Functional and Molecular Characterization of the Halomicrobium sp. IBSBa Inulosucrase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Cultivation Conditions
2.2. Cloning of HmcIsc
2.3. Expression and Purification of HmcIsc
2.4. SDS-PAGE Analysis
2.5. Protein and Enzyme Activity Assays
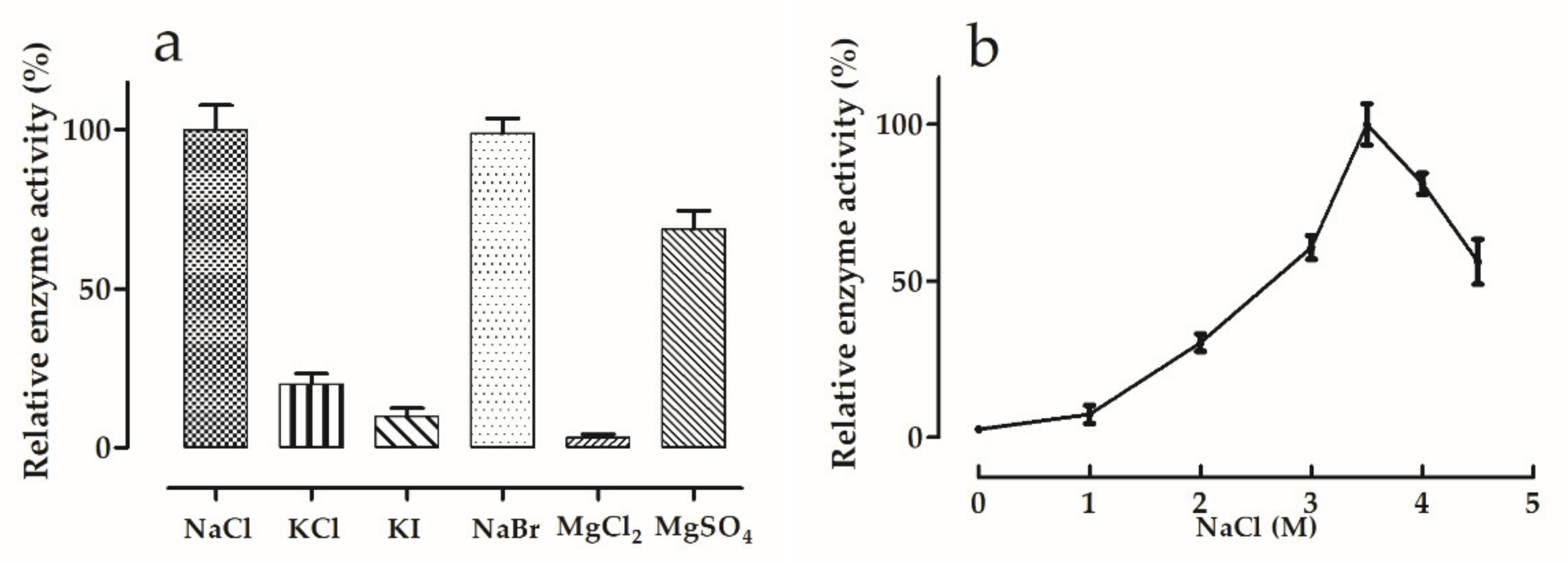
2.6. Effects of Salt Type and Salt Concentration
2.7. Effects of pH and Temperature
2.8. Effects of Various Substances
2.9. Enzyme Activity over Time
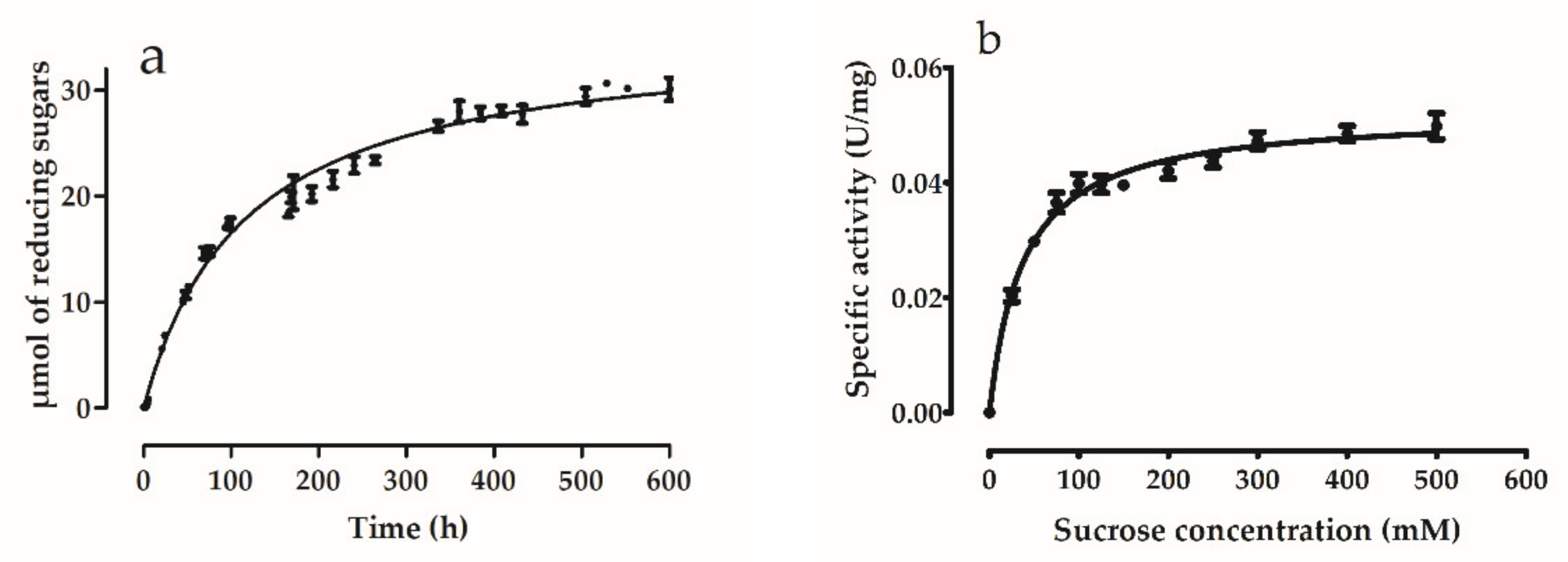
2.10. Enzyme Kinetics
2.11. Fructan Production and Purification
2.12. NMR Characterization of the Fructan
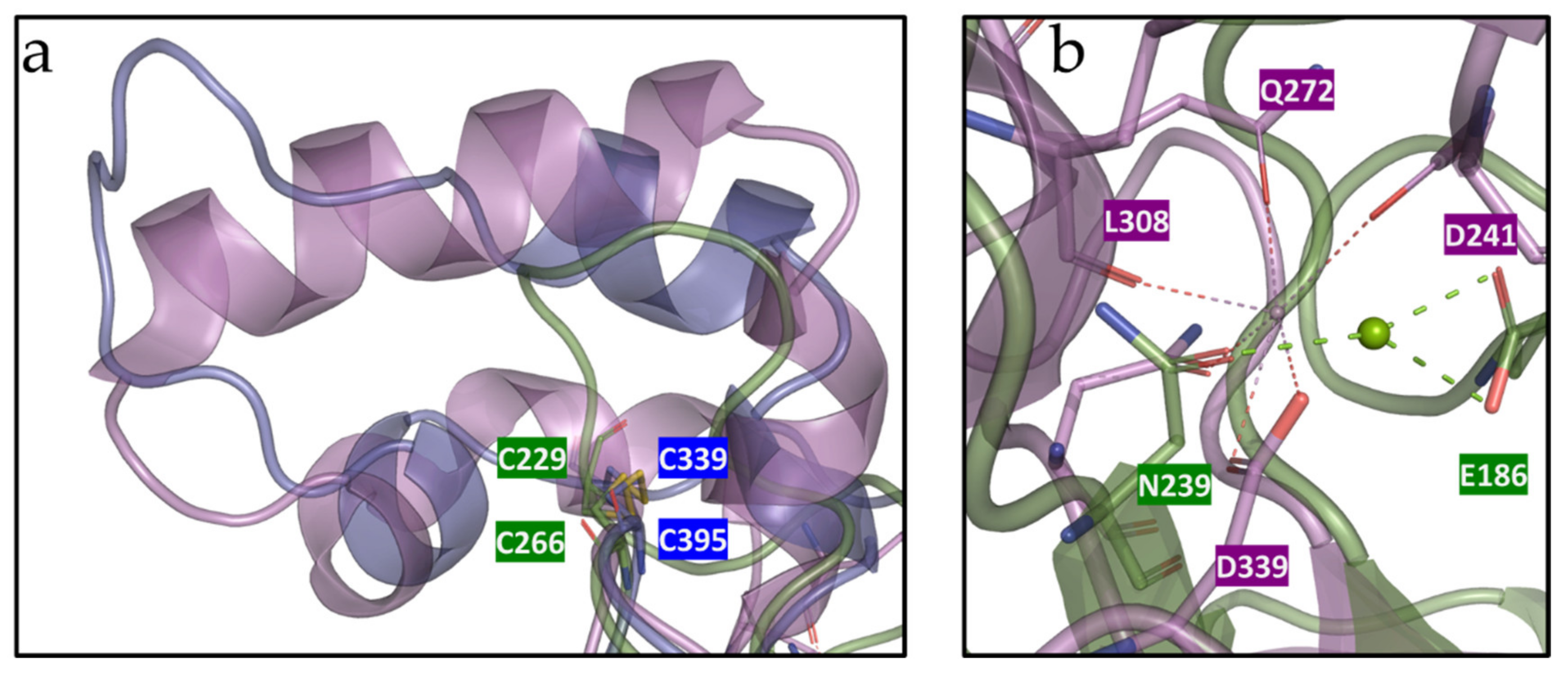
2.13. Molecular and Structural Analyses
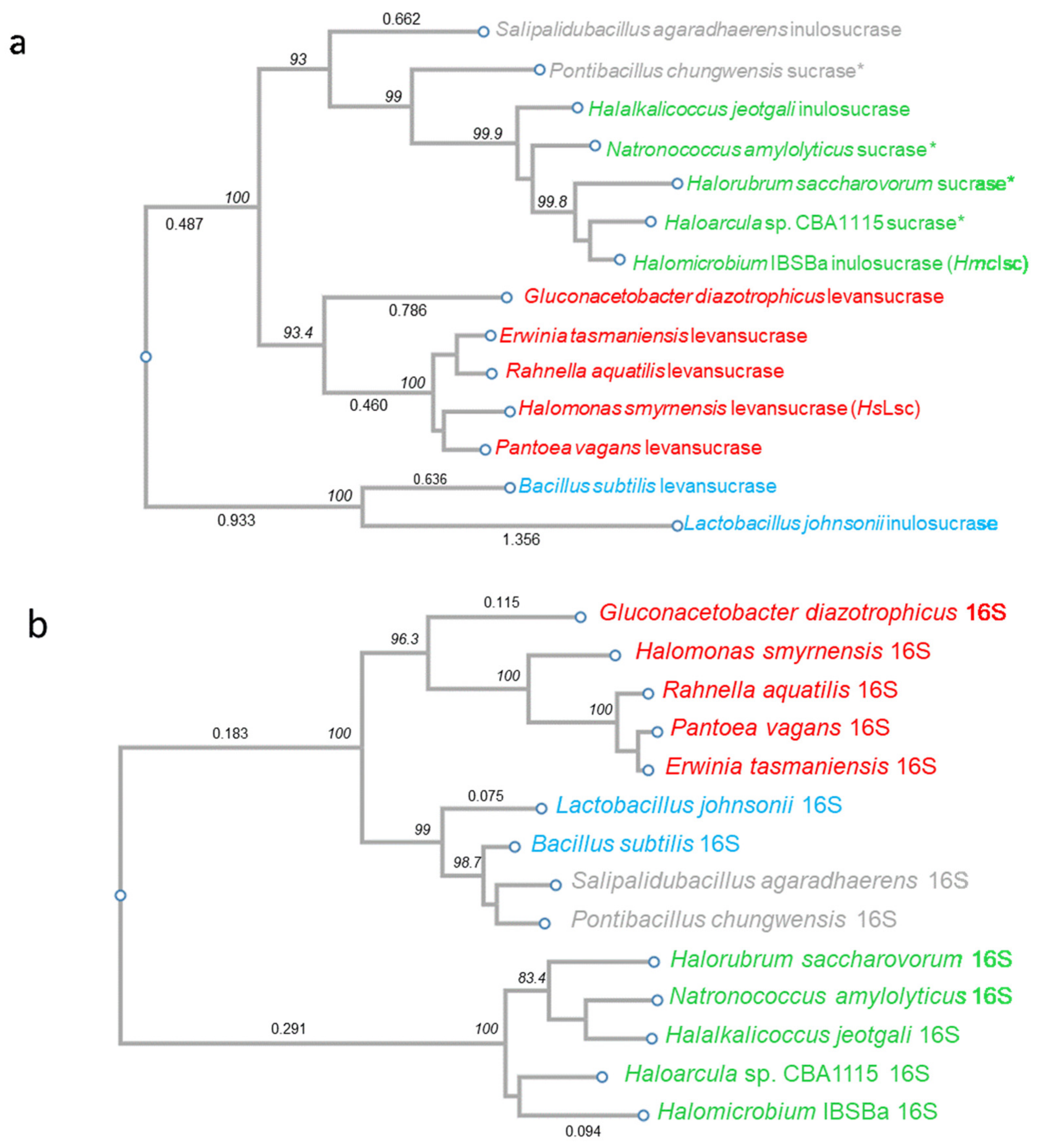
2.14. Phylogenetic Analysis
3. Results and Discussion
3.1. ORF Analysis of the Halomicrobium sp. IBSBa GH68 Member
3.2. Phylogenetic Analysis: Evidence in Favour of HGT
3.3. Cloning, Heterologous Expression and Purification
3.4. Product Identification through NMR
3.5. Enzymatic Characterization
3.5.1. Effects of pH and Temperature
3.5.2. Effects of Salt Type and Concentration
3.5.3. Effects of Various Substances
3.5.4. Enzyme Kinetics
3.6. Molecular and Structural Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blöchl, E.; Rachel, R.; Burggraf, S.; Hafenbradl, D.; Jannasch, H.W.; Stetter, K.O. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113 °C. Extremophiles 1997, 1, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Bolhuis, H.; Palm, P.; Wende, A.; Falb, M.; Rampp, M.; Rodriguez-Valera, F.; Pfeiffer, F.; Oesterhelt, D. The genome of the square archaeon Haloquadratum walsbyi: Life at the limits of water activity. BMC Genom. 2006, 7, 169. [Google Scholar] [CrossRef] [Green Version]
