Gut Digestive Function and Microbiome after Correction of Experimental Dysbiosis in Rats by Indigenous Bifidobacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Rat Model of Antibiotic-Associated Dysbiosis
2.3. Autoprobiotic Strains of Bifidobacteria
2.4. Design of the Study
2.5. Microbiome Study
2.5.1. Metagenome Analysis
2.5.2. OTU Generation
2.6. Biochemical Analysis
2.7. Statistical Analysis
3. Results
3.1. Health Status and Body Weight of Rats
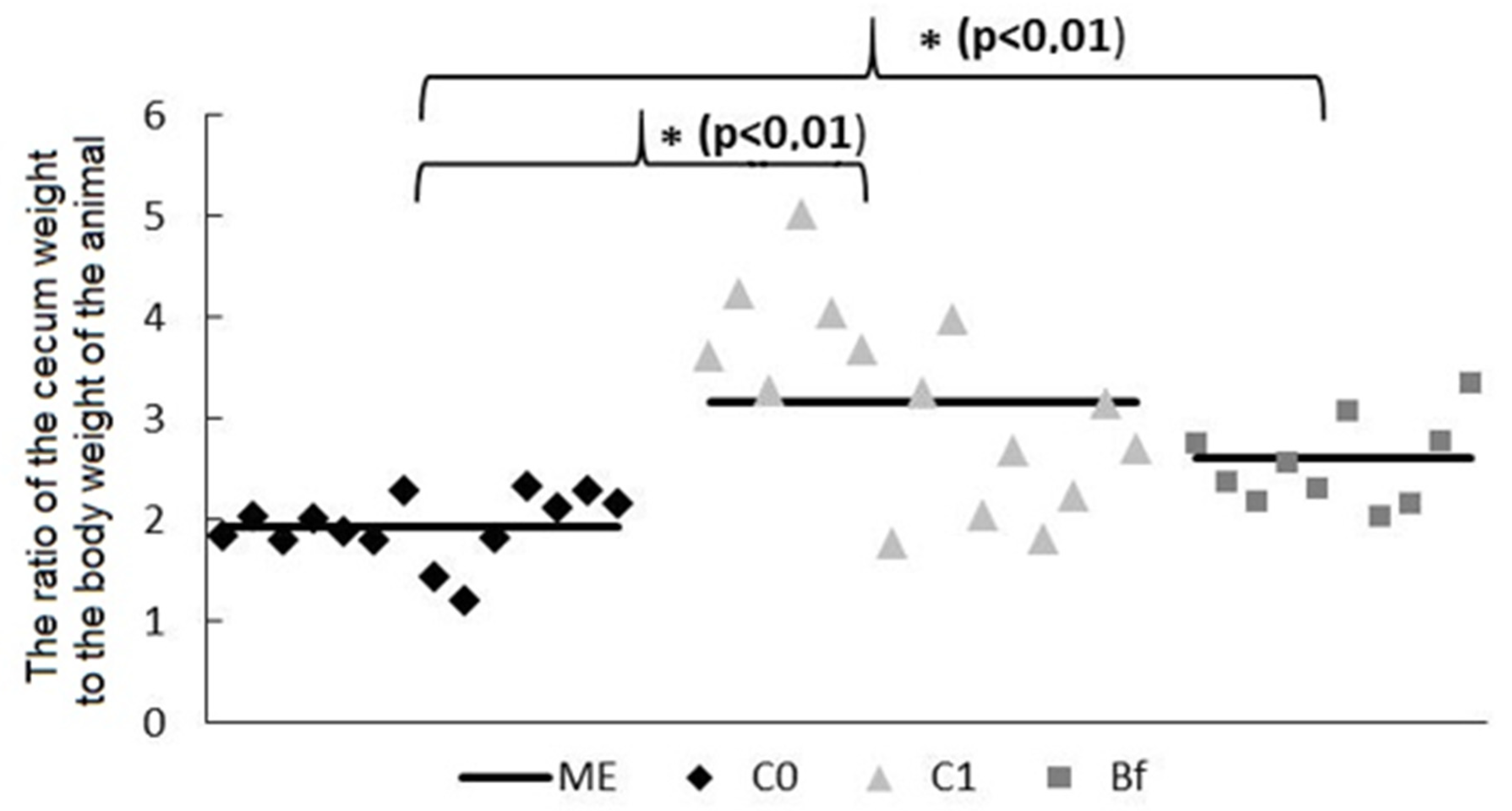
3.2. Mass of Mucosa and Chyme in Intestinal Segments
3.3. Microbiota Study by qPCR
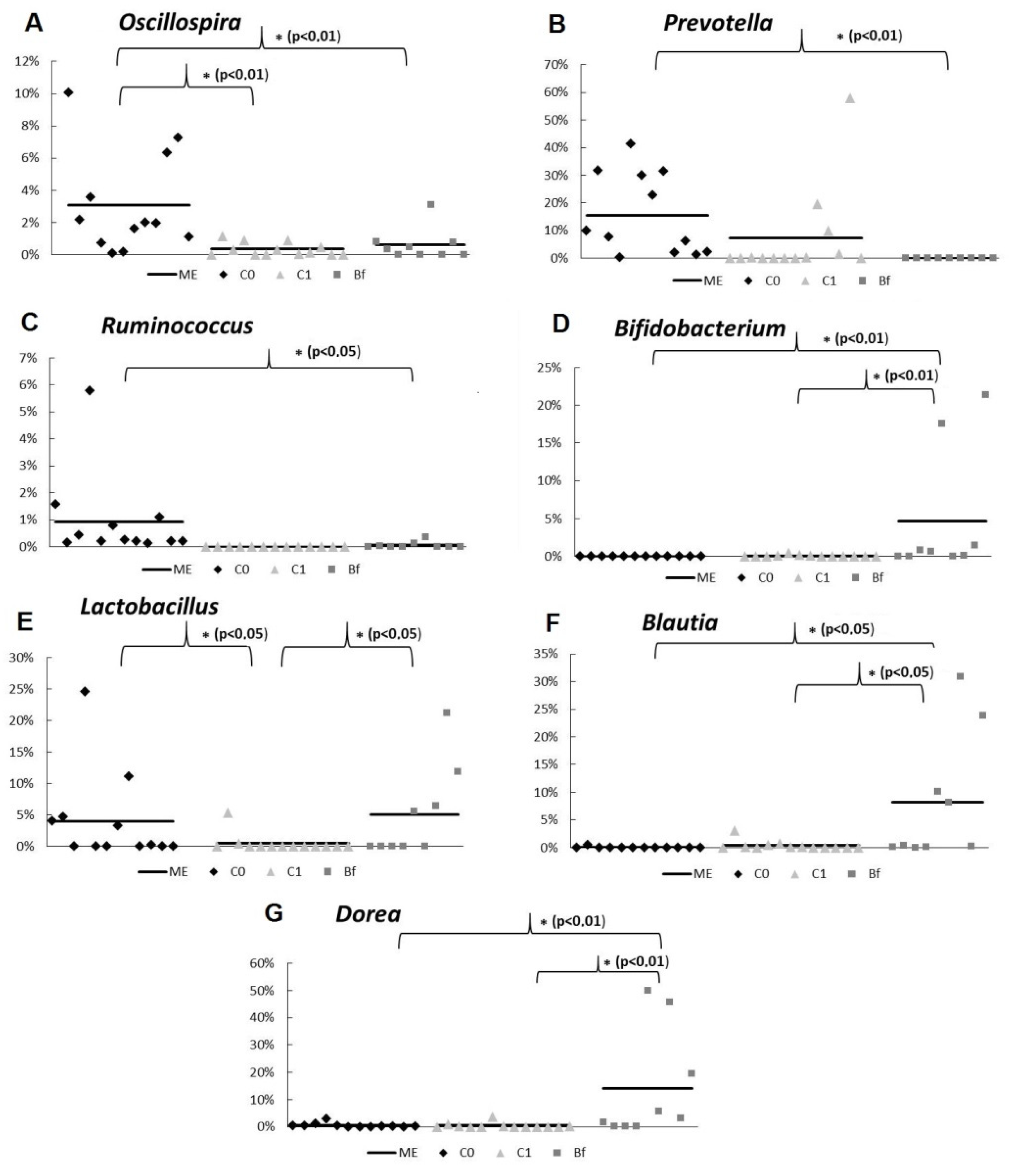
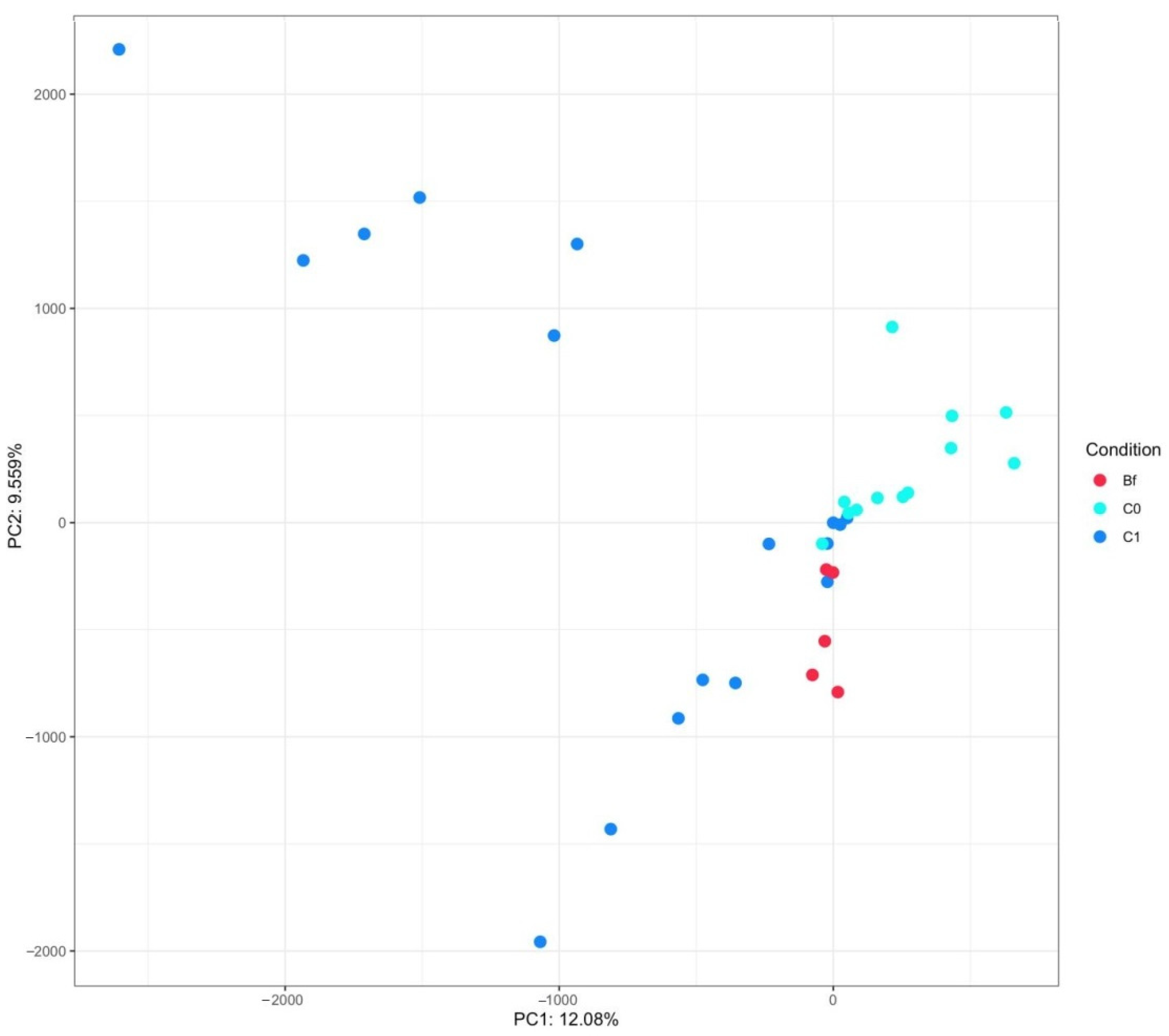
3.4. Metagenome Analysis of Microbiota
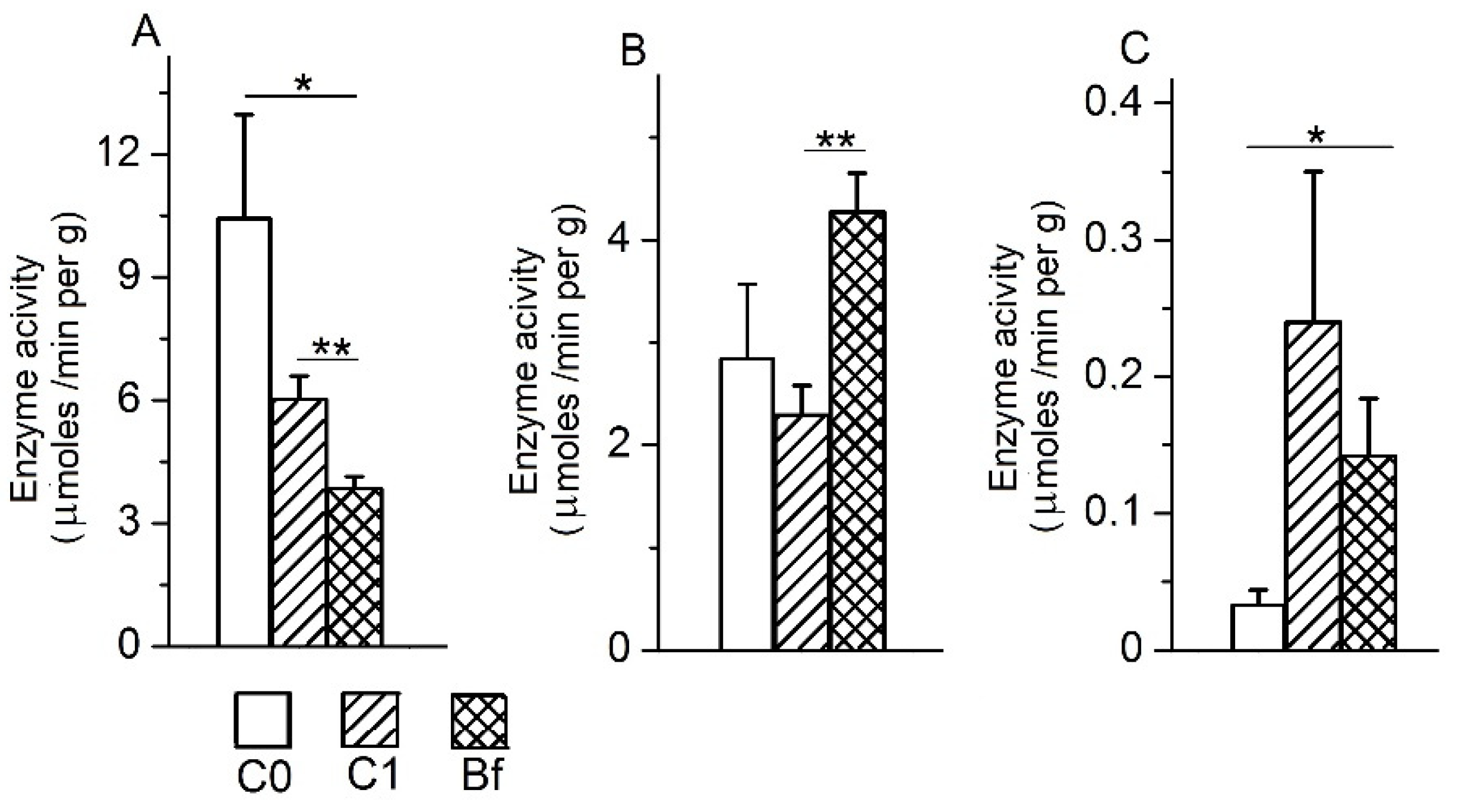
3.5. The Activity of Intestinal Enzymes in the Mucosa of Various Parts of the Intestine
3.6. The Activity of Intestinal Enzymes in the Chyme of the Colon
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ugolev, A.M. Adequate Nutrition Theory and Trophology; Nauka: Leningrad, Russia, 1991; p. 272. [Google Scholar]
- Claus, S.P.; Ellero, S.L.; Berger, B.; Krause, L.; Bruttin, A.; Molina, J.; Paris, A.; Want, E.J.; de Waziers, I.; Cloarec, O.; et al. Colonization-induced host-gut microbial metabolic interaction. mBio 2011, 2, e00271-10. [Google Scholar] [CrossRef] [Green Version]
- Lallès, J.P. Microbiota-host interplay at the gut epithelial level, health and nutrition. J. Anim. Sci. Biotechnol. 2016, 8, 7–66. [Google Scholar] [CrossRef] [PubMed]
