The Anaerobic Fungi: Challenges and Opportunities for Industrial Lignocellulosic Biofuel Production
Abstract
:1. Introduction
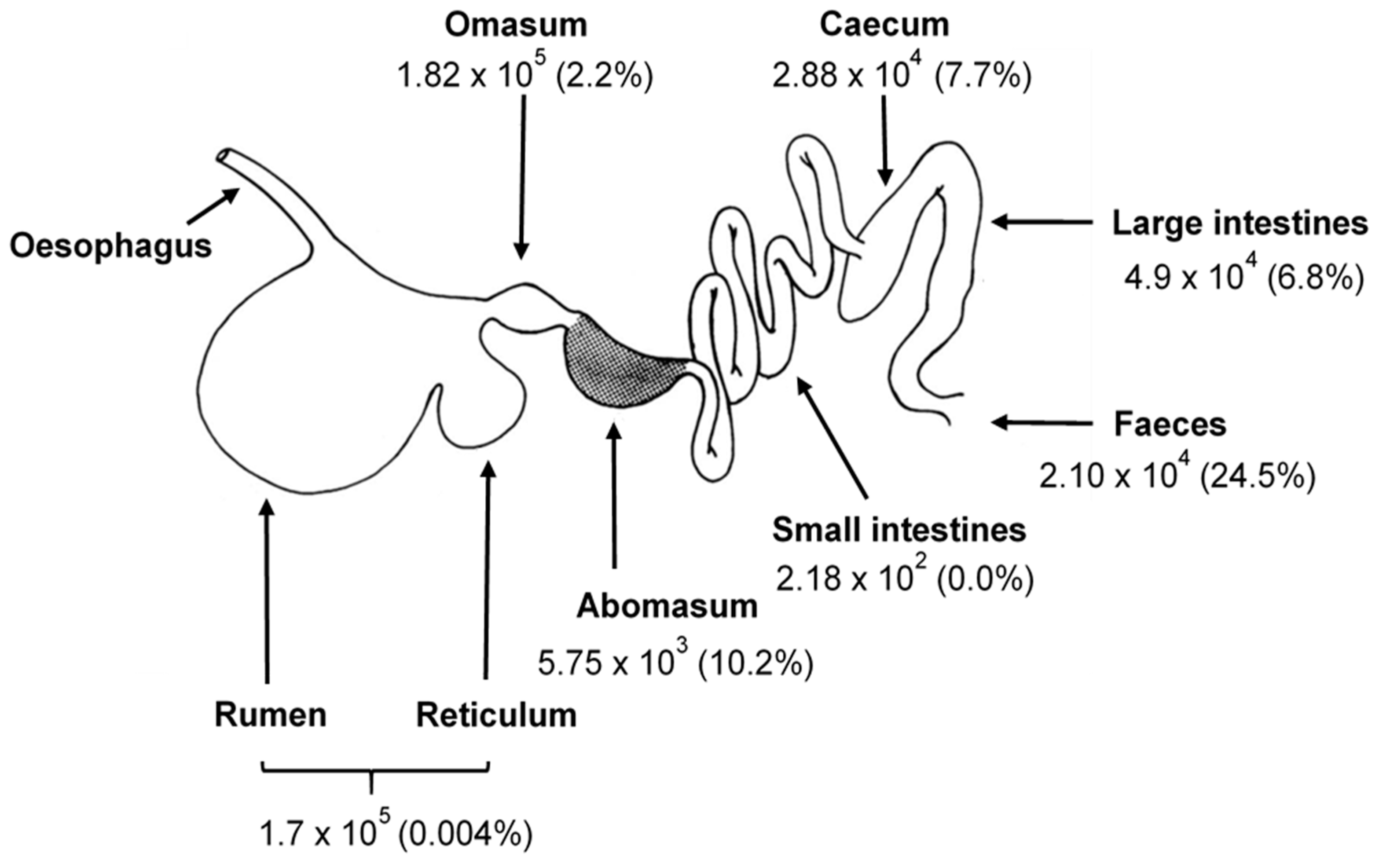
2. The Gastrointestinal Tract of Herbivores
2.1. Rumen Function
2.2. The Relative Functional Role of Anaerobic Fungi
2.3. Life Cycle and Niche of Anaerobic Fungi
3. Process Engineering and Genetic Engineering
3.1. Process Engineering
3.1.1. Anaerobic Fungi in Anaerobic Digestion (AD)
3.1.2. Bioreactor Design and Habitat Engineering
3.1.3. Solid Substrate Fermentation
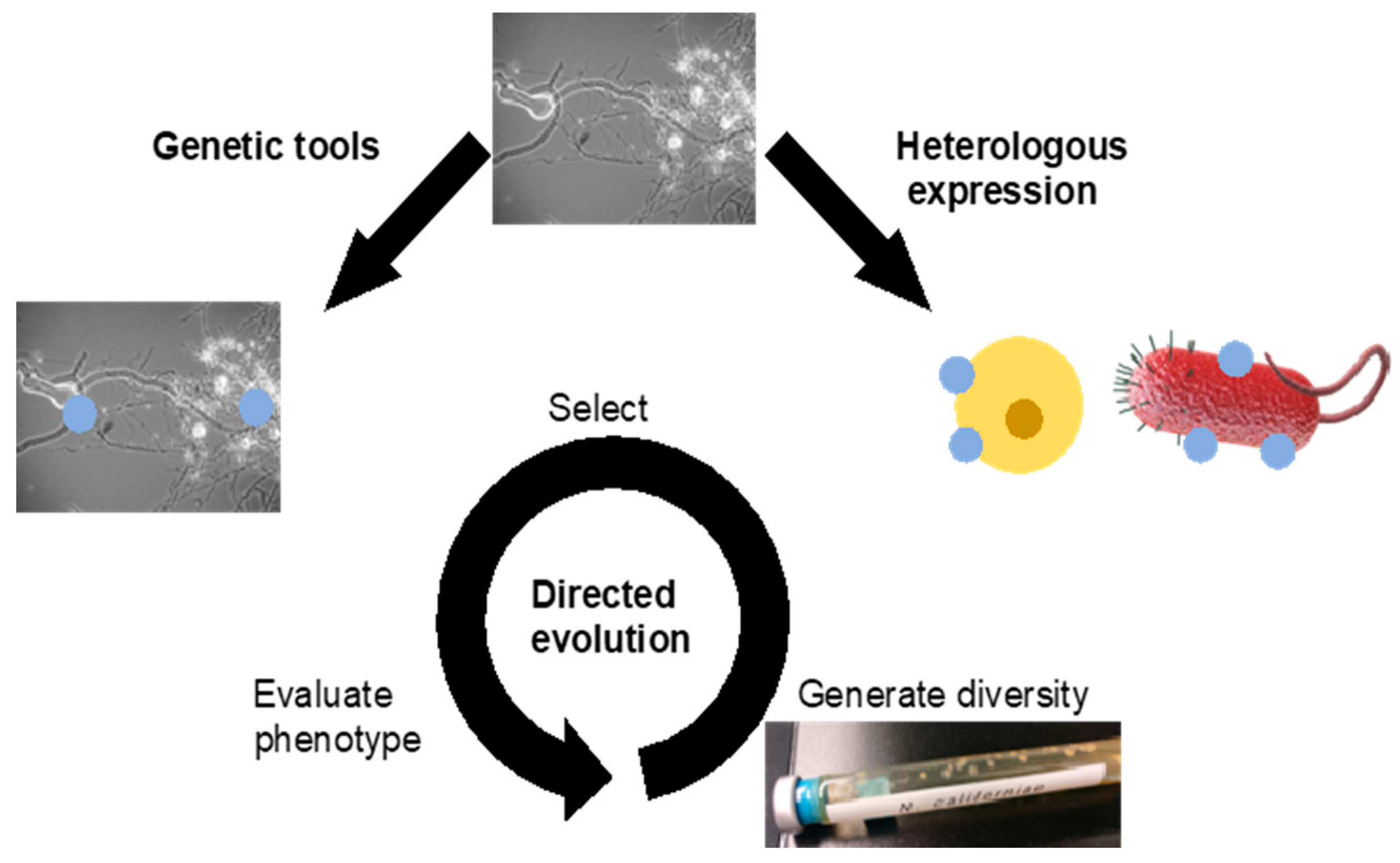
3.2. Genetic Engineering
3.2.1. Transformation
3.2.2. Heterologous Expression
4. Emerging Opportunities for Industrial Biofuel Production
4.1. Biological Pretreatment
4.2. Consolidated Biofuel Production
4.2.1. Bioethanol Production
| Fungal Isolate | Substrate | Ethanol Yield [µmol g−L] * | Reference |
|---|---|---|---|
| Piromyces sp., isolate E2 | Cellobiose | 50 | [124] |
| Piromyces sp., isolate E2 | Glucose | 80 | [124] |
| Piromyces sp., isolate E2 | Fructose | 80 | [124] |
| Piromyces sp., isolate E2 | Mannose | 80 | [124] |
| Piromyces sp., isolate E2 | Lactose | 14.77 | [124] |
| Piromyces sp., isolate F1 | Xylose | 1920 | [129] |
| Piromyces sp., isolate E2 | Xylose | 113 | [124] |
| Piromyces sp., isolate E2 | Xylan | 84 | [124] |
| N. frontalis | Cellulose | 2310 | [5] |
| N. frontalis | Cellulose | 3750 | [130] |
| Piromyces sp., isolate E2 | Cellulose | 157 | [124] |
| Piromyces sp., isolate E2 | Wheat straw | 695 | [124] |
| Piromyces sp., isolate E2 | Wheat bran | 891 | [124] |
| Piromyces sp., isolate E2 | Starch | 157 | [124] |
| Pecoramyces ruminantium | Switch grass ** | 540 | [118] |
| P. ruminantium | Energy cane ** | 510 | [118] |
| P. ruminantium | Sorghum ** | 560 | [118] |
| P. ruminantium | Mixed prairie ** | 490 | [118] |
| P. ruminantium | Corn stover ** | 1030 | [118] |
4.2.2. Dark Fermentation
| Fungal Isolate | Substrate | H2 Yield [μmoL g−1] * | Reference |
|---|---|---|---|
| Piromyces sp., isolate E2 | Cellobiose | 54 | [124] |
| Neocallimastix sp., isolate R1 ** | Glucose | 3464 | [105] |
| Piromyces sp., isolate F1 | Glucose | ≈377 *** | [138] |
| Piromyces sp., isolate E2 | Glucose | 70 | [124] |
| Piromyces sp., isolate E2 | Fructose | 161 | [124] |
| Piromyces sp., isolate E2 | Lactose | 106 | [124] |
| Piromyces sp., isolate E2 | Mannose | 88 | [124] |
| Piromyces sp., isolate E2 | Xylose | 106 | [124] |
| Neocallimastix sp., isolate R1 ** | Xylose | 8020 | [105] |
| N. frontalis | Cellulose | 2177 | [5] |
| Sphaeromonas communis | Cellulose | 2880 | [143] |
| Neocallimastix sp.,isolate N1 | Cellulose | 2520 | [66] |
| Neocallimastix sp., isolate N2 | Cellulose | 2600 | [66] |
| Piromyces sp., isolate E2 | Cellulose | 2220 | [66] |
| Piromyces sp., isolate R1 | Cellulose | 2460 | [66] |
| Piromyces sp., isolate E2 | Cellulose | 159 | [124] |
| N. frontalis | Xylan | ≈2381 *** | [141] |
| Piromyces sp., isolate E2 | Wheat Straw | 2261 | [124] |
| Piromyces sp., isolate E2 | Wheat bran | 1370 | [124] |
| Piromyces sp., isolate E2 | Bagasse | 1957 | [124] |
| N. frontalis. | Poplar wood chips | 1984 *** | [149] |
| Piromyces sp., isolate E2 | Xylan | 134 | [124] |
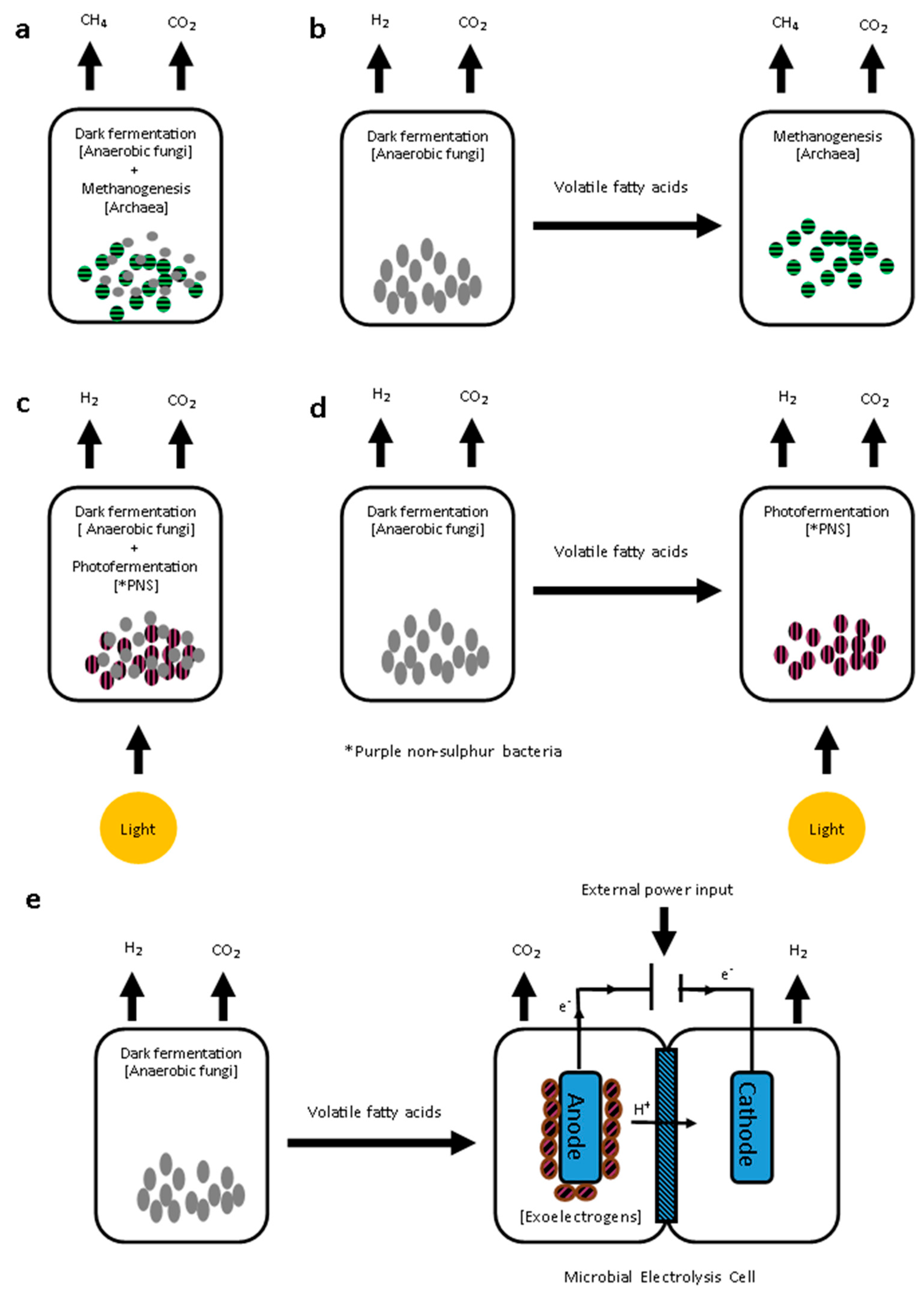
4.3. Biofuel Production from Dark Fermentation Products
4.3.1. Integration of Dark Fermentation with Biomethane Production
4.3.2. Integration of Dark Fermentation with Additional Biohydrogen Production
Photofermentation
Microbial Electrolysis Cells (MEC)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Yang, B.; Wyman, C.E. Pretreatment: The key to unlocking low-cost cellulosic ethanol. Biofuels Bioprod. Biorefining 2008, 2, 26–40. [Google Scholar] [CrossRef]
- Chen, H.; Liu, J.; Chang, X.; Chen, D.; Xue, Y.; Liu, P.; Lin, H.; Han, S. A review on the pretreatment of lignocellulose for high-value chemicals. Fuel Process. Technol. 2017, 160, 196–206. [Google Scholar] [CrossRef]
- Flad, V.; Young, D.; Seppälä, S.; Hooker, C.; Youssef, N.; Podmirseg, S.M.; Nagler, M.; Reilly, M.; Li, Y.; Fliegerová, K.; et al. 17 The Biotechnological Potential of Anaerobic Gut Fungi. In Genetics and Biotechnology; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 413–437. [Google Scholar]
