Novel Mode Engineering for β-Alanine Production in Escherichia coli with the Guide of Adaptive Laboratory Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Media and Culture Conditions
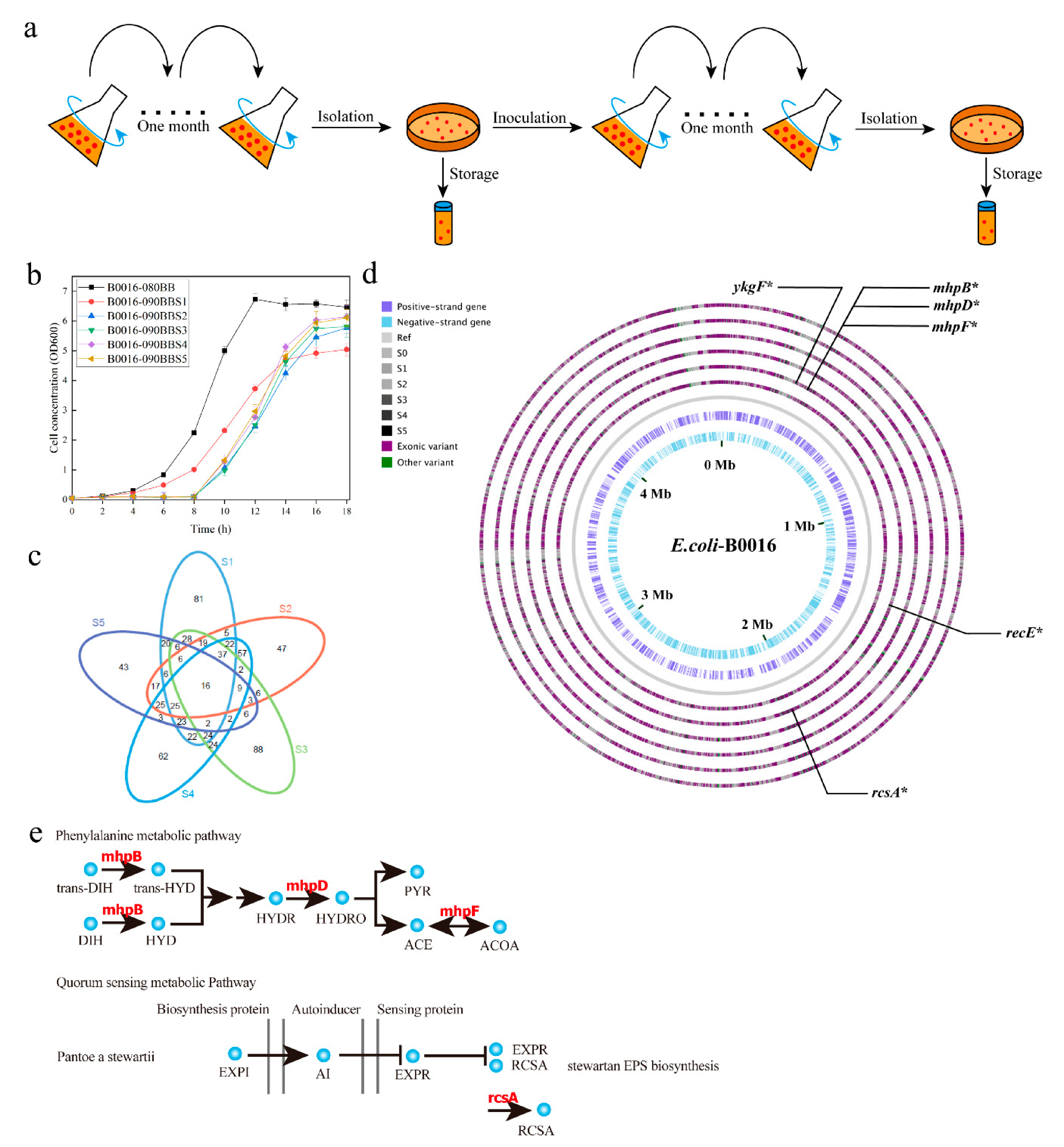
2.3. Adaptive Laboratory Evolution
2.3.1. Continuous Adaptive Laboratory Evolution
2.3.2. Marginal Effect-Assisted Adaptive Laboratory Evolution
2.4. Measurement of β-Alanine and L-Aspartic Acid
3. Results
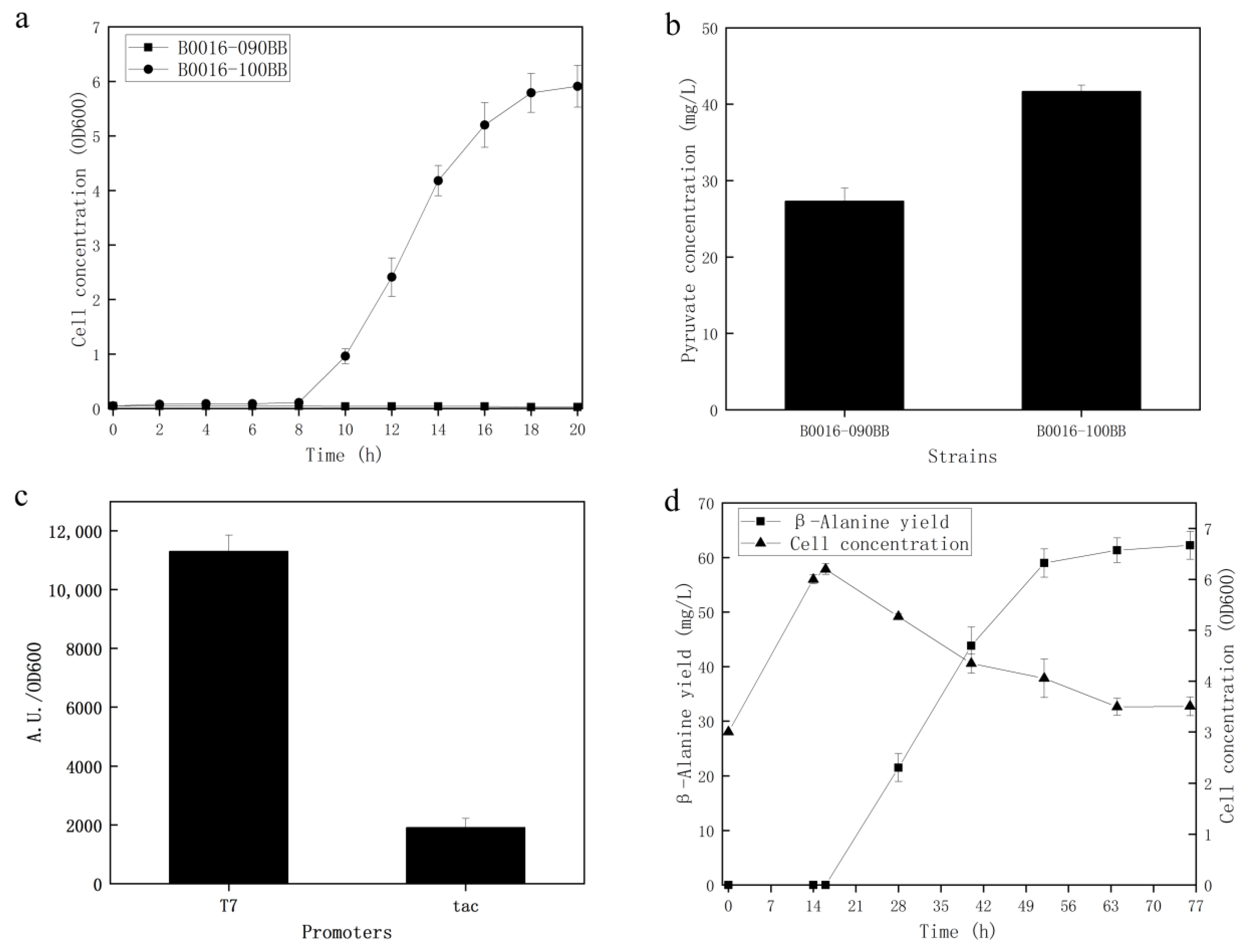
3.1. Evolution of PPC Deficient E. Coli Strains
3.2. Establishment of the GML by Whole Genome Sequencing of the Adapted Mutant Pool
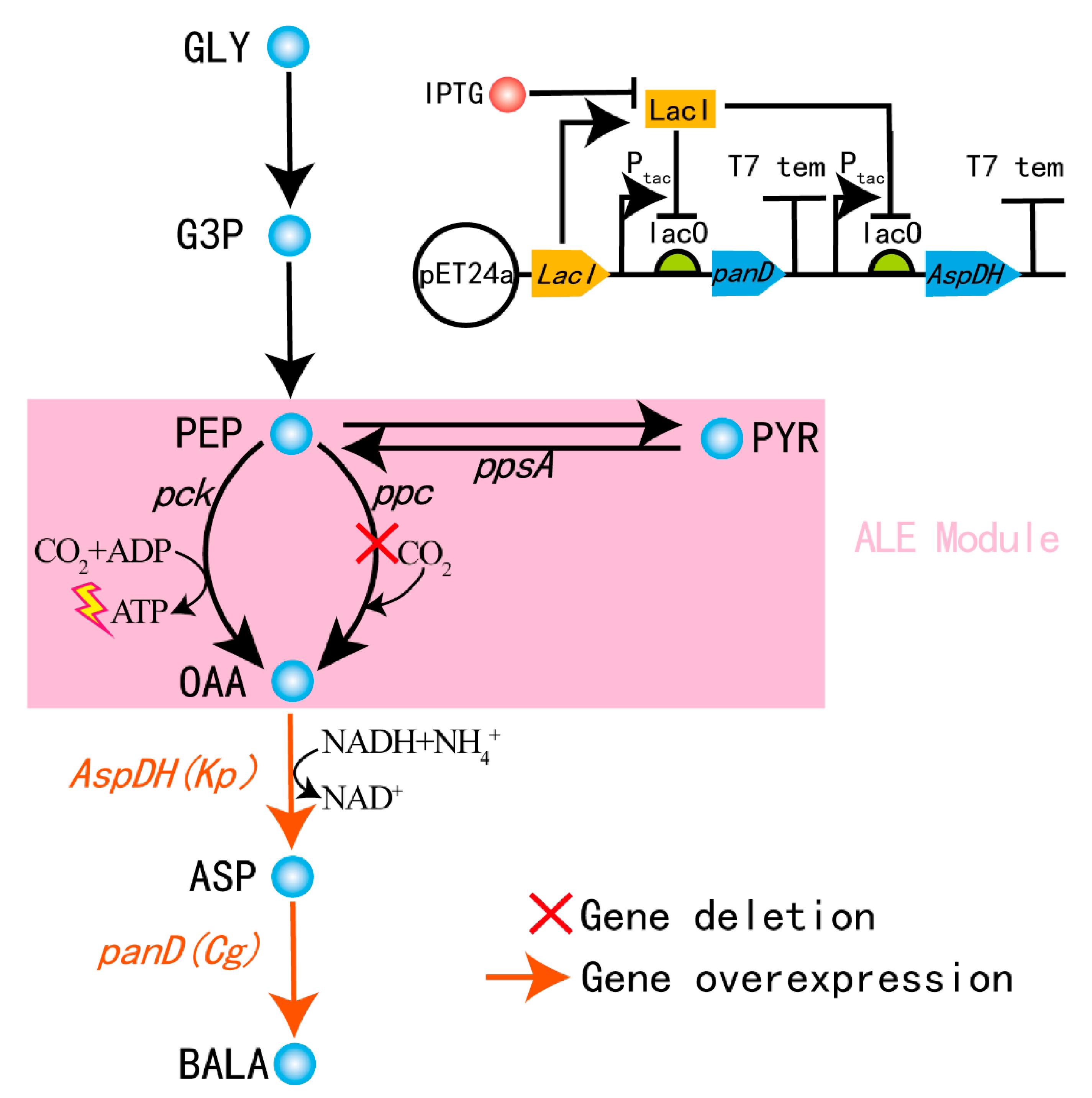
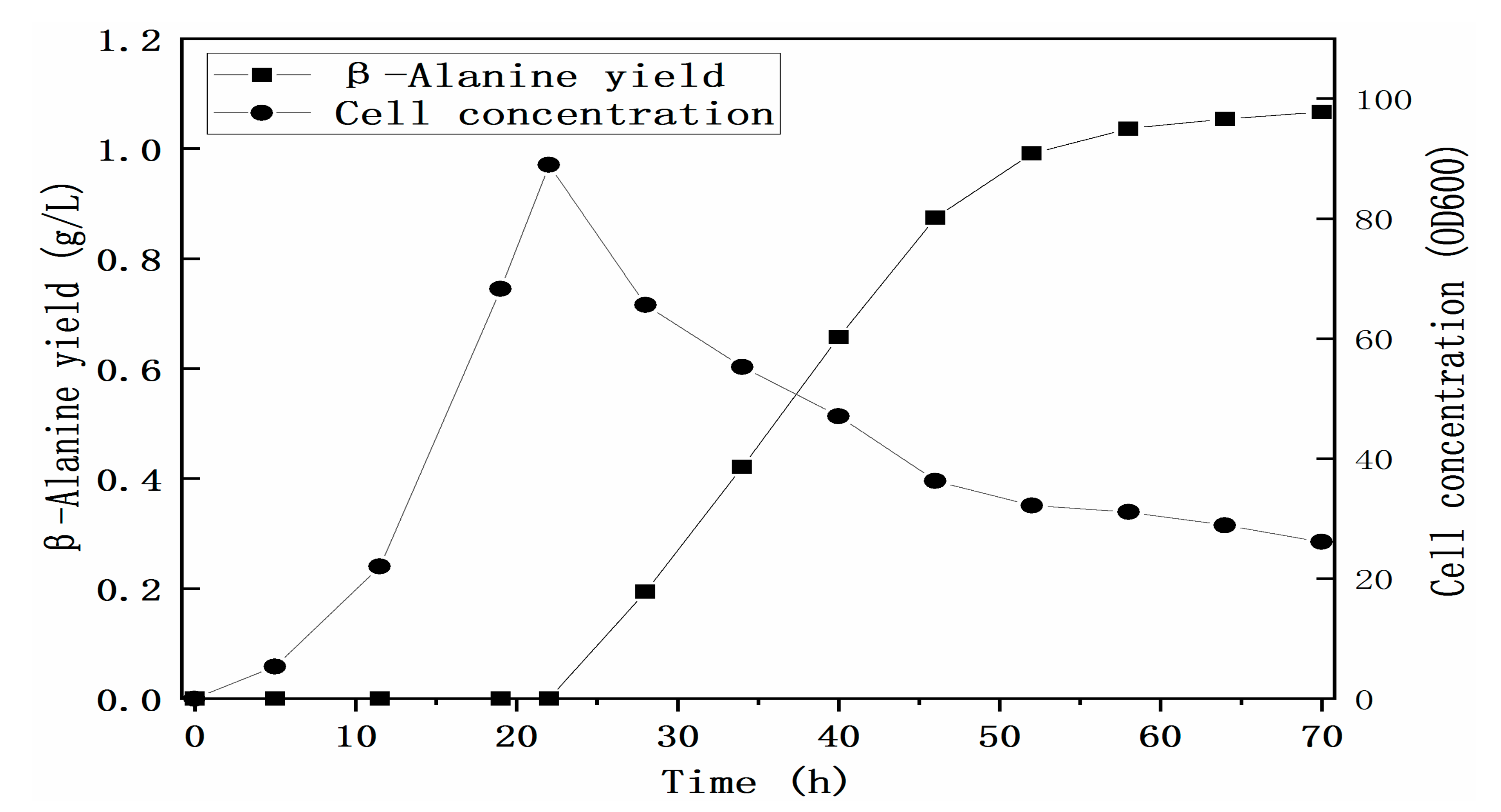
3.3. Construction of the Cell Factory for the Biosynthesis of β-Alanine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Borodina, I.; Kildegaard, K.R.; Jensen, N.B.; Blicher, T.H.; Maury, J.; Sherstyk, S.; Schneider, K.; Lamosa, P.; Herrgård, M.J.; Rosenstand, I.; et al. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metab. Eng. 2015, 27, 57–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.W.; Lee, J.; Ko, Y.-S.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of 3-aminopropionic acid. Metab. Eng. 2015, 30, 121–129. [Google Scholar] [CrossRef]
- Piao, X.; Wang, L.; Lin, B.; Chen, H.; Liu, W.; Tao, Y. Metabolic engineering of Escherichia coli for production of L-aspartate and its derivative β-alanine with high stoichiometric yield. Metab. Eng. 2019, 54, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Ma, Q.; Liu, X.; Fan, X.; Men, J.; Wu, H.; Jiang, S.; Tian, D.; Xiong, B.; Xie, X. High-yield