Transcriptome Analysis on the Mechanism of Ethylicin Inhibiting Pseudomonas syringae pv. actinidiae on Kiwifruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bioassay Methods
2.3. Time-Concentration Dependence Measurement of Ethylicin on Psa Growth
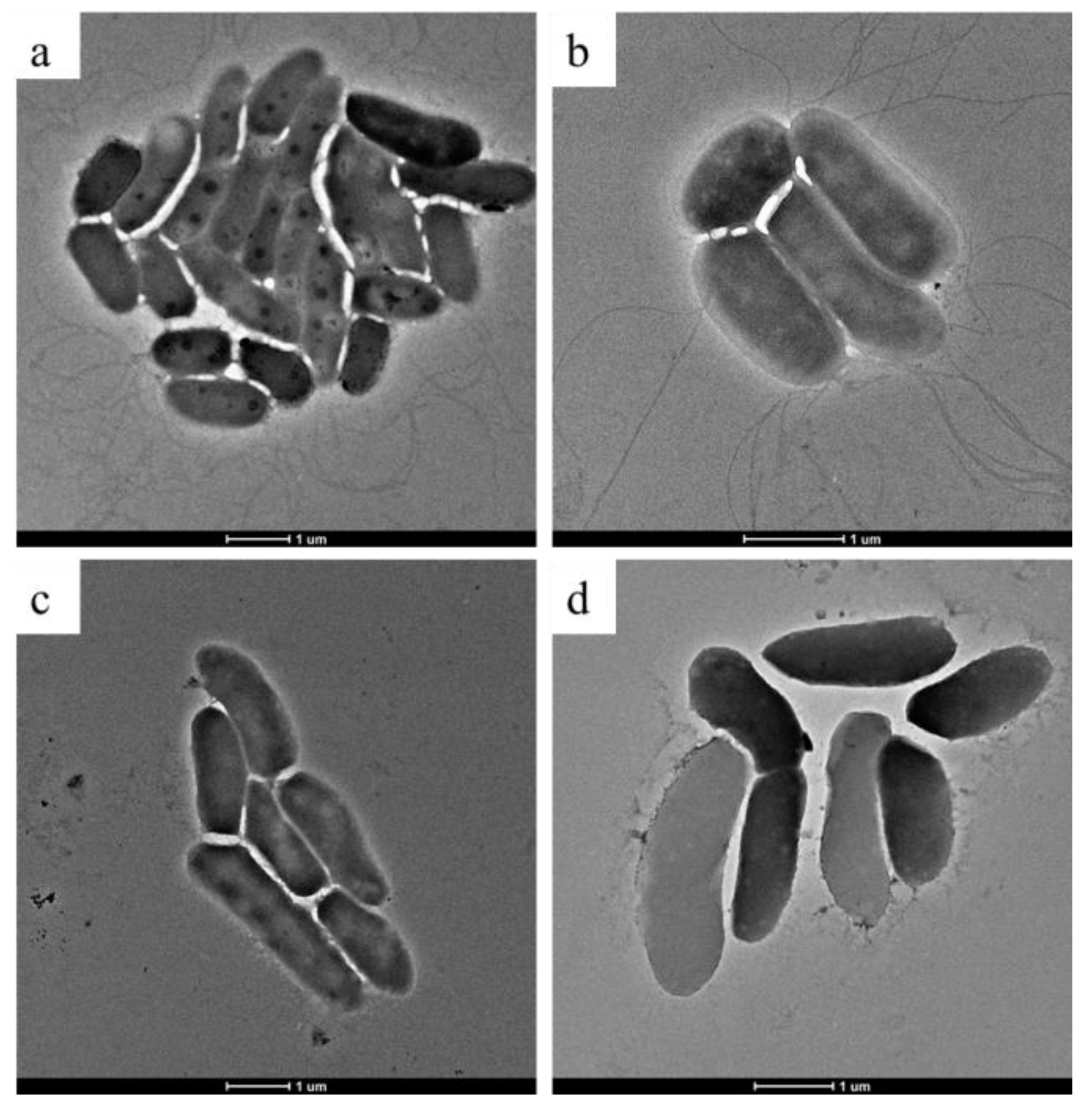
2.4. Morphological Analysis by Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM)
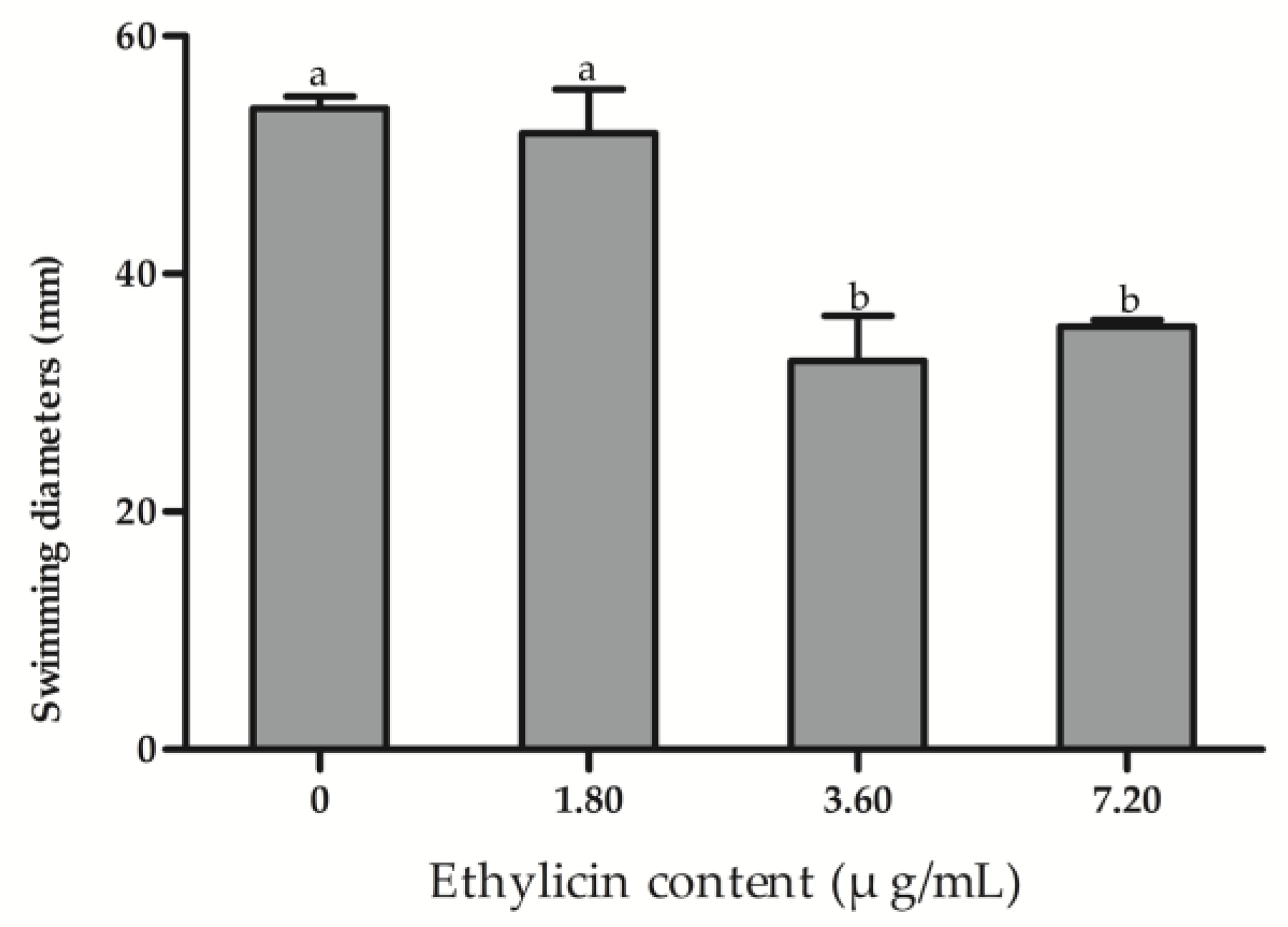
2.5. Bacteria Swimming
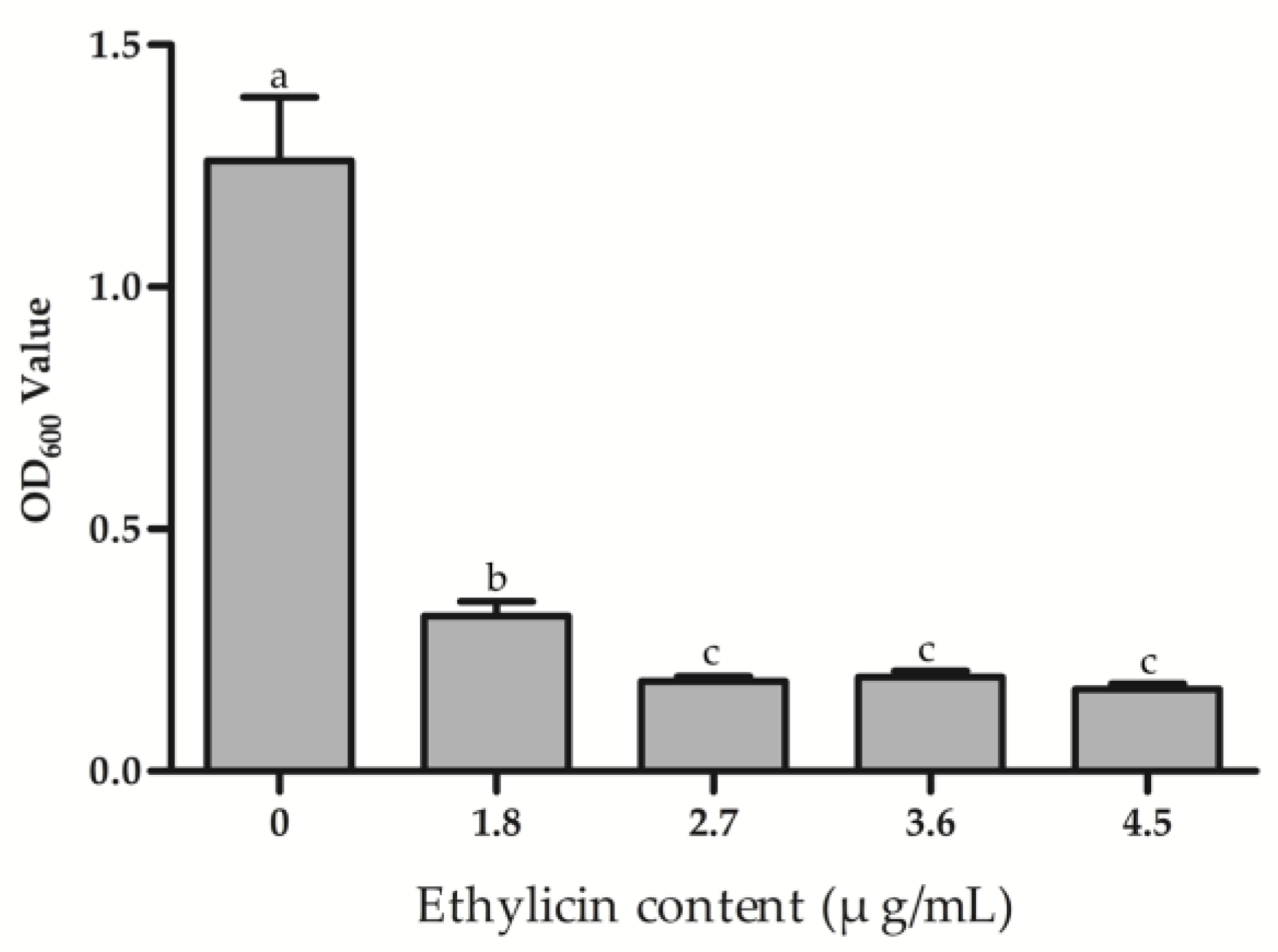
2.6. Effect of Ethylicin on Psa Biofilm Formation
2.7. Psa RNA Extraction, cDNA Library Construction, and Sequencing
2.8. RNA-Seq Data Analysis
2.9. Quantitative RT-PCR
2.10. Statistical Analysis
3. Results
3.1. Bioassay Results
3.2. Psa Growth Concentration–Time Dependence on Ethylicin
3.3. SEM and TEM Results
3.4. Effect of Ethylicin on Psa Biofilm Formation and Bacterial Migration
3.5. Transcriptome Results
3.5.1. Quality Control of Sequencing Data
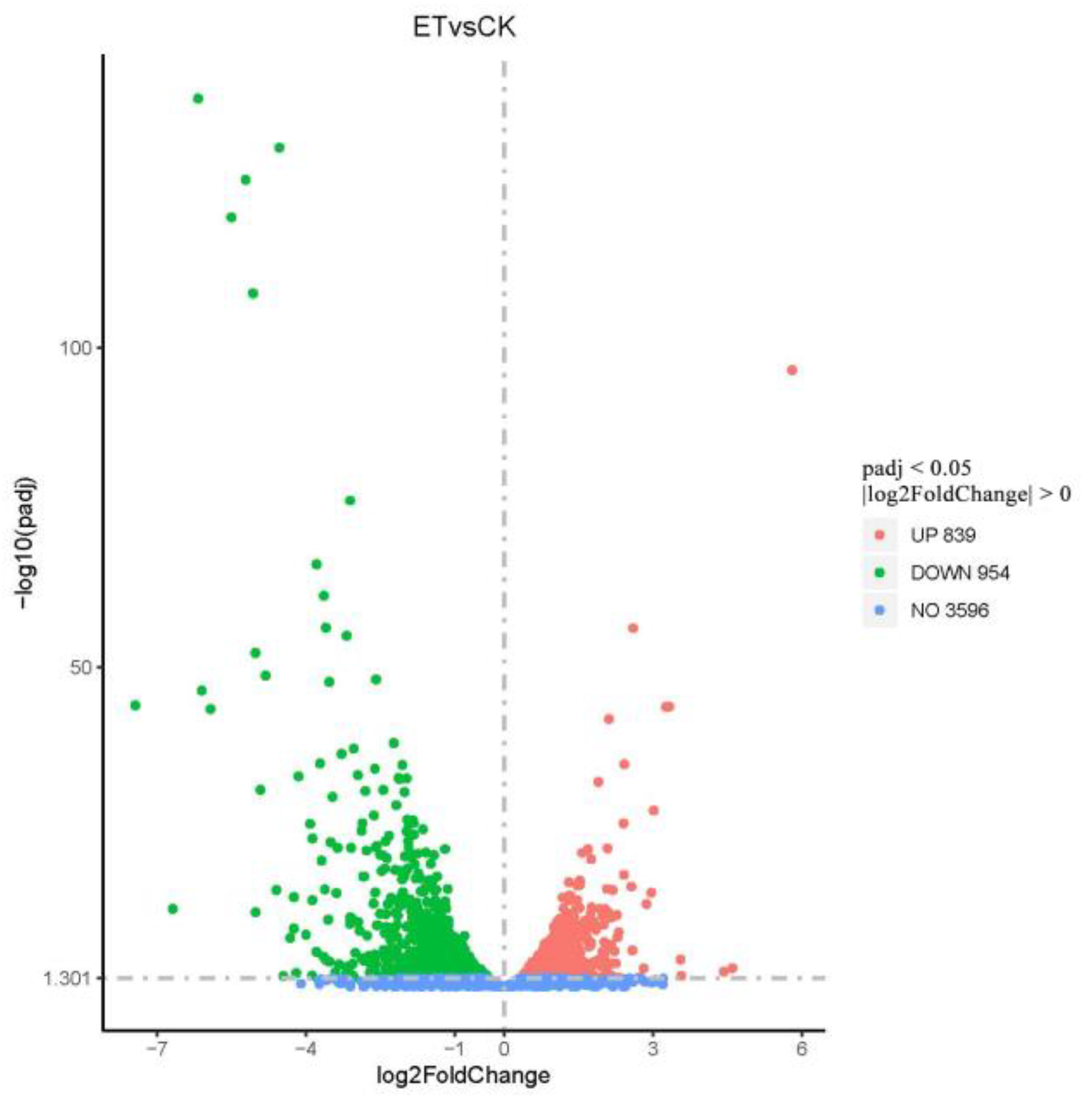
3.5.2. Differential Gene Expression Analysis
3.5.3. Gene Ontology Annotation
3.5.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Annotation
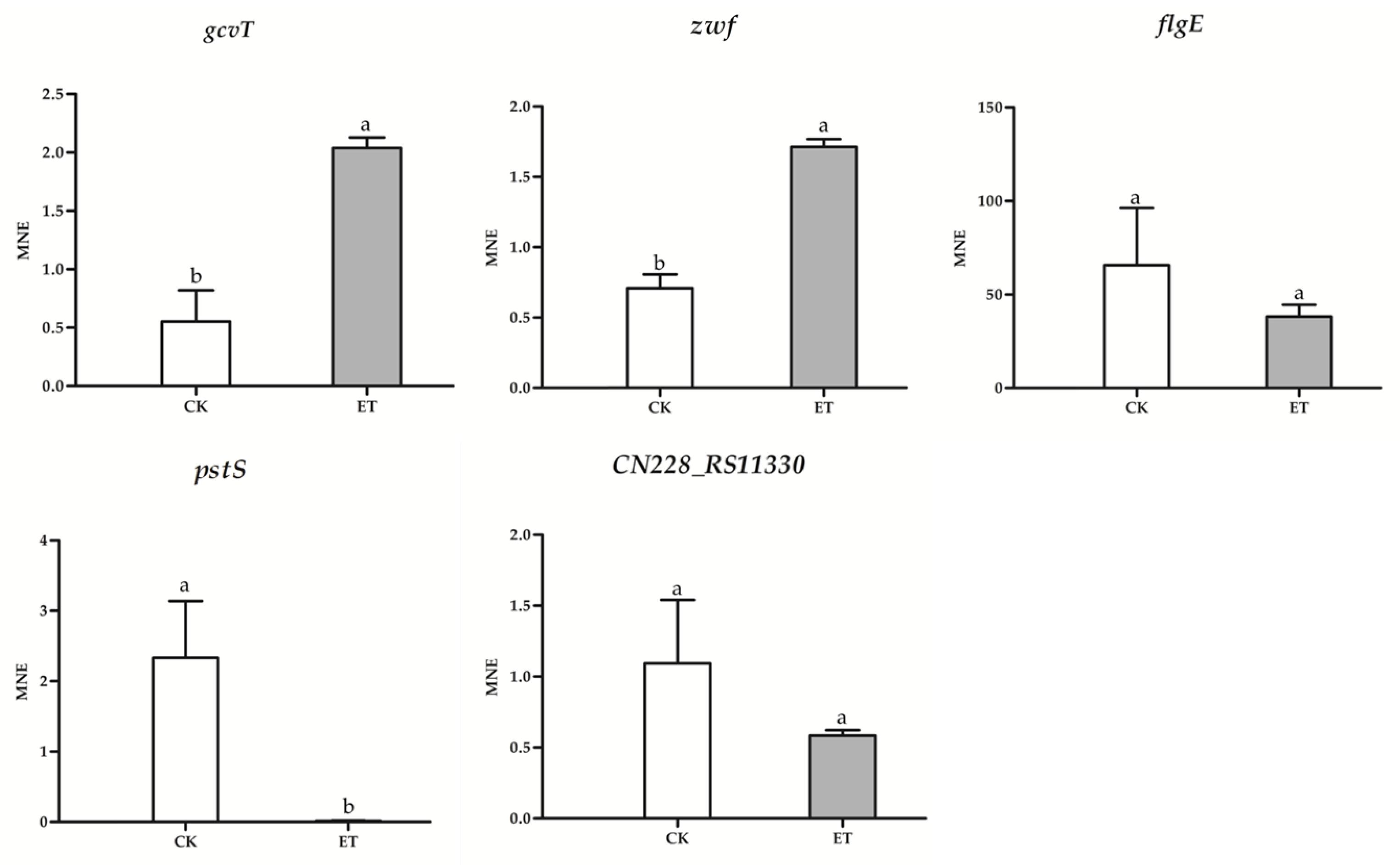
3.6. qRT-PCR Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghods, S.; Sims, I.M.; Moradali, M.F.; Rehm, B.H.A. Bactericidal compounds controlling growth of the plant pathogen Pseudomonas syringae pv. actinidiae, which forms biofilms composed of a novel exopolysaccharide. Appl. Environ. Microbiol. 2015, 81, 4026–4036. [Google Scholar] [CrossRef] [Green Version]