- Solomon, K.V.; Haitjema, C.H.; Henske, J.K.; Gilmore, S.P.; Borges-Rivera, D.; Lipzen, A.; Brewer, H.M.; Purvine, S.O.; Wright, A.T.; Theodorou, M.K.; et al. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 2016, 351, 1192–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauchop, T.; Mountfort, D.O. Cellulose Fermentation by a Rumen Anaerobic Fungus in Both the Absence and the Presence of Rumen Methanogens. Appl. Environ. Microbiol. 1981, 42, 1103–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joblin, K.N.; Campbell, G.P.; Richardson, A.J.; Stewart, C.S. Fermentation of barley straw by anaerobic rumen bacteria and fungi in axenic culture and in co-culture with methanogens. Lett. Appl. Microbiol. 1989, 9, 195–197. [Google Scholar] [CrossRef]
- Orpin, C.G. Studies on the rumen flagellate Neocallimastix frontalis. J. Gen. Microbiol. 1975, 91, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Orpin, C.G. Studies on the rumen flagellate Sphaeromonas communis. J. Gen. Microbiol. 1976, 94, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Orpin, C.G. The rumen flagellate Piromonas communis: Its life history and invasion of plant material in the rumen. J. Gen. Microbiol. 1977, 99, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Braune, R. Untersuchungen über die im Wiederkäuermagen vorkommenden Protozoen. Arch. Protistenkd. 1913, 32, 111–170. [Google Scholar]
- Hsiung, T.-S. A Monograph on the Protozoa of the Large Intestine of the Horse. Master’s Thesis, Iowa State University, Ames, IA, USA, 1930. [Google Scholar]
- Weissenberg, R. Callimastix cyclopis, ng, n. sp., ein geisseltragendes Protozoon aus dem Serum von Cyclops. Sitz. Ges. Naturfr Freunde 1912, 5, 299–305. [Google Scholar]
- Weissenberg, R. The development and affinities of Callimastix cyclopis Weissenberg, a parasitic microorganism from the serum of Cyclops. Proc. Am. Soc. Protozool. 1950, 1, 4–5. [Google Scholar]
- Liebentanz, E. Die parasitischen Protozoen des Wiederkäuermayens. Arch. Protistenkd. 1910, 19, 19–80. [Google Scholar]
- Vavra, J.; Joyon, L. Étude sur la morphologie, le cycle évolutif et la position systématique de Callimastix cyclopsis Weissenberg 1912. Pro-Tislologica 1966, 2, 15–16. [Google Scholar]
- Li, J.; Heath, I.B.; Packer, L. The phylogenetic relationships of the anaerobic chytridiomycetous gut fungi (Neocallimasticaceae) and the Chytridiomycota. II. Cladistic analysis of structural data and description of Neocallimasticales ord.nov. Can. J. Bot. 1993, 71, 393–407. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef] [PubMed]
- Stabel, M.; Hanafy, R.A.; Schweitzer, T.; Greif, M.; Aliyu, H.; Flad, V.; Young, D.; Lebuhn, M.; Elshahed, M.S.; Ochsenreither, K.; et al. Aestipascuomyces dupliciliberans gen. Nov, sp. nov., the first cultured representative of the uncultured SK4 clade from aoudad sheep and alpaca. Microorganisms 2020, 8, 1734. [Google Scholar] [CrossRef]
- Gleason, F.H.; Kagami, M.; Lefevre, E.; Sime-Ngando, T. The ecology of chytrids in aquatic ecosystems: Roles in food web dynamics. Fungal Biol. Rev. 2008, 22, 17–25. [Google Scholar] [CrossRef]
- Davies, D.R.; Theodorou, M.K.; Lawrence, M.I.G.; Trinci, A.P.J. Distribution of anaerobic fungi in the digestive tract of cattle and their survival in faeces. J. Gen. Microbiol. 1993, 139, 1395–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, D.R.; Theodorou, M.K.; Brooks, A.E.; Trinci, A.P.J. Influence of drying on the survival of anaerobic fungi in rumen digesta and faeces of cattle. FEMS Microbiol. Lett. 1993, 106, 59–63. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J.; et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef]
- James, T.Y.; Letcher, P.M.; Longcore, J.E.; Mozley-Standridge, S.E.; Porter, D.; Powell, M.J.; Griffith, G.W.; Vilgalys, R. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 2006, 98, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Youssef, N.; Couger, M.; Hanafy, R.; Elshahed, M.; Stajich, J.E. Comparative genomics and divergence time estimation of the anaerobic fungi in herbivorous mammals. bioRxiv 2018, 401869. [Google Scholar] [CrossRef]