- Schleper, C.; Pühler, G.; Klenk, H.P.; Zillig, W. Picrophilus oshimae and Picrophilus torridae fam. Nov. gen. nov., sp. nov., Two Species of Hyperacidophilic, Thermophilic, Heterotrophic, Aerobic Archaea. Int. J. Syst. Bact. 1996, 46, 814–816. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Liu, Y.; Pan, J.; Wang, F.; Li, M. Perspectives on Cultivation Strategies of Archaea. Microb. Ecol. 2020, 79, 770–784. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Gouy, M. Adaptation to Environmental Temperature is a Major Determinant of Molecular Evolutionary Rates in Archaea. Mol. Biol. Evol. 2011, 28, 2661–2674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrel, G.; Brugere, J.F.; Gribaldo, S.; Schmitz, R.A.; Mossl-Eichinger, C. The host-associated archeome. Nat. Rev. Microbiol. 2020, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Borrel, G.; McCann, A.; Deane, J.; Neto, M.C.; Lynch, D.B.; Brugère, J.F.; O’Toole, P.W. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 2017, 11, 2059–2074. [Google Scholar] [CrossRef] [Green Version]
- Taffner, J.; Erlacher, A.; Bragina, A.; Berg, C.; Moissl-Eichinger, C.; Berg, G. What is the Role of Archaea in Plants? New Insights from the Vegetation of Alpine Bogs. mSphere 2018, 3, e00122-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayangbenro, A.S.; Babalola, O.O. Reclamation of arid and semi-arid soils: The role of plant growth-promoting archaea and bacteria. Curr. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Williams, T.J.; Allen, M.A.; DeMaere, M.Z.; Kyrpides, N.C.; Tringe, S.G.; Woyke, T.; Cavicchioli, R. Microbial ecology of an Antartic hypersaline lake: Genomic assessment of ecophysiology among dominant haloarchaea. ISME J. 2014, 8, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Korzhenkov, A.A.; Toschakov, S.V.; Bargiela, R.; Gibbard, H.; Ferrer, M.; Teplyuk, A.V.; Jones, D.L.; Kublanov, I.V.; Golyshin, P.N.; Golyshina, O.V. Archaea dominate the microbial community in an ecosystem with low-to-moderate temperature and extreme acidity. Microbiome 2019, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Williams, T.J.; Allen, M.A.; Liao, Y.; Raftery, M.L.; Cavicchioli, R. Sucrose Metabolism in Haloarchaea: Reassessment Using Genomics, Proteomics, and Metagenomics. Appl. Environ. Microbiol. 