- Opgenorth, D.C.; Lai, M.; Sorrell, M.; White, J.B. Pseudomonas canker of kiwifruit. Plant Dis. 1983, 67, 1283–1284. [Google Scholar] [CrossRef] [Green Version]
- Serizawa, S.; Ichikawa, T.; Takikawa, Y.; Tsuyumu, S.; Goto, M. Occurrence of bacterial canker of kiwifruit in Japan: Description of symptoms, isolation of the pathogen and screening of bactericides. Ann. Phytopath. Soc. Japan 1989, 55, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Yuichi, T.; Setsuo, S.; Takeshi, I.; Shinji, T.; Masao, G. Pseudomonas syringae pv. actinidae pv. nov.: The causal bacterium of canker of kiwifruit in Japan. Ann. Phytopath. Soc. Japan 1989, 55, 437–444. [Google Scholar]
- Cheng, H.Y.; Li, Y.; Wan, S.K.; Zhang, J.; Pang, Q.; Li, G.; Xing, J.H. Pathogenic identification of kiwifruit bacterial canker in Anhui. J. Anhui Agric. Univ. 1995, 22, 219–223. [Google Scholar]
- Scortichini, M. Occurrence of Pseudomonas syringae pv. actinidiae on kiwifruit in Italy. Plant Pathol. 1994, 43, 1035–1038. [Google Scholar] [CrossRef]
- Mazarei, M.; Mostofipour, P. First report of bacterial canker of kiwifruit in Iran. Plant Pathol. 1994, 43, 1055–1056. [Google Scholar] [CrossRef]
- Balestra, G.M.; Renzi, M.; Mazzaglia, A. First report of bacterial canker of Actinidia deliciosa caused by Pseudomonas syringae pv. actinidiae in Portugal. New Dis. Rep. 2010, 22, 10. [Google Scholar] [CrossRef] [Green Version]
- Everett, K.R.; Taylor, R.K.; Romberg, M.K.; Rees-George, J.; Fullerton, R.A.; Vanneste, J.L.; Manning, M.A. First report of Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker in new zealand. Australas. Plant Dis. Notes 2011, 6, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Jae, H.L.; Jung, H.K.; Gyoung, H.K.; Jae, S.J.; Jae, S.H.; Young, J.K. Comparative analysis of Korean and Japanese strains of Pseudomonas syringae pv. actinidiae causing bacterial canker of kiwifruit. Plant Pathol. J. 2005, 21, 119–126. [Google Scholar]
- Altimira, F.; Yáñez, C.; Bravo, G.; González, M.; Rojas, L.A.; Seeger, M. Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central chile. BMC Microbiol. 2012, 12, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Cellini, A.; Fiorentini, L.; Buriani, G.; Yu, J.; Donati, I.; Cornish, D.A.; Novak, B.; Costa, G.; Vanneste, J.L.; Spinelli, F. Elicitors of the salicylic acid pathway reduce incidence of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidae. Ann. Appl. Biol. 2014, 165, 441–453. [Google Scholar] [CrossRef]
- Monchiero, M.; Gullino, M.L.; Pugliese, M.; Spadaro, D.; Garibaldi, A. Efficacy of different chemical and biological products in the control of Pseudomonas syringae pv. actinidiae on kiwifruit. Australas. Plant Pathol. 2015, 44, 13–23. [Google Scholar] [CrossRef]
- Long, Y.H.; Yin, X.H.; Wang, M.; Wu, X.M.; Li, R.Y.; Tian, X.L.; Li, M. Effects of sulfur on kiwifruit canker caused by Pseudomonas syringae pv. actinidae. Bangladesh J. Bot. 2017, 46, 1183–1192. [Google Scholar]
- Donati, I.; Buriani, G.; Cellini, A.; Mauri, S.; Costa, G.; Spinelli, F. New insights on the bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). J. Berry Res. 2014, 4, 53–67. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.L.; Zhang, H.; Ding, R.Y.; Wang, W.Z.; Wang, W.B.; Li, H.D. Determination and dynamics of ethylicin residues in cotton-field ecosystem. Bull. Environ. Contam. Toxicol. 2012, 89, 853–856. [Google Scholar] [CrossRef]