- Hobson, P.N.; Wallace, R.J.; Bryant, M.P. Microbial ecology and activities in the rumen: Part I. Crit. Rev. Microbiol. 1982, 9, 165–225. [Google Scholar] [CrossRef] [PubMed]
- Mambrini, M.; Peyraud, J.L. Retention time of feed particles and liquids in the stomachs and intestines of dairy cows. Direct measurement and calculations based on faecal collection. Reprod. Nutr. Dev. 1997, 37, 427–442. [Google Scholar] [CrossRef] [Green Version]
- Bartocci, S.; Amici, A.; Verna, M.; Terramoccia, S.; Martillotti, F. Solid and fluid passage rate in buffalo, cattle and sheep fed diets with different forage to concentrate ratios. Livest. Prod. Sci. 1997, 52, 201–208. [Google Scholar] [CrossRef]
- Van Weyenberg, S.; Sales, J.; Janssens, G.P.J. Passage rate of digesta through the equine gastrointestinal tract: A review. Livest. Sci. 2006, 99, 3–12. [Google Scholar] [CrossRef]
- Hobson, P.N. Rumen micro-organisms. Prog. Ind. Microbiol. 1971, 9, 41–77. [Google Scholar] [PubMed]
- Ulyatt, M.J.; Dellow, D.W.; John, A.; Reid, C.S.W.; Waghorn, G.C. Contribution of chewing during eating and rumination to the clearance of digesta from the rumen reticulum. In Control of Digestion and Metabolism in Ruminants. In Proceedings of the 6th International Symposium on Ruminant Physiology, Banff, AB, Canada, 14–16 September 1984; pp. 498–515. [Google Scholar]
- Ulyatt, M.J.; Baldwin, R.L.; Koong, L.J. The basis of nutritive value—A modelling approach. Proc. N. Z. Soc. Anim. Prod. 1976, 36, 140–149. [Google Scholar]
- Nishida, A.H.; Ochman, H. Rates of gut microbiome divergence in mammals. Mol. Ecol. 2018, 27, 1884–1897. [Google Scholar] [CrossRef]
- Hume, I.D. Optimal digestive strategies in mammalian herbivores. Physiol. Zool. 1989, 62, 1145–1163. [Google Scholar] [CrossRef]
- Soest, P.J. Van Nutritional Ecology of the Ruminant, 2nd ed.; Cornell University Press: Ithaca, NY, USA, 1994; ISBN 9780801427725. [Google Scholar]
- France, J.; Dhanoa, M.S.; Theodorou, M.K.; Lister, S.J.; Davies, D.R.; Isac, D. A model to interpret gas accumulation profiles associated with in vitro degradation of ruminant feeds. J. Theor. Biol. 1993, 163, 99–111. [Google Scholar] [CrossRef]
- Peng, X.; Wilken, S.E.; Lankiewicz, T.S.; Gilmore, S.P.; Brown, J.L.; Henske, J.K.; Swift, C.L.; Salamov, A.; Barry, K.; Grigoriev, I.V.; et al. Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat. Microbiol. 2021, 1–13. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Gilmore, S.P.; Henske, J.K.; Solomon, K.V.; De Groot, R.; Kuo, A.; Mondo, S.J.; Salamov, A.A.; LaButti, K.; Zhao, Z.; et al. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017, 2, 17087. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.E.; Kingston-Smith, A.H.; Jimenez, H.R.; Huws, S.A.; Skøt, K.P.; Griffith, G.W.; McEwan, N.R.; Theodorou, M.K. Dynamics of initial colonization of nonconserved perennial ryegrass by anaerobic fungi in the bovine rumen. FEMS Microbiol. Ecol. 2008, 66, 537–545. [Google Scholar] [CrossRef]
- Theodorou, M.K.; Gill, M.; King-Spooner, C.; Beever, D.E. Enumeration of anaerobic chytridiomycetes as thallus-forming units: Novel method for quantification of fibrolytic fungal populations from the digestive tract ecosystem. Appl. Environ. Microbiol. 1990, 56, 1073–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, S.E.; Griffith, G.G.; Milne, A.; Theodorou, M.K.; Trinci, A.P.J. The life cycle and growth kinetics of an anaerobic rumen fungus. J. Gen. Microbiol. 1987, 133, 1815–1827. [Google Scholar] [CrossRef] [Green Version]
- Joblin, K.N. Isolation, Enumeration, and Maintenance of Rumen Anaerobic Fungi in Roll Tubes. Appl. Environ. Microbiol. 1981, 42, 1119–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leedle, J.A.; Bryant, M.P.; Hespell, R.B. Diurnal variations in bacterial numbers and fluid parameters in ruminal contents of animals fed low- or high-forage diets. Appl. Environ. Microbiol. 1982, 44, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodorou, M.K.; Beever, D.E.; Haines, M.J.; Brooks, A. The effect of a fungal probiotic on intake and performance of early weaned calvesle. Anim. Prod. 1990, 50, 577. [Google Scholar]
