/microorganisms9020393
Chicago/Turabian StyleFernandes, Gabriela Cabral, Elwi Guillermo Machado Sierra, Paul Brear, Mariana Rangel Pereira, and Eliana G. M. Lemos. 2021. "From Data Mining of Chitinophaga sp. Genome to Enzyme Discovery of a Hyperthermophilic Metallocarboxypeptidase" Microorganisms 9, no. 2: 393. https://doi.org/10.3390/microorganisms9020393