- Lu, X.; Li, C.P.; Shi, T.; Huang, G.X. Bactericide Screening against Xanthomonas axonopodis pv. manihotis. Plant Dis. Pests 2014, 5, 1–3. [Google Scholar]
- Zhang, S.M.; Liu, S.; Zhang, J.K.; Reiter, R.J.; Wang, Y.; Qiu, D.; Luo, X.M.; Khalid, A.R.; Wang, H.Y.; Feng, L.; et al. Synergistic anti-oomycete effect of melatonin with a biofungicide against oomycetic black shank disease. J. Pineal. Res. 2018, 65, e12492. [Google Scholar] [CrossRef]
- Zhang, S.M.; Zhang, M.Q.; Khalid, A.R.; Li, L.X.; Chen, Y.; Dong, P.; Wang, H.Y.; Ren, M.Z. Ethylicin prevents potato late blight by disrupting protein biosynthesis of phytophthora infestans. Pathogens 2020, 9, 299. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Liu, P.; Zhang, Y.L.; Ma, L.G.; Qi, K.; Li, C.S.; Qi, J.S. Toxicity of several biological agents to wheat root rot. J. Triticeae Crop. 2018, 38, 366–371. [Google Scholar]
- Fu, R.T.; Ke, Y.X.; Chen, C.; Wang, J.; Gong, X.S.; Lu, D.H.; Liao, Y. Toxicity test and field control effect of ethylicin against several plant pathogens. Agrochemicals 2018, 57, 611–613. [Google Scholar]
- Buchenauer, H.; Rohner, E. Effect of triadimefonand and triadimenol on growth of various plant as well as on species gibberellin content and sterol metabolism in shoots of barley. Pest. Biochem. Physiol. 1981, 15, 58–70. [Google Scholar] [CrossRef]
- Hu, R.P.; Shi, J.; Lin, L.J.; Yang, D.S.; Ye, H.L.; Chen, C.; Fan, Z.H.; Chen, S.; Chen, Q.D. Efficient bactericides for kiwifruit canker: Screening and field application evaluation. Chin. Agric. Sci. Bull. 2020, 36, 112–116. [Google Scholar]