2019, 85, e02935-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardavid, R.E.; Khristo, P.; Oren, A. Interrelationships between Dunaliella and halophilic prokaryotes in saltern crystallizer ponds. Extremophiles 2008, 12, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Rascovan, N.; Maldonado, J.; Vazquez, M.P.; Farías, M.E. Metagenomic study of red biofilms from Diamante Lake reveals ancient arsenic bioenergetics in haloarchaea. ISME J. 2016, 10, 299–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutlu, M.B.; Martinez-Garcia, M.; Santos, F.; Peña, A.; Guven, K.; Antón, J. Prokaryotic diversity in Tuz Lake, a hypersaline environment in Inland Turkey. FEMS Microbiol. Ecol. 2008, 65, 474–483. [Google Scholar] [CrossRef]
- Kırtel, O.; Lescrinier, E.; Van den Ende, W.; Toksoy Öner, E. Discovery of fructans in Archaea. Carb. Pol. 2019, 220, 149–156. [Google Scholar] [CrossRef]
- Méheust, R.; Watson, A.K.; Lapointe, F.-L.; Papke, R.T.; Lopez, P.; Bapteste, E. Hundreds of novel composite genes and chimeric genes with bacterial origins contributed to haloarchaeal evolution. Genome Biol. 2018, 19, 75. [Google Scholar] [CrossRef] [Green Version]
- Dogsa, I.; Brloznik, M.; Stopar, D.; Mandic-Mulec, I. Exopolymer Diversity and the Role of Levan in Bacillus subtilis Biofilms. PLoS ONE 2013, 8, e62044. [Google Scholar] [CrossRef]
- Laue, H.; Schenk, A.; Li, H.; Lambertsen, L.; Neu, T.R.; Molin, S.; Ullrich, M.S. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology 2006, 152, 2909–2918. [Google Scholar] [CrossRef] [Green Version]