production of L-valine in engineered Escherichia coli by a novel two-stage fermentation. Metab. Eng. 2020, 62, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhu, X.; Xu, H.; Zhang, X. Construction of an energy-conserving glycerol utilization pathways for improving anaerobic succinate production in Escherichia coli. Metab. Eng. 2019, 56, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, L.; Shi, Z.; Du, G.; Chen, J. ATP in current biotechnology: Regulation, applications and perspectives. Biotechnol. Adv. 2009, 27, 94–101. [Google Scholar] [CrossRef]
- Zhang, X.; Jantama, K.; Moore, J.C.; Jarboe, L.R.; Shanmugam, K.T.; Ingram, L.O. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc. Natl. Acad. Sci. USA 2009, 106, 20180. [Google Scholar] [CrossRef] [Green Version]
- Krebs, A.; Bridger, W.A. The kinetic properties of phosphoenolpyruvate carboxykinase of Escherichia coli. Can. J. Biochem. 1980, 58, 309–318. [Google Scholar] [CrossRef]
- Kai, Y.; Matsumura, H.; Inoue, T.; Terada, K.; Nagara, Y.; Yoshinaga, T.; Kihara, A.; Tsumura, K.; Izui, K. Three-dimensional structure of phosphoenolpyruvate carboxylase: A proposed mechanism for allosteric inhibition. Proc. Natl. Acad. Sci. USA 1999, 96, 823. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Jantama, K.; Shanmugam, K.T.; Ingram, L.O. Reengineering Escherichia coli for Succinate Production in Mineral Salts Medium. Appl. Environ. Microbiol. 2009, 75, 7807. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Feng, X.; Ding, Y.; Gao, W.; Xian, M.; Wang, J.; Zhao, G. Characterization and directed evolution of propionyl-CoA carboxylase and its application in succinate biosynthetic pathway with two CO2 fixation reactions. Metab. Eng. 2020, 62, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Bruland, N.; Voß, I.; Brämer, C.; Steinbüchel, A. Unravelling the C3/C4 carbon metabolism in Ralstonia eutropha H16. J. Appl. Microbiol. 2009, 109, 79–90. [Google Scholar] [CrossRef]
- Ren, L.; Sun, X.; Zhang, L.; Huang, H.; Zhao, Q. Exergy analysis for docosahexaenoic acid production by fermentation and strain improvement by adaptive laboratory evolution for Schizochytrium sp. Bioresour. Technol. 2020, 298, 122562. [Google Scholar] [CrossRef]
- Liu, R.; Chen, Y.; Tian, Z.; Mao, Z.; Cheng, H.; Zhou, H.; Wang, W. Enhancing microbial community performance on acid resistance by modified adaptive laboratory evolution. Bioresour. Technol. 2019, 287, 121416. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, H.; Li, Z.; Ye, Q. Enhanced succinate production from glycerol by engineered Escherichia coli strains. Bioresour. Technol. 2016, 218, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liu, Z.; Cui, W.; Zhou, Z. Establishment of Bioprocess for Synthesis of Nicotinamide by Recombinant Escherichia coli Expressing High-Molecular-Mass Nitrile Hydratase. Appl. Biochem. Biotechnol. 2017, 182, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Ren, C.; Xu, Y. Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb. Cell Factories 2019, 18, 108. [Google Scholar] [CrossRef]