- Gordon, G.L.R.; Phillips, M.W. The role of anaerobic gut fungi in ruminants. Nutr. Res. Rev. 1998, 11, 133–168. [Google Scholar] [CrossRef] [Green Version]
- Gordon, G.L.R.; Phillips, M.W. Removal of anaerobic fungi from the rumen of sheep by chemical treatment and the effect on feed consumption and in vivo fibre digestion. Lett. Appl. Microbiol. 1993, 17, 220–223. [Google Scholar] [CrossRef]
- Theodorou, M.K.; Zhu, W.-Y.; Rickers, A.; Nielsen, B.B.; Gull, K.; Trinci, A.P.J. Biochemistry and Ecology of Anaerobic Fungi. In Human and Animal Relationships; Springer: Berlin/Heidelberg, Germany, 1996; pp. 265–295. [Google Scholar]
- France, J.; Theodorou, M.K.; Davies, D. Use of zoospore concentrations and life cycle parameters in determining the population of anaerobic fungi in the rumen ecosystem. J. Theor. Biol. 1990, 147, 413–422. [Google Scholar] [CrossRef]
- Orpin, C.G. On the induction of zoosporogenesis in the rumen phycomycetes Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J. Gen. Microbiol. 1977, 101, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Trinci, A.P.J.; Davies, D.R.; Gull, K.; Lawrence, M.I.; Bonde Nielsen, B.; Rickers, A.; Theodorou, M.K. Anaerobic fungi in herbivorous animals. Mycol. Res. 1994, 98, 129–152. [Google Scholar] [CrossRef]
- Orpin, C.G.; Bountiff, L. Zoospore chemotaxis in the rumen phycomycete Neocallimastix frontalis. J. Gen. Microbiol. 1978, 104, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Orpin, C.G.; Greenwood, Y. The role of haems and related compounds in the nutrition and zoosporogenesis of the rumen chytridiomycete Neocallimastix frontalis H8. J. Gen. Microbiol. 1986, 132, 2179–2185. [Google Scholar] [CrossRef] [Green Version]
- Wubah, D.A.; Kim, D.S.H. Chemoattraction of anaerobic ruminal fungi zoospores to selected phenolic acids. Microbiol. Res. 1996, 151, 257–262. [Google Scholar] [CrossRef]
- Nielsen, B.B.; Zhu, W.Y.; Trinci, A.P.J.; Theodorou, M.K. Demonstration of zoosporangia of anaerobic fungi on plant residues recovered from faeces of cattle. Mycol. Res. 1995, 99, 471–474. [Google Scholar] [CrossRef]
- Gleason, F.H.; Schmidt, S.K.; Marano, A.V. Can zoosporic true fungi grow or survive in extreme or stressful environments? Extremophiles 2010, 14, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Dal Pont, J.P. Process Engineering and Industrial Management; John Wiley & Sons: Hoboken, NJ, USA, 2013; ISBN 9781848213265. [Google Scholar]
- Lockhart, R.J.; Van Dyke, M.I.; Beadle, I.R.; Humphreys, P.; McCarthy, A.J. Molecular biological detection of anaerobic gut fungi (Neocallimastigales) from landfill sites. Appl. Environ. Microbiol. 2006, 72, 5659–5661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dollhofer, V.; Callaghan, T.M.; Griffith, G.W.; Lebuhn, M.; Bauer, J. Presence and transcriptional activity of anaerobic fungi in agricultural biogas plants. Bioresour. Technol. 2017, 235, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Young, D.; Dollhofer, V.; Callaghan, T.M.; Reitberger, S.; Lebuhn, M.; Benz, J.P. Isolation, identification and characterization of lignocellulolytic aerobic and anaerobic fungi in one- and two-phase biogas plants. Bioresour. Technol. 2018, 268, 470–479. [Google Scholar] [CrossRef]
- Fliegerová, K.; Mrázek, J.; Hoffmann, K.; Zábranská, J.; Voigt, K. Diversity of anaerobic fungi within cow manure determined by ITS1 analysis. Folia Microbiol. 2010, 55, 319–325. [Google Scholar] [CrossRef]
- Drake, H.; Ivarsson, M. The role of anaerobic fungi in fundamental biogeochemical cycles in the deep biosphere. Fungal Biol. Rev. 2018, 32, 20–25. [Google Scholar] [CrossRef]
- McGranaghan, P.; Davies, J.C.; Griffith, G.W.; Davies, D.R.; Theodorou, M.K. The survival of anaerobic fungi in cattle faeces. FEMS Microbiol. Ecol. 1999, 29, 293–300. [Google Scholar] [CrossRef]
- Theodorou, M.K.; King-Spooner, C.; Beever, D.E. Presence or Absence of Anaerobic Fungi in Landfill Refuse; Energy Technology Support Unit, Department of Energy Publications: London, UK, 1989; pp. 1–40.