- Bezzate, S.; Aymerich, S.; Chambert, R.; Czarnes, S.; Berge, O.; Heulin, T. Disruption of the Paenibacillus polymyxa levansucrase gene impairs its ability to aggregate soil in the wheat rhizosphere. Environ. Microb. 2001, 2, 333–342. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, F.; Liu, H.; Kolter, R.; Losick, R.; Guo, J. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microb. 2012, 15, 848–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, N.; Wegmann, U.; Dertli, E.; Mulholland, F.; Collins, S.R.A.; Waldron, K.W.; Bongaerts, R.J.; Mayer, M.J.; Narbad, A. Spontaneous mutation reveals influence of exopolysaccharide on Lactobacillus johnsonii surface characteristics. PLoS ONE 2013, 8, e59957. [Google Scholar] [CrossRef] [Green Version]
- Abaramak, G.; Kırtel, O.; Toksoy Öner, E. Fructanogenic halophiles: A new perspective on extremophiles. In Physiological and Biotechnological Aspects of Extremophiles; Academic Press: Cambridge, MA, USA, 2020; pp. 123–130. [Google Scholar] [CrossRef]
- Versluys, M.; Kirtel, O.; Toksoy Öner, E.; van den Ende, W. The fructan syndrome: Evolutionary aspects and common themes among plants and microbes. Plant Cell Envrion. 2018, 41, 16–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kırtel, O.; Menendez, C.; Versluys, M.; Van den Ende, W.; Hernandez, L.; Toksoy Öner, E. Levansucrase from Halomonas smyrnensis AAD6(T): First halophilic GH-J clan enzyme recombinantly expressed, purified, and characterized. Appl. Microbiol. Biotechnol. 2018, 102, 9207–9220. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Garces-Carrera, S.; Whitworth, R.J.; Fellers, J.P.; Park, Y.; Chen, M.-S. A Horizontal Gene Transfer Led to the Acquisition of a Fructan Metabolic Pathway in Gall Midge. Adv. Biosys. 2020, 4, 1900275. [Google Scholar] [CrossRef]
- Miyazaki, T.; Oba, N.; Park, E.Y. Structural insight into the substrate specificity of Bombyx mori β-fructofuranosidase belonging to the glycoside hydrolase family 32. Insect Biochem. Mol. Biol. 2020, 127, 103494. [Google Scholar] [CrossRef]