- Schwentner, A.; Feith, A.; Münch, E.; Busche, T.; Rückert, C.; Kalinowski, J.; Takors, R.; Blombach, B. Metabolic engineering to guide evolution—Creating a novel mode for L-valine production with Corynebacterium glutamicum. Metab. Eng. 2018, 47, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, R.; Zhang, Y.; Yang, Y.; Lin, Y.; Yan, Y. Developing a pyruvate-driven metabolic scenario for growth-coupled microbial production. Metab. Eng. 2019, 55, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Solomon, K.V.; Sanders, T.M.; Prather, K.L. A dynamic metabolite valve for the control of central carbon metabolism. Metab. Eng. 2012, 14, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Huang, B.; Wu, H.; Li, Z.; Ye, Q. Efficient anaerobic production of succinate from glycerol in engineered Escherichia coli by using dual carbon sources and limiting oxygen supply in preceding aerobic culture. Bioresour. Technol. 2017, 231, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Zhao, C.; Zhang, T.; Zhu, H.; Lin, Z.; Tao, W.; Zhang, Y.; Li, Y. A systematically chromosomally engineered Escherichia coli efficiently produces butanol. Metab. Eng. 2017, 44, 284–292. [Google Scholar] [CrossRef]
- Singhvi, M.; Zendo, T.; Iida, H.; Gokhale, D.; Sonomoto, K. Stimulation of d- and l-lactate dehydrogenases transcriptional levels in presence of diammonium hydrogen phosphate resulting to enhanced lactic acid production by Lactobacillus strain. J. Biosci. Bioeng. 