- Dollhofer, V.; Dandikas, V.; Dorn-In, S.; Bauer, C.; Lebuhn, M.; Bauer, J. Accelerated biogas production from lignocellulosic biomass after pre-treatment with Neocallimastix frontalis. Bioresour. Technol. 2018, 264, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, A.; Dottorini, G.; Massini, G.; Mazzurco Miritana, V.; Signorini, A.; Lembo, G.; Fabbricino, M. Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour. Technol. 2018, 260, 364–373. [Google Scholar] [CrossRef]
- Vinzelj, J.; Joshi, A.; Insam, H.; Podmirseg, S.M. Employing anaerobic fungi in biogas production: Challenges & opportunities. Bioresour. Technol. 2020, 300, 122687. [Google Scholar]
- Teunissen, M.J.; Kets, E.P.W.; Op den Camp, H.J.M.; Huis in’t Veld, J.H.J.; Vogels, G.D. Effect of coculture of anaerobic fungi isolated from ruminants and non-ruminants with methanogenic bacteria on cellulolytic and xylanolytic enzyme activities. Arch. Microbiol. 1992, 157, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, S.P.; Lankiewicz, T.S.; Wilken, S.E.; Brown, J.L.; Sexton, J.A.; Henske, J.K.; Theodorou, M.K.; Valentine, D.L.; O’Malley, M.A. Top-Down Enrichment Guides in Formation of Synthetic Microbial Consortia for Biomass Degradation. ACS Synth. Biol. 2019, 8, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Wubah, D.A.; Fuller, M.S.; Akin, D.E. Resistant Body Formation in Neocallimastix Sp., an Anaerobic Fungus from the Rumen of a Cow. Mycologia 1991, 83, 40–47. [Google Scholar] [CrossRef]
- Hungate, R.E. The Rumen Bacteria. In Rumen and Its Microbes; Elsevier: Amsterdam, The Netherlands, 1966; pp. 8–90. [Google Scholar]
- Bryant, M.P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 1972, 25, 1324–1328. [Google Scholar] [CrossRef]
- Hungate, R.E.; Macy, J. The roll-tube method for cultivation of strict anaerobes. Bull. Ecol. Res. Comm. 1973, 17, 123–126. [Google Scholar]
- Miller, T.L.; Wolin, M.J. A Serum Bottle Modification of the Hungate Technique for Cultivating Obligate Anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [CrossRef] [PubMed]
- Theodorou, M.K.; Davies, D.R.; Nielsen, B.B.; Lawrence, M.I.G.; Trinci, A.P.J. Determination of growth of anaerobic fungi on soluble and cellulosic substrates using a pressure transducer. Microbiology 1995, 141, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Wilken, S.E.; Monk, J.M.; Leggieri, P.A.; Lawson, C.A.; Lankiewicz, T.S.; Seppälä, S.; Daum, C.; Jenkins, J.; Lipzen, A.; Mondo, S.J.; et al. Experimentally validated reconstruction and analysis of a genome-scale metabolic model of an anaerobic Neocallimastigomycota fungus. mSystems 2021, in press. [Google Scholar]
- Zhu, W.Y.; Theodorou, M.K.; Longland, A.C.; Nielsen, B.B.; Dijkstra, J.; Trinci, A.P.J. Growth and survival of anaerobic fungi in batch and continuous-flow cultures. Anaerobe 1996, 2, 29–37. [Google Scholar] [CrossRef]
- Zhu, W.Y.; Theodorou, M.K.; Nielsen, B.B.; Trinci, A.P.J. Dilution rate increases production of plant cell-wall degrading enzymes by anaerobic fungi in continuous-flow culture. Anaerobe 1997, 3, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Lonsane, B.K.; Ghildyal, N.P.; Budiatman, S.; Ramakrishna, S. V Engineering aspects of solid state fermentation. Enzyme Microb. Technol. 1985, 7, 258–265. [Google Scholar] [CrossRef]
- Soccol, C.R.; da Costa, E.S.F.; Letti, L.A.J.; Karp, S.G.; Woiciechowski, A.L.; de Vandenberghe, L.P.S. Recent developments and innovations in solid state fermentation. Biotechnol. Res. Innov. 2017, 1, 52–71. [Google Scholar] [CrossRef]
- Moo-Young, A.R.; Tengerdy, R.P.; Moreira, M. Principles of solid-substrate fermentation. Filam. Fungi 1983, 4, 117–144. [Google Scholar]
- Tengerdy, R.P. Solid substrate fermentation. Trends Biotechnol. 1985, 3, 96–99. [Google Scholar] [CrossRef]
- Tengerdy, R.P.; Szakacs, G. Bioconversion of lignocellulose in solid substrate fermentation. Biochem. Eng. J. 2003, 13, 169–179. [Google Scholar] [CrossRef]
- Duff, S.J.B.; Murray, W.D. Bioconversion of forest products industry waste cellulosics to fuel ethanol: A review. Bioresour. Technol. 1996, 55, 1–33. [Google Scholar] [CrossRef]
- Himmel, M.E.; Ruth, M.F.; Wyman, C.E. Cellulase for commodity products from cellulosic biomass. Curr. Opin. Biotechnol. 1999, 10, 358–364. [Google Scholar] [CrossRef]
- Rayner, A.D.M.; Boddy, L. Fungal Communities in the Decay of Wood. In Advances in Microbial Ecology; Springer: Boston, MA, USA, 1988; pp. 115–166. [Google Scholar]
- Kim, S.W.; Kang, S.W.; Lee, J.S. Cellulase and xylanase production by Aspergillus niger KKS in various bioreactors. Bioresour. Technol. 1997, 59, 63–67. [Google Scholar] [CrossRef]
- Ishida, H.; Hata, Y.; Ichikawa, E.; Kawato, A.; Suginami, K.; Imayasu, S. Regulation of the glucoamylase-encoding gene (glaB), expressed in solid- state culture (koji) of Aspergillus oryzae. J. Ferment. Bioeng. 1998, 86, 301–307. [Google Scholar] [CrossRef]
- Arora, S.; Rani, R.; Ghosh, S. Bioreactors in solid state fermentation technology: Design, applications and engineering aspects. J. Biotechnol. 2018, 269, 16–34. [Google Scholar] [CrossRef]
- Farinas, C.S. Developments in solid-state fermentation for the production of biomass-degrading enzymes for the bioenergy sector. Renew. Sustain. Energy Rev. 2015, 52, 179–188. [Google Scholar] [CrossRef]
- Youssef, N.H.; Couger, M.B.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Prade, R.A.; Najar, F.Z.; Atiyeh, H.K.; Wilkins, M.R.; Elshahed, M.S. The genome of the anaerobic fungus orpinomyces sp. strain c1a reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmore, S.P.; Lillington, S.P.; Haitjema, C.H.; de Groot, R.; O’Malley, M.A. Designing chimeric enzymes inspired by fungal cellulosomes. Synth. Syst. Biotechnol. 2020, 5, 23–32. [Google Scholar] [CrossRef]
- Gilmore, S.P.; Henske, J.K.; O’Malley, M.A. Driving biomass breakdown through engineered cellulosomes. Bioengineered 2015, 6, 204–208. [Google Scholar] [CrossRef] [Green Version]
- Haitjema, C.H.; Solomon, K.V.; Henske, J.K.; Theodorou, M.K.; O’Malley, M.A. Anaerobic gut fungi: Advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol. Bioeng. 2014. [Google Scholar] [CrossRef] [PubMed]
- Podolsky, I.A.; Seppälä, S.; Lankiewicz, T.S.; Brown, J.L.; Swift, C.L.; O’Malley, M.A. Harnessing Nature’s Anaerobes for Biotechnology and Bioprocessing. Annu. Rev. Chem. Biomol. Eng. 2019, 10, 105–128. [Google Scholar] [CrossRef]
- Wilken, S.E.; Swift, C.L.; Podolsky, I.A.; Lankiewicz, T.S.; Seppälä, S.; O’Malley, M.A. Linking “omics” to function unlocks the biotech potential of non-model fungi. Curr. Opin. Syst. Biol. 2019, 14, 9–17. [Google Scholar] [CrossRef]
- Durand, R.; Rascle, C.; Fischer, M.; Fèvre, M. Transient expression of the β-glucuronidase gene after biolistic transformation of the anaerobic fungus Neocallimastix frontalis. Curr. Genet. 1997, 31, 158–161. [Google Scholar] [CrossRef]