- Tojo, R.; Suárez, A.; Clemente, M.G.; de los Reyes-Gavilán, C.G.; Margolles, A.; Gueimonde, M.; Ruas-Madiedo, P. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World J. Gastroenterol. 2014, 20, 15163–15176. [Google Scholar] [CrossRef]
- Wang, M.; Monaco, M.H.; Donovan, S.M. Impact of early gut microbiota on immune and metabolic development and function. Semin. Fetal Neonatal. Med. 2016, 21, 380–387. [Google Scholar] [CrossRef]
- Ling, X.; Linglong, P.; Weixia, D.; Hong, W. Protective Effects of bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of Caco-2 monolayers and in a Rat NEC model. PLoS ONE 2016, 11, e0161635. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Odamak, T.; Mitsuyama, E.; Hirosuke, S.; Xiao, J.-Z.; Osawa, R. Age-related changes in the composition of gut Bifidobacterium species. Curr. Microbiol. 2017, 8, 987–995. [Google Scholar] [CrossRef]
- Bahmani, S.; Azarpira, N.; Moazamian, E. Anti-colon cancer activity of Bifidobacterium metabolites on colon cancer cell line SW742. Turk. J. Gastroenterol. 2019, 30, 835–842. [Google Scholar] [CrossRef]
- Engevik, M.A.; Luk, B.; Chang-Graha, A.L.; Hall, A.; Herrmann, B.; Ruan, W.; Endres, B.T.; Shi, Z.; Garey, K.W.; Hyser, J.M.; et al. Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. mBio 2019, 10, e01087-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and their health-promoting effects. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Ermolenko, E.; Gromova, L.; Lavrenova, N.; Kotyleva, M.; Dmitrieva, Y.; Alekseeva, A.; Karaseva, A.; Kramskaya, T.; Lapidus, A.; Sepp, A.; et al. Effects of autoprobiotic consortium and fecal transplant on the digestive system and intestinal microbiota in the correction of experimental dysbiosis. Gastroenterol. Hepatol. Open Access 2020, 11, 198–206. [Google Scholar]