2017, 124, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Pinchuk, G.E.; Rodionov, D.A.; Yang, C.; Li, X.; Osterman, A.L.; Dervyn, E.; Geydebrekht, O.V.; Reed, S.B.; Romine, M.F.; Collart, F.R.; et al. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc. Natl. Acad. Sci. USA 2009, 106, 2874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, K.; Bentley, W.E. Synthetic Biology for Manipulating Quorum Sensing in Microbial Consortia. Trends Microbiol. 2020, 28, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nat. Cell Biol. 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed]
- McBride, S.G.; Strickland, M.S. Quorum sensing modulates microbial efficiency by regulating bacterial investment in nutrient acquisition enzymes. Soil Biol. Biochem. 2019, 136, 107514. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, P.; Sarmah, B.K.; Nandi, S.P. Quorum sensing: Its role in microbial social networking. Res. Microbiol. 2020, 171, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Dong, X.; Grenier, D.; Wang, K.; Wang, Y. Research progress of bacterial quorum sensing receptors: Classification, structure, function and characteristics. Sci. Total. Environ. 2021, 763, 143031. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Abdelsamad, A.; Wassermann, T.; Porse, A.; Becker, J.; Sommer, M.O.A.; Høiby, N.; Ciofu, O. The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: Beyond selection of antibiotic resistance. NPJ Biofilms Microbiomes 2020, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Tian, X. Quorum Sensing and Bacterial Social Interactions in Biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef]