- Zhao, Y.L.; Huang, X.; Liu, L.W.; Wang, P.Y.; Long, Q.S.; Tao, Q.Q.; Li, Z.; Yang, S. Identification of racemic and chiral carbazole derivatives containing an isopropanolamine linker as prospective surrogates against plant pathogenic bacteria: In vitro and in vivo assays and quantitative proteomics. J. Agric. Food Chem. 2019, 67, 7512–7525. [Google Scholar] [CrossRef]
- Liu, H.W.; Ji, Q.T.; Ren, G.G.; Wang, F.; Su, F.; Wang, P.Y.; Zhou, X.; Wu, Z.B.; Li, Z.; Yang, S. Antibacterial functions and proposed modes of action of novel 1,2,3,4-tetrahydro-β-carboline derivatives that possess an attractive 1,3-diaminopropan-2-ol pattern against rice bacterial blight, kiwifruit bacterial canker, and citrus bacterial canker. J. Agric. Food Chem. 2020, 68, 12558–12568. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, C.; Laborda, P.; Zhao, Y.C.; Palmer, I.; Fu, Z.Q.; Qiu, J.P.; Liu, F.Q. Melatonin treatment inhibits the growth of Xanthomonas oryzae pv. oryzae. Front. Microbiol. 2018, 9, 2280. [Google Scholar] [CrossRef] [Green Version]
- Tao, Q.Q.; Liu, L.W.; Wang, P.Y.; Long, Q.S.; Zhao, Y.L.; Jin, L.H.; Xu, W.M.; Chen, Y.; Li, Z.; Yang, S. Synthesis and in vitro and in vivo biological activity evaluation and quantitative proteome profiling of oxadiazoles bearing flexible heterocyclic patterns. J. Agric. Food Chem. 2019, 67, 7626–7639. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Feng, Y.M.; Qi, P.Y.; Shao, W.B.; Wu, Z.B.; Liu, L.W.; Wang, Y.; Ma, H.D.; Wang, P.Y.; Li, Z.; et al. Synthesis and docking study of N-(Cinnamoyl)-N’-(substituted)acryloyl hydrazide derivatives containing pyridinium moieties as a novel class of filamentous temperature-sensitive protein Z inhibitors against the intractable Xanthomonas oryzae pv. oryzae infections in Rice. J. Agric. Food Chem. 2020, 68, 8132–8142. [Google Scholar] [PubMed]
- Lovato, A.; Pignatti, A.; Vitulo, N.; Vandelle, E.; Polverari, A. Inhibition of virulence-related traits in Pseudomonas syringae pv. actinidiae by gunpowder green tea extracts. Front. Microbiol. 2019, 10, 2362. [Google Scholar] [CrossRef]
- Ni, P.; Wang, L.; Deng, B.H.; Jiu, S.T.; Ma, C.; Zhang, C.X.; Almeida, A.; Wang, D.P.; Xu, W.P.; Wang, S.P. Combined application of bacteriophages and carvacrol in the control of Pseudomonas syringae pv. actinidiae planktonic and biofilm forms. Microorganisms 2020, 8, 837. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression of RNA-Seq data at the gene level-the DESeq package. Heidelb. Ger. Eur. Mol. Biol. Lab. (EMBL) 2012, 10, f1000. [Google Scholar]
- Duan, Q.D.; Zhou, M.X.; Zhu, L.Q.; Zhu, G.Q. Flagella and bacterial pathogenicity. J. Basic Microb. 2012, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chaban, B.; Hughes, H.V.; Beeby, M. The flagellum in bacterial pathogens: For motility and a whole lot more. Semin. Cell Dev. Biol. 2015, 46, 91–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemon, K.P.; Higgins, D.E.; Kolter, R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 2007, 189, 4418–4424. [Google Scholar] [CrossRef] [Green Version]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef]