- Ozaki, T.; Abe, N.; Kimura, K.; Suzuki, A.; Kaneko, J. Genomic analysis of Bacillus subtilis lytic bacteriophage ΦNIT1 capable of obstructing natto fermentation carrying genes for the capsule-lytic soluble enzymes poly-γ-glutamate hydrolase and levanase. Biosci. Biotech. Biochem. 2017, 81, 1. [Google Scholar] [CrossRef] [Green Version]
- Hill, A.; Chen, L.; Mariage, A.; Petit, J.-L.; de Barrdinis, V.; Karbourne, S. Discovery of new levansucrase enzymes with interesting properties and improved catalytic activity to produce levan and fructooligosaccharides. Cat. Sci. Techol. 2019, 9, 2931. [Google Scholar] [CrossRef]
- Ghauri, K.; Ali, H.; Munawar, N.; Ghauri, M.A.; Anwar, M.A. Glycoside hydrolase family 68 gene of halophilic archaeon Halalkalicoccus jeotgali B3T codes for an inulosucrase enzyme. Biocat. Biotrans. 2020. [Google Scholar] [CrossRef]
- Van den Ende, W. Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 2013, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Soto, M.E.; Porras-Dominguez, J.R.; Seibel, J.; Lopez Munguia, A. A close look at the structural features and reaction condition that modulate the synthesis of low and high molecular weight fructans by levansucrases. Carbohydr. Pol. 2019, 219, 130–142. [Google Scholar] [CrossRef]
- Ortiz-Soto, M.E.; Porras-Dominguez, J.R.; Rodriguez-Alegria, M.E.; Morales-Moreno, L.A.; Diaz-Vilchis, A.; Rudino-Pinera, E.; Beltran-Hernandez, N.E.; Rivera, H.M.; Seibel, J.; Lopez Munguia, A. Implications of the mutation S164A on Bacillus subtilis levansucrase product specificity and insights into protein interactions acting upon levan synthesis. Int. J. Biol. Macromol. 2020, 161, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Pijning, T.; Anwar, M.A.; Böger, M.; Dobruchowska, J.M.; Leemhuis, H.; Slavko Kralj, S.; Dijkhuizen, L.; Dijkstra, B.W. Crystal Structure of Inulosucrase from Lactobacillus: Insights into the Substrate Specificity and Product Specificity of GH68 Fructansucrases. J. Mol. Biol. 2011, 412, 80–93. [Google Scholar] [CrossRef]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Vogt, L.M.; Meyer, D.; Pullens, G.; Faas, M.; Smelt, M.; Venema, K.; Ramasamy, U.; Schols, H.A.; De