- Murarka, A.; Dharmadi, Y.; Yazdani, S.S.; Gonzalez, R. Fermentative Utilization of Glycerol by Escherichia coli and Its Implications for the Production of Fuels and Chemicals. Appl. Environ. Microbiol. 2008, 74, 1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Zhu, Y.; Li, H.; Chen, L.; Chen, W.; Cui, M.; Han, L.; Hou, W.; Li, D. Alanine mother liquor as a nitrogen source for docosahexaenoic acid production by Schizochytrium sp. B4D1. Electron. J. Biotechnol. 2018, 35, 10–17. [Google Scholar] [CrossRef]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef]
- Schmidt-Rohr, K. Oxygen Is the High-Energy Molecule Powering Complex Multicellular Life: Fundamental Corrections to Traditional Bioenergetics. ACS Omega 2020, 5, 2221–2233. [Google Scholar] [CrossRef] [PubMed]
- Gawryluk, R.M.; Stairs, C.W. Diversity of Electron Transport Chains in Anaerobic Protists. Biochim. Biophys. Acta (BBA) Bioenerg. 2020, 1862, 148334. [Google Scholar] [CrossRef]
| Strains | Relevant Characteristics | Source or Reference |
|---|---|---|
| B0016-082BB | B0016-080BB, panD:: CgpanD | Lab stock |
| B0016-082B | B0016-082BB, aspA::FRT | This study |
| B0016-090BB | B0016-082BB, ppc::FRT | This study |
| B0016-090BBS1 | B0016-090BB, ALE mutant | This study |
| B0016-090BBS2 | B0016-090BB, ALE mutant | This study |
| B0016-090BBS3 | B0016-090BB, ALE mutant | This study |
| B0016-090BBS4 | B0016-090BB, ALE mutant | This study |