- Ermolenko, E.I.; Donets, V.N.; Dmitrieva, Y.V.; Ilyasov, Y.Y.; Suvorova, M.A.; Gromova, L.V. Influence of probiotic enterococci on functional characteristics of rat bowel under disbiosis induced by antibiotics. Bull. St. Petersburg State Univ. 2009, 11, 157–167. (In Russian) [Google Scholar]
- Tarasova, E.; Yermolenko, E.; Donets, V.; Sundukova, Z.; Bochkareva, A.; Borschev, I.; Suvorova, M.; Ilyasov, I.; Simanenkov, V.; Suvorov, A. The influence of probiotic Enterococcus faecium strain L5 on the microbiota and cytokines expression in rats with dysbiosis induced by antibiotics. Benef. Microbes 2010, 1, 265–270. [Google Scholar] [CrossRef]
- Ermolenko, E.; Gromova, L.; Borschev, Y.; Voeikova, A.; Karaseva, A.; Ermolenko, K.; Gruzdkov, A.; Suvorov, A. Influence of different probiotic lactic acid bacteria on microbiota and metabolism of rats with dysbiosis. Biosci. Microb. Food Health 2013, 32, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borshchev, Y.; Gromova, L.V.; Ermolenko, E.I.; Grefner, N.M.; Borshov, I.Y.; Gruzdkov, A.A. Responses of peptide hydrolases of the small and large intestines in rats on the administration of antibiotics. Rossiiskii fiziologicheskii zhurnal imeni IM Sechenova 2012, 98, 724–733. (In Russian) [Google Scholar]
- Gert, H.H.; Niels-Christiansen, L.-L.; Immerdal, L.; Nystrøm, B.T.; Danielsen, E.M. Intestinal alkaline phosphatase: Selective endocytosis from the enterocyte brush border during fat absorption. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G1325–G1332. [Google Scholar]