Vos, P. Immunological Properties of Inulin-Type Fructans. Crit. Rev. Food Sci. Nutr. 2015, 55, 414–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janse van Rensburg, H.C.; Takács, Z.; Freynschlag, F.; Toksoy Öner, E.; Jonak, C.; van den Ende, W. Fructans prime ROS dynamics and Botrytis cinerea resistance in Arabidopsis. Antioxidants 2020, 9, 805. [Google Scholar] [CrossRef] [PubMed]
- Toksoy Öner, E.; Hernández, L.; Combie, J. Review of Levan polysaccharide: From a century of past experiences to future prospects. Biotech. Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Fuchs, A. Current and potential food and non-food applications of fructans. Biochem. Soc. Trans. 1991, 19, 555–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beganovic, J.; Pavunc, A.L.; Gjuracic, K.; Spoljarec, M.; Suskovic, J. Improved Sauerkraut Production with Probiotic Strain Lactobacillus plantarum L4 and Leuconostoc mesenteroides LMG 7954. J. Food Sci. 2011, 76, 2. [Google Scholar] [CrossRef]
- Kırtel, O.; Versluys, M.; Van den Ende, W.; Toksoy Öner, E. Fructans of the saline world. Biotechnol. Adv. 2018, 36, 1524–1539. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Miller, G.L. Modified DNS method for reducing sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Prot. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; Gonzalez-Outeirino, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, J. Development and Validation of a Genetic Algorithm for Flexible Docking. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Hall, T. BioEdit Version 7.0. 0. Distributed by the Author. 2014. Available online: www.mbio.ncsu.edu/BioEdit/bioedit (accessed on 27 February 2021).