- Lawrence, J.R.; Delaquis, P.J.; Korber, D.R.; Caldwell, D.E. Behaviour of Pseudomonas fluorescens within the hydrodynamic boundary layers of surface microenvironments. Microb. Ecol. 1987, 14, 1–14. [Google Scholar] [CrossRef]
- Molina-Henares, M.A.; Ramos-González, M.I.; Daddaoua, A.; Fernández-Escamilla, A.M.; Espinosa-Urgel, M. Fleq of Pseudomonas putida kt2440 is a multimeric cyclic diguanylate binding protein that differentially regulates expression of biofilm matrix components. Res. Microbiol. 2016, 1–10. [Google Scholar] [CrossRef]
- Jiménez-Fernández, A.; López-Sánchez, A.; Jiménez-Díaz, L.; Navarrete, B.; Calero, P.; Platero, A.I.; Govantes, F. Complex interplay between fleQ, cyclic diguanylate and multiple σ factors coordinately regulates flagellar motility and biofilm development in Pseudomonas putida. PLoS ONE 2016, 11, e0163142. [Google Scholar] [CrossRef]
- Baker, P.; Whitfield, G.B.; Hill, P.J.; Little, D.J.; Pestrak, M.J.; Robinson, H.; Wozniak, D.J.; Howell, P.L. Characterization of the Pseudomonas aeruginosa glycoside hydrolase pslg reveals that its levels are critical for psl polysaccharide biosynthesis and biofilm formation. J. Biol. Chem. 2015, 290, 28374–28387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Merritt, P.M.; Danhorn, T.; Fuqua, C. Motility and chemotaxis in agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 2007, 189, 8005–8014. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Bactericide | Treatment (μg/mL) | Inhibition Rate (%) | Regression Equation | R | EC50 (μg/mL) |
|---|---|---|---|---|---|