- Henske, J.K.; Wilken, S.E.; Solomon, K.V.; Smallwood, C.R.; Shutthanandan, V.; Evans, J.E.; Theodorou, M.K.; O’Malley, M.A. Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose. Biotechnol. Bioeng. 2018, 115, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Weld, R.J.; Plummer, K.M.; Carpenter, M.A.; Ridgway, H.J. Approaches to functional genomics in filamentous fungi. Cell Res. 2006, 16, 31–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teunissen, M.J.; Op Den Camp, H.J.M.; Orpin, C.G.; Huis, J.H.; Vogels, G.D. Comparison of growth characteristics of anaerobic fungi isolated from ruminant and non-ruminant herbivores during cultivation in a defined medium. J. Gen. Microbiol. 1991, 137, 1401–1408. [Google Scholar] [CrossRef] [Green Version]
- Gleason, F.H.; Lilje, O. Structure and function of fungal zoospores: Ecological implications. Fungal Ecol. 2009, 2, 53–59. [Google Scholar] [CrossRef]
- Calkins, S.; Elledge, N.C.; Hanafy, R.A.; Elshahed, M.S.; Youssef, N. A fast and reliable procedure for spore collection from anaerobic fungi: Application for RNA uptake and long-term storage of isolates. J. Microbiol. Methods 2016, 127, 206–213. [Google Scholar] [CrossRef] [Green Version]
- Calkins, S.S.; Elledge, N.C.; Mueller, K.E.; Marek, S.M.; Couger, M.B.; Elshahed, M.S.; Youssef, N.H. Development of an RNA interference (RNAi) gene knockdown protocol in the anaerobic gut fungus Pecoramyces ruminantium strain C1A. PeerJ 2018, 6, e4276. [Google Scholar] [CrossRef] [Green Version]
- Mello, C.C.; Conte, D. Revealing the world of RNA interference. Nature 2004, 431, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Swafford, A.J.M.; Hussey, S.P.; Fritz-Laylin, L.K. High-efficiency electroporation of chytrid fungi. Sci. Rep. 2020, 10, 15145. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Lowe, S.E.; Theodorou, M.K.; Trinci, A.P.J. Growth and fermentation of an anaerobic rumen fungus on various carbon sources and effect of temperature on development. Appl. Environ. Microbiol. 1987, 53, 1210–1215. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Xia, L. Heterologous expression of an endo-β-1,4-glucanase gene from the anaerobic fungus Orpinomyces PC-2 in Trichoderma reesei. World J. Microbiol. Biotechnol. 2011, 27, 2913–2920. [Google Scholar] [CrossRef]
- Wilken, S.E.; Seppälä, S.; Lankiewicz, T.S.; Saxena, M.; Henske, J.K.; Salamov, A.A.; Grigoriev, I.V.; O’Malley, M.A. Genomic and proteomic biases inform metabolic engineering strategies for anaerobic fungi. Metab. Eng. Commun. 2020, 10, e00107. [Google Scholar] [CrossRef] [PubMed]
- Seppälä, S.; Yoo, J.I.; Yur, D.; O’Malley, M.A. Heterologous transporters from anaerobic fungi bolster fluoride tolerance in Saccharomyces cerevisiae. Metab. Eng. Commun. 2019, 9, e00091. [Google Scholar] [CrossRef]
- O’Malley, M.A.; Theodorou, M.K.; Kaiser, C.A. Evaluating expression and catalytic activity of anaerobic fungal fibrolytic enzymes native topiromyces sp E2 inSaccharomyces cerevisiae. Environ. Prog. Sustain. Energy 2012, 31, 37–46. [Google Scholar] [CrossRef]
- Hooker, C.A.; Lee, K.Z.; Solomon, K. V Leveraging anaerobic fungi for biotechnology. Curr. Opin. Biotechnol. 2019, 59, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.S.; Nugent, B.; Mullins, E.; Doohan, F.M. Fungal-mediated consolidated bioprocessing: The potential of Fusarium oxysporum for the lignocellulosic ethanol industry. AMB Express 2016, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabed, H.M.; Akter, S.; Yun, J.; Zhang, G.; Awad, F.N.; Qi, X.; Sahu, J.N. Recent advances in biological pretreatment of microalgae and lignocellulosic biomass for biofuel production. Renew. Sustain. Energy Rev. 2019, 105, 105–128. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, H.; Huang, H.; Liu, Y. Evaluation of biological pretreatment with white rot fungi for the enzymatic hydrolysis of bamboo culms. Int. Biodeterior. Biodegrad. 2007, 60, 159–164. [Google Scholar] [CrossRef]
- López-Abelairas, M.; Álvarez Pallín, M.; Salvachúa, D.; Lú-Chau, T.; Martínez, M.J.; Lema, J.M. Optimisation of the biological pretreatment of wheat straw with white-rot fungi for ethanol production. Bioprocess Biosyst. Eng. 2013, 36, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Hatakka, A.I. Pretreatment of wheat straw by white-rot fungi for enzymic saccharification of cellulose. Eur. J. Appl. Microbiol. Biotechnol. 1983, 18, 350–357. [Google Scholar] [CrossRef]
- Lalak, J.; Kasprzycka, A.; Martyniak, D.; Tys, J. Effect of biological pretreatment of Agropyron elongatum “BAMAR” on biogas production by anaerobic digestion. Bioresour. Technol. 2016, 200, 194–200. [Google Scholar] [CrossRef]
- Morrison, J.M.; Elshahed, M.S.; Youssef, N.H. Defined enzyme cocktail from the anaerobic fungus Orpinomyces sp. strain C1A effectively releases sugars from pretreated corn stover and switchgrass. Sci. Rep. 2016, 6, 29217. [Google Scholar] [CrossRef] [Green Version]
- Ranganathan, A.; Smith, O.P.; Youssef, N.H.; Struchtemeyer, C.G.; Atiyeh, H.K.; Elshahed, M.S. Utilizing Anaerobic Fungi for Two-stage Sugar Extraction and Biofuel Production from Lignocellulosic Biomass. Front. Microbiol. 2017, 8, 635. [Google Scholar] [CrossRef] [Green Version]
- Kamra, D.; Singh, B. Anaerobic Gut Fungi. In Developments in Fungal Biology and Applied Mycology; Springer: Singapore, 2017; pp. 125–134. ISBN 978-981-10-4767-1. [Google Scholar]
- Dollhofer, V.; Podmirseg, S.M.; Callaghan, T.M.; Griffith, G.W.; Fliegerová, K. Anaerobic Fungi and Their Potential for Biogas Production. Adv. Biochem. Eng. Biotechnol. 2015, 151, 41–61. [Google Scholar] [CrossRef]
- Sharma, H.K.; Xu, C.; Qin, W. Biological Pretreatment of Lignocellulosic Biomass for Biofuels and Bioproducts: An Overview. Waste Biomass Valorization 2019, 10, 235–251. [Google Scholar] [CrossRef]
- Henske, J.K.; Gilmore, S.P.; Haitjema, C.H.; Solomon, K.V.; O’Malley, M.A. Biomass-degrading enzymes are catabolite repressed in anaerobic gut fungi. AIChE J. 2018, 64, 4263–4270. [Google Scholar] [CrossRef]
- Solomon, K.V.; Henske, J.K.; Gilmore, S.P.; Lipzen, A.; Grigoriev, I.V.; Thompson, D.; O’Malley, M.A. Catabolic repression in early-diverging anaerobic fungi is partially mediated by natural antisense transcripts. Fungal Genet. Biol. 2018, 121, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, M.J.; DE Kort, G.V.M.; Op Den Camp, H.J.M.; Tveld, J.H.H.I. Production of cellulolytic and xylanolytic enzymes during growth of the anaerobic fungus Piromyces sp. on different substrates. Microbiology 1992, 138, 1657–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Singh, A.; Himmel, M.E. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr. Opin. Biotechnol. 2009, 20, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K.; Murakami, M.; Nakashimada, Y.; Nishio, N. Efficient production of cellulolytic and xylanolytic enzymes by the rumen anaerobic fungus, Neocallimastix frontalis, in a repeated batch culture. J. Biosci. Bioeng. 2001, 91, 153–158. [Google Scholar] [CrossRef]
- Stevenson, D.M.; Weimer, P.J. Isolation and characterization of a Trichoderma strain capable of fermenting cellulose to ethanol. Appl. Microbiol. Biotechnol. 2002, 59, 721–726. [Google Scholar] [CrossRef]
- Ali, S.S.; Nugent, B.; Mullins, E.; Doohan, F.M. Insights from the fungus Fusarium oxysporum point to high affinity glucose transporters as targets for enhancing ethanol production from lignocellulose. PLoS ONE 2013, 8, e54701. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jin, W.; Mu, C.; Cheng, Y.; Zhu, W. Indigenously associated methanogens intensified the metabolism in hydrogenosomes of anaerobic fungi with xylose as substrate. J. Basic Microbiol. 2017, 57, 933–940. [Google Scholar] [CrossRef]