- Lynes, M.; Narisawa, S.; Millán, J.L.; Widmaier, E.P. Interactions between CD36 and global intestinal alkaline phosphatase in mouse small intestine and effects of high-fat diet. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R1738–R1747. [Google Scholar] [CrossRef] [Green Version]
- Estaki, M.; DeCoffe, D.; Gibson, D.L. Interplay between intestinal alkaline phosphatase, diet, gut microbes and immunity. World J. Gastroenterol. 2014, 20, 15650–15656. [Google Scholar] [CrossRef] [PubMed]
- Lallès, J.-P.; Orozco-Solís, R.; Bolaños-Jiménez, F.; de Coppet, P.; Le Dréan, G.; Segain, J.-P. Perinatal undernutrition alters intestinal alkaline phosphatase and its main transcription factors KLF4 and Cdx1 in adult offspring fed a high-fat diet. J. Nutr. Biochem. 2012, 23, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Lallès, J.-P. Luminal ATP: The missing link between intestinal alkaline phosphatase, the gut microbiota, and inflammation? Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G824–G825. [Google Scholar]
- Joshi, S.; Chen, L.; Winter, M.B.; Lin, Y.-L.; Yang, Y.; Shapovalova, M.; Smith, P.M.; Liu, C.; Li, F.; LeBeau, A.M. The rational design of therapeutic peptides for aminopeptidase N using a substrate-based approach. Sci. Rep. 2017, 7, 1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, W.; Girbig, F.; Corsiero, D.; Pfenninger, A.; Frick, W.; Jähne, G.; Rhein, M.; Wendler, W.; Lottspeich, F.; Hochleitner, E.O.; et al. Aminopeptidase N (CD13) is a molecular target of the cholesterol absorption inhibitor ezetimibe in the enterocyte