| Ethylicin | 50 | 99.59 ± 0.30 | y = 3.4788x + 4.1133 | 0.9478 | 1.80 ± 0.06 |
| 10 | 99.03 ± 0.17 | ||||
| 5 | 98.46 ± 0.10 | ||||
| Kocide 3000 | 100 | 94.60 ± 0.94 | y = 3.9823x − 1.2784 | 0.9930 | 37.72 ± 2.12 |
| 50 | 74.72 ± 3.24 | ||||
| Copper abietate | 100 | 44.15 ± 5.16 | y = 0.8925x + 3.0393 | 0.9964 | 157.35 ± 5.88 |
| 50 | 32.67 ± 2.22 |
| Treatment | 14 Days after Inoculation | ||
|---|---|---|---|
| Morbidity (%) | Lesion Length (mm) | Control Efficiency (%) | |
| (a) Blank control * | 0 | / | / |
| (b) Psa | 100 | 6.27 ± 1.41a | / |
| (c) 200 μg/mL Kocide 3000 | 100 | 2.19 ± 0.87b | 65.00% ± 13.92%b |
| (d) 200 μg/mL Ethylicin | 100 | 0.56 ± 0.24 b | 91.01% ± 3.87% a |
| (e) 500 μg/mL Kocide 3000 | 100 | 0.66 ± 0.26b | 89.47% ± 4.10%a |
| (f) 500 μg/mL Ethylicin | 100 | 0.5 ± 0.16 b | 92.02% ± 2.51% a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Ren, X.; Cao, G.; Zhou, X.; Jin, L. Transcriptome Analysis on the Mechanism of Ethylicin Inhibiting Pseudomonas syringae pv. actinidiae on Kiwifruit. Microorganisms 2021, 9, 724. https://doi.org/10.3390/microorganisms9040724
Liu T, Ren X, Cao G, Zhou X, Jin L. Transcriptome Analysis on the Mechanism of Ethylicin Inhibiting Pseudomonas syringae pv. actinidiae on Kiwifruit. Microorganisms. 2021; 9(4):724. https://doi.org/10.3390/microorganisms9040724
Chicago/Turabian StyleLiu, Tao, Xiaoli Ren, Guangyun Cao, Xia Zhou, and Linhong Jin. 2021. "Transcriptome Analysis on the Mechanism of Ethylicin Inhibiting Pseudomonas syringae pv. actinidiae on Kiwifruit" Microorganisms 9, no. 4: 724. https://doi.org/10.3390/microorganisms9040724