- Nakashimada, Y.; Srinivasan, K.; Murakami, M.; Nishio, N. Direct conversion of cellulose to methane by anaerobic fungus Neocallimastix frontalis and defined methanogens. Biotechnol. Lett. 2000, 22, 223–227. [Google Scholar] [CrossRef]
- Dincer, I.; Acar, C. Review and evaluation of hydrogen production methods for better sustainability. Int. J. Hydrog. Energy 2015, 40, 11094–11111. [Google Scholar] [CrossRef]
- IEA. The Future of Hydrogen; IEA: Paris, France, 2019. [Google Scholar]
- Hawkes, F.R.; Hussy, I.; Kyazze, G.; Dinsdale, R.; Hawkes, D.L. Continuous dark fermentative hydrogen production by mesophilic microflora: Principles and progress. Int. J. Hydrog. Energy 2007, 32, 172–184. [Google Scholar] [CrossRef]
- Ohta, S.; Miyamoto, K.; Miura, Y. Hydrogen Evolution as a Consumption Mode of Reducing Equivalents in Green Algal Fermentation. Plant Physiol. 1987, 83, 1022–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackstein, J.H.P.; Baker, S.E.; van Hellemond, J.J.; Tielens, A.G.M. Hydrogenosomes of Anaerobic Fungi: An Alternative Way to Adapt to Anaerobic Environments BT—Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes. In Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes; Tachezy, J., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 159–175. ISBN 978-3-030-17941-0. [Google Scholar]
- Van Niel, E.W.J.; Claassen, P.A.M.; Stams, A.J.M. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 2003, 81, 255–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hungate, R.E.; Smith, W.; Bauchop, T.; Yu, I.; Rabinowitz, J.C. Formate as an intermediate in the bovine rumen fermentation. J. Bacteriol. 1970, 102, 389–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Jin, W.; Cheng, Y.; Zhu, W. Effect of the Associated Methanogen Methanobrevibacter thaueri on the Dynamic Profile of End and Intermediate Metabolites of Anaerobic Fungus Piromyces sp. F1. Curr. Microbiol. 2016, 73, 434–441. [Google Scholar] [CrossRef]
- Mountfort, D.O.; Asher, R.A.; Bauchop, T. Fermentation of Cellulose to Methane and Carbon Dioxide by a Rumen Anaerobic Fungus in a Triculture with Methanobrevibacter sp. Strain RA1 and Methanosarcina barkeri. Appl. Environ. Microbiol. 1982, 44, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Wood, T.M.; Wilson, C.A.; McCrae, S.I.; Joblin, K.N. A highly active extracellular cellulase from the anaerobic rumen fungus Neocallimastix frontalis. FEMS Microbiol. Lett. 1986, 34, 37–40. [Google Scholar] [CrossRef]
- Joblin, K.N.; Naylor, G.E.; Williams, A.G. Effect of Methanobrevibacter smithii on Xylanolytic Activity of Anaerobic Ruminal Fungi. Appl. Environ. Microbiol. 1990, 56, 2287–2295. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.F.; Edwards, J.E.; Allison, G.G.; Zhu, W.-Y.; Theodorou, M.K. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour. Technol. 2009, 100, 4821–4828. [Google Scholar] [CrossRef] [Green Version]
- Marvin-Sikkema, F.D.; Richardson, A.J.; Stewart, C.S.; Gottschal, J.C.; Prins, R.A. Influence of hydrogen-consuming bacteria on cellulose degradation by anaerobic fungi. Appl. Environ. Microbiol. 1990, 56, 3793–3797. [Google Scholar] [CrossRef] [Green Version]
- Swift, C.L.; Brown, J.L.; Seppälä, S.; O’Malley, M.A. Co-cultivation of the anaerobic fungus Anaeromyces robustus with Methanobacterium bryantii enhances transcription of carbohydrate active enzymes. J. Ind. Microbiol. Biotechnol. 2019, 46, 1427–1433. [Google Scholar] [CrossRef]
- Joblin, K.N.; Williams, A.G. Effect of cocultivation of ruminal chytrid fungi with Methanobrevibacter smithii on lucerne stem degradation and extracellular fungal enzyme activities. Lett. Appl. Microbiol. 1991, 12, 121–124. [Google Scholar] [CrossRef]
- Ahring, B.K.; Westermann, P. Product inhibition of butyrate metabolism by acetate and hydrogen in a thermophilic coculture. Appl. Environ. Microbiol. 1988, 54, 2393–2397. [Google Scholar] [CrossRef] [Green Version]
- Cazier, E.A.; Trably, E.; Steyer, J.P.; Escudie, R. Biomass hydrolysis inhibition at high hydrogen partial pressure in solid-state anaerobic digestion. Bioresour. Technol. 2015, 190, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mah, R.A.; Kuenen, J.G.; Quayle, J.R.; Quayle, J.R.; Bull, A.T. Methanogenesis and methanogenic partnerships. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982, 297, 599–616. [Google Scholar] [CrossRef]
- Joblin, K.N.; Naylor, G.E. Fermentation of woods by rumen anaerobic fungi. FEMS Microbiol. Lett. 1989, 65, 119–122. [Google Scholar] [CrossRef]
- Webb, J.; Theodorou, M.K. Neocallimastix hurleyensis sp.nov., an anaerobic fungus from the ovine rumen. Can. J. Bot. 1991, 69, 1220–1224. [Google Scholar] [CrossRef]
- Brookman, J.L.; Mennim, G.; Trinci, A.P.J.; Theodorou, M.K.; Tuckwell, D.S. Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS1 and 18S rRNAThe GenBank accession numbers for the sequences determined in this work are given in Methods. Microbiology 2000, 146, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Niño-Navarro, C.; Chairez, I.; Torres-Bustillos, L.; Ramírez-Muñoz, J.; Salgado-Manjarrez, E.; Garcia-Peña, E.I. Effects of fluid dynamics on enhanced biohydrogen production in a pilot stirred tank reactor: CFD simulation and experimental studies. Int. J. Hydrog. Energy 2016, 41, 14630–14640. [Google Scholar] [CrossRef]
- Mizuno, O.; Dinsdale, R.; Hawkes, F.R.; Hawkes, D.L.; Noike, T. Enhancement of hydrogen production from glucose by nitrogen gas sparging. Bioresour. Technol. 2000, 73, 59–65. [Google Scholar] [CrossRef]
- Cho, S.-K.; Jeong, M.-W.; Choi, Y.-K.; Shin, J.; Shin, S.G. Effects of low-strength ultrasonication on dark fermentative hydrogen production: Start-up performance and microbial community analysis. Appl. Energy 2018, 219, 34–41. [Google Scholar] [CrossRef]
- Ramírez-Morales, J.E.; Tapia-Venegas, E.; Campos, J.L.; Ruiz-Filippi, G. Operational behavior of a hydrogen extractive membrane bioreactor (HEMB) during mixed culture acidogenic fermentation. Int. J. Hydrog. Energy 2019, 44, 25565–25574. [Google Scholar] [CrossRef]
- Kisielewska, M.; Dębowski, M.; Zieliński, M. Improvement of biohydrogen production using a reduced pressure fermentation. Bioprocess Biosyst. Eng. 2015, 38, 1925–1933. [Google Scholar] [CrossRef] [PubMed]
- Massanet-Nicolau, J.; Jones, R.J.; Guwy, A.; Dinsdale, R.; Premier, G.; Mulder, M.J.J. Maximising biohydrogen yields via continuous electrochemical hydrogen removal and carbon dioxide scrubbing. Bioresour. Technol. 2016, 218, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Han, S.-K.; Kim, S.-H.; Shin, H.-S. Effect of gas sparging on continuous fermentative hydrogen production. Int. J. Hydrog. Energy 2006, 31, 2158–2169. [Google Scholar] [CrossRef]
- Saady, N.M.C. Homoacetogenesis during hydrogen production by mixed cultures dark fermentation: Unresolved challenge. Int. J. Hydrog. Energy 2013, 38, 13172–13191. [Google Scholar] [CrossRef]
- Reilly, M.; Dinsdale, R.; Guwy, A. Mesophilic biohydrogen production from calcium hydroxide treated wheat straw. Int. J. Hydrog. Energy 2014, 39, 16891–16901. [Google Scholar] [CrossRef]