brush border membrane. J. Biol. Chem. 2005, 280, 1306–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bank, U.; Tadje, J.; Täger, M.; Wolke, C.; Bukowska, A.; Ittenson, A.; Reinhold, D.; Helmuth, M.; Ansorge, S.; Shakespeare, A.; et al. Inhibition of alanyl-aminopeptidase on CD4+CD25+ regulatory T-cells enhances expression of FoxP3 and TGF-beta1 and ameliorates acute colitis in mice. Int. J. Mol. Med. 2007, 20, 483–492. [Google Scholar] [PubMed]




| Forward | 5′gcgtgcttaacacatgcaagtc3′ |
| Reverse | 5′cacccgtttccaggagctatt3′ |
| Oligonucleotide sequences of the TaqMan probe | 5′tcacgcattactcacccgttcgcc3′ |
| Groups | Treatment (1–3 Days) | Treatment (4–8 Days) | Analysis of Samples |
|---|---|---|---|
| C0 | Distilled water | PBS | Fecal samples were harvested 7 days before the start of administration of antibiotics for indigenous bifidobacteria strains’ isolation and preparation of autoprobiotic. Fecal samples harvested on days 0 and 9 of experiments were used for microbiota study. |
| C1 | Ampicillin + metronidazole | PBS | Fecal samples harvested on days 0 and 9 of experiments were used for microbiota study. Samples of mucosa and chyme were taken from various parts of the intestine on ninth day of experiments for analysis of the activities of digestive enzymes. |
| Bf | Ampicillin + metronidazole | Autoprobiotic Bifidobacterium spp. |
| V3-V4 16S Region Sequencing Primers V4 16S Region Sequencing Primers | Amplicon Size bp | |
|---|---|---|
| Forward (341) | tcgtcggcagcgtcagatgtgtataagagacagcctacgggnggcwgcag | 464 |