| B0016-090BBS5 | B0016-090BB, ALE mutant | This study |
| B0016-100BB | B0016-090BB, recE::FRT, mhpF::FRT, ykgF::FRT, mhpB:: mhpB *, mhpD:: mhpD *, rcsA:: rcsA * | This study |
| B0016-200BB | B0016-100BB, aspA::FRT | This study |
| B0016-082B/pET24a-panD-AspDH | B0016-082B with pPL451-panD-AspDH plasmid | This study |
| B0016-200BB/pET24a-panD-AspDH | B0016-200BB with pPL451-panD-AspDH plasmid | This study |
| Genes | Mutations | Amino Acid Mutations |
|---|---|---|
| recE | 1737–1737 del a, 1739–1741 del | Frameshift mutation |
| mhpB | A107C b | E36A d |
| mhpD | T278C | I93T |
| mhpF | T179C, G189C, 76–77 ins GC c, 116–117 del, 120–121 ins AA | Frameshift mutation |
| ykgF | A19G, T27A,33–35 del, 37–38 ins T, 39–40 ins GT, C94G, | Frameshift mutation |
| rcsA | T209A | L70H |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Zhou, L.; Yin, M.; Zhou, Z. Novel Mode Engineering for β-Alanine Production in Escherichia coli with the Guide of Adaptive Laboratory Evolution. Microorganisms 2021, 9, 600. https://doi.org/10.3390/microorganisms9030600
Xu J, Zhou L, Yin M, Zhou Z. Novel Mode Engineering for β-Alanine Production in Escherichia coli with the Guide of Adaptive Laboratory Evolution. Microorganisms. 2021; 9(3):600. https://doi.org/10.3390/microorganisms9030600
Chicago/Turabian StyleXu, Jian, Li Zhou, Meng Yin, and Zhemin Zhou. 2021. "Novel Mode Engineering for β-Alanine Production in Escherichia coli with the Guide of Adaptive Laboratory Evolution" Microorganisms 9, no. 3: 600. https://doi.org/10.3390/microorganisms9030600