- Pan, X.; Angelidaki, I.; Alvarado-Morales, M.; Liu, H.; Liu, Y.; Huang, X.; Zhu, G. Methane production from formate, acetate and H2/CO2; focusing on kinetics and microbial characterization. Bioresour. Technol. 2016, 218, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Gong, H.; Liu, X.; Giwa, A.S.; Wang, K. Evaluation of bacterial association in methane generation pathways of an anaerobic digesting sludge via metagenomic sequencing. Arch. Microbiol. 2020, 202, 31–41. [Google Scholar] [CrossRef]
- Luo, G.; Xie, L.; Zhou, Q.; Angelidaki, I. Enhancement of bioenergy production from organic wastes by two-stage anaerobic hydrogen and methane production process. Bioresour. Technol. 2011, 102, 8700–8706. [Google Scholar] [CrossRef] [Green Version]
- Massanet-Nicolau, J.; Dinsdale, R.; Guwy, A.; Shipley, G. Use of real time gas production data for more accurate comparison of continuous single-stage and two-stage fermentation. Bioresour. Technol. 2013, 129, 561–567. [Google Scholar] [CrossRef]
- Guwy, A.J.; Dinsdale, R.M.; Kim, J.R.; Massanet-Nicolau, J.; Premier, G. Fermentative biohydrogen production systems integration. Bioresour. Technol. 2011, 102, 8534–8542. [Google Scholar] [CrossRef] [PubMed]
- Gest, H.; Kamen, M.D. Photoproduction of molecular hydrogen by Rhodospirillum rubrum. Science 1949, 109, 558 LP–559. [Google Scholar] [CrossRef]
- Akhlaghi, N.; Najafpour-Darzi, G. A comprehensive review on biological hydrogen production. Int. J. Hydrog. Energy 2020, 45, 22492–22512. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Harwood, C.S. Photobiological production of hydrogen gas as a biofuel. Curr. Opin. Biotechnol. 2010, 21, 244–251. [Google Scholar] [CrossRef]
- Hassan, A.H.S.; Mietzel, T.; Brunstermann, R.; Schmuck, S.; Schoth, J.; Küppers, M.; Widmann, R. Fermentative hydrogen production from low-value substrates. World J. Microbiol. Biotechnol. 2018, 34, 176. [Google Scholar] [CrossRef] [PubMed]
- Laurinavichene, T.V.; Belokopytov, B.F.; Laurinavichius, K.S.; Khusnutdinova, A.N.; Seibert, M.; Tsygankov, A.A. Towards the integration of dark- and photo-fermentative waste treatment. 4. Repeated batch sequential dark- and photofermentation using starch as substrate. Int. J. Hydrog. Energy 2012, 37, 8800–8810. [Google Scholar] [CrossRef]
- Nath, K.; Kumar, A.; Das, D. Hydrogen production by Rhodobacter sphaeroides strain O.U.001 using spent media of Enterobacter cloacae strain DM11. Appl. Microbiol. Biotechnol. 2005, 68, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Odom, J.M.; Wall, J.D. Photoproduction of h(2) from cellulose by an anaerobic bacterial coculture. Appl. Environ. Microbiol. 1983, 45, 1300–1305. [Google Scholar] [CrossRef] [Green Version]
- Özgür, E.; Mars, A.E.; Peksel, B.; Louwerse, A.; Yücel, M.; Gündüz, U.; Claassen, P.A.M.; Eroğlu, İ. Biohydrogen production from beet molasses by sequential dark and photofermentation. Int. J. Hydrog. Energy 2010, 35, 511–517. [Google Scholar] [CrossRef]
- Argun, H.; Kargi, F.; Kapdan, I.K. Light fermentation of dark fermentation effluent for bio-hydrogen production by different Rhodobacter species at different initial volatile fatty acid (VFA) concentrations. Int. J. Hydrog. Energy 2008, 33, 7405–7412. [Google Scholar] [CrossRef]
- Liu, H.; Grot, S.; Logan, B.E. Electrochemically Assisted Microbial Production of Hydrogen from Acetate. Environ. Sci. Technol. 2005, 39, 4317–4320. [Google Scholar] [CrossRef] [PubMed]
- Kadier, A.; Simayi, Y.; Kalil, M.S.; Abdeshahian, P.; Hamid, A.A. A review of the substrates used in microbial electrolysis cells (MECs) for producing sustainable and clean hydrogen gas. Renew. Energy 2014, 71, 466–472. [Google Scholar] [CrossRef]
- Cheng, S.; Logan, B.E. Sustainable and efficient biohydrogen production via electrohydrogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 18871–18873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, E.; Omar Mohamed, H.; Park, S.-G.; Obaid, M.; Al-Qaradawi, S.Y.; Castaño, P.; Chon, K.; Chae, K.-J. A review on self-sustainable microbial electrolysis cells for electro-biohydrogen production via coupling with carbon-neutral renewable energy technologies. Bioresour. Technol. 2021, 320, 124363. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Luo, H.; Liu, G.; Zhang, R.; Li, J.; Fu, S. Improved Hydrogen Production in the Microbial Electrolysis Cell by Inhibiting Methanogenesis Using Ultraviolet Irradiation. Environ. Sci. Technol. 2014, 48, 10482–10488. [Google Scholar] [CrossRef] [PubMed]
- Lalaurette, E.; Thammannagowda, S.; Mohagheghi, A.; Maness, P.-C.; Logan, B.E. Hydrogen production from cellulose in a two-stage process combining fermentation and electrohydrogenesis. Int. J. Hydrog. Energy 2009, 34, 6201–6210. [Google Scholar] [CrossRef]
- Li, X.-H.; Liang, D.-W.; Bai, Y.-X.; Fan, Y.-T.; Hou, H.-W. Enhanced H2 production from corn stalk by integrating dark fermentation and single chamber microbial electrolysis cells with double anode arrangement. Int. J. Hydrog. Energy 2014, 39, 8977–8982. [Google Scholar] [CrossRef]
- Lu, L.; Ren, N.; Xing, D.; Logan, B.E. Hydrogen production with effluent from an ethanol–H2-coproducing fermentation reactor using a single-chamber microbial electrolysis cell. Biosens. Bioelectron. 2009, 24, 3055–3060. [Google Scholar] [CrossRef]
- Werner, C.M.; Katuri, K.P.; Hari, A.R.; Chen, W.; Lai, Z.; Logan, B.E.; Amy, G.L.; Saikaly, P.E. Graphene-Coated Hollow Fiber Membrane as the Cathode in Anaerobic Electrochemical Membrane Bioreactors—Effect of Configuration and Applied Voltage on Performance and Membrane Fouling. Environ. Sci. Technol. 2016, 50, 4439–4447. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Prévoteau, A.; Rabaey, K. A novel tubular microbial electrolysis cell for high rate hydrogen production. J. Power Sources 2017, 356, 484–490. [Google Scholar] [CrossRef]
- Call, D.; Logan, B.E. Hydrogen Production in a Single Chamber Microbial Electrolysis Cell Lacking a Membrane. Environ. Sci. Technol. 2008, 42, 3401–3406. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saye, L.M.G.; Navaratna, T.A.; Chong, J.P.J.; O’Malley, M.A.; Theodorou, M.K.; Reilly, M. The Anaerobic Fungi: Challenges and Opportunities for Industrial Lignocellulosic Biofuel Production. Microorganisms 2021, 9, 694. https://doi.org/10.3390/microorganisms9040694
Saye LMG, Navaratna TA, Chong JPJ, O’Malley MA, Theodorou MK, Reilly M. The Anaerobic Fungi: Challenges and Opportunities for Industrial Lignocellulosic Biofuel Production. Microorganisms. 2021; 9(4):694. https://doi.org/10.3390/microorganisms9040694
Chicago/Turabian StyleSaye, Luke M. G., Tejas A. Navaratna, James P. J. Chong, Michelle A. O’Malley, Michael K. Theodorou, and Matthew Reilly. 2021. "The Anaerobic Fungi: Challenges and Opportunities for Industrial Lignocellulosic Biofuel Production" Microorganisms 9, no. 4: 694. https://doi.org/10.3390/microorganisms9040694
APA StyleSaye, L. M. G., Navaratna, T. A., Chong, J. P. J., O’Malley, M. A., Theodorou, M. K., & Reilly, M. (2021). The Anaerobic Fungi: Challenges and Opportunities for Industrial Lignocellulosic Biofuel Production. Microorganisms, 9(4), 694. https://doi.org/10.3390/microorganisms9040694