| Reverse (785) | gtctcgtgggctcggagatgtgtataagagacaggactachvgggtatctaatcc | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gromova, L.V.; Ermolenko, E.I.; Sepp, A.L.; Dmitrieva, Y.V.; Alekseeva, A.S.; Lavrenova, N.S.; Kotyleva, M.P.; Kramskaya, T.A.; Karaseva, A.B.; Suvorov, A.N.; et al. Gut Digestive Function and Microbiome after Correction of Experimental Dysbiosis in Rats by Indigenous Bifidobacteria. Microorganisms 2021, 9, 522. https://doi.org/10.3390/microorganisms9030522
Gromova LV, Ermolenko EI, Sepp AL, Dmitrieva YV, Alekseeva AS, Lavrenova NS, Kotyleva MP, Kramskaya TA, Karaseva AB, Suvorov AN, et al. Gut Digestive Function and Microbiome after Correction of Experimental Dysbiosis in Rats by Indigenous Bifidobacteria. Microorganisms. 2021; 9(3):522. https://doi.org/10.3390/microorganisms9030522
Chicago/Turabian StyleGromova, Lyudmila V., Elena I. Ermolenko, Anastasiya L. Sepp, Yulia V. Dmitrieva, Anna S. Alekseeva, Nadezhda S. Lavrenova, Mariya P. Kotyleva, Tatyana A. Kramskaya, Alena B. Karaseva, Alexandr N. Suvorov, and et al. 2021. "Gut Digestive Function and Microbiome after Correction of Experimental Dysbiosis in Rats by Indigenous Bifidobacteria" Microorganisms 9, no. 3: 522. https://doi.org/10.3390/microorganisms9030522
APA StyleGromova, L. V., Ermolenko, E. I., Sepp, A. L., Dmitrieva, Y. V., Alekseeva, A. S., Lavrenova, N. S., Kotyleva, M. P., Kramskaya, T. A., Karaseva, A. B., Suvorov, A. N., & Gruzdkov, A. A. (2021). Gut Digestive Function and Microbiome after Correction of Experimental Dysbiosis in Rats by Indigenous Bifidobacteria. Microorganisms, 9(3), 522. https://doi.org/10.3390/microorganisms9030522