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Bai, Y.; Pijning, T.; Van Leeuwen, S.S.; Dijkhuizen, L. Structure–function relationships of family GH70 glucansucrase and 4,6-α-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell. Mol. Life Sci. 2016, 73, 2681–2706. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Tsirigos, K.D.; Casper Kaae Sønderby, C.K.; Thomas Nordahl Petersen, T.N.; Ole Winther, O.; Søren Brunak, S.; Gunnar von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotech. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Bagos, P.G.; Nikolau, E.P.; Liakopoulos, T.D.; Tsirigos, K.D. Combined prediction of Tat and Sec signal peptides with Hidden Markov Models. Bioinformatics 2010, 26, 2811–2817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonin, P.; Lombard, C.; Huguet, A.; Kish, A. Improved Isolation of SlaA and SlaB S-layer proteins in Sulfolobus acidocaldarius. Extremophiles 2020, 24, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.-L.; Zhou, P.-J.; Oren, A.; Liu, S.-J. Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 2009, 13, 31–37. [Google Scholar] [CrossRef]
- Van den Ende, W.; Coopman, M.; Clerens, S.; Vergauwen, R.; Le Roy, K.; Lammens, W.; Van Laere, A. Unexpected presence of graminan- and levan-type fructans in the evergreen frost-hardy eudicot Pachysandra terminalis (Buxaceae). Purification, cloning and functional analysis of a 6-SST/6-SFT enzyme. Plant Physiol. 2011, 155, 603–614. [Google Scholar] [CrossRef] [Green Version]
- Berghuis, B.A.; Yu, F.B.; Schulz, F.; Blainey, P.C.; Woyke, T.; Quake, S.R. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc. Natl. Acad. Sci. USA 2019, 116, 5037–5044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, P.S.; Borrel, G.; Gribaldo, S. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc. Natl. Acad. Sci. USA 2018, 115, E1166–E1173. [Google Scholar] [CrossRef] [Green Version]
- Lunn, J.E. Evolution of sucrose synthesis. Plant Physiol. 2002, 128, 1490–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inthanavong, L.; Tian, F.; Khodadadi, M.; Karbourne, S. Properties of Geobacillus stearothermophilus levansucrase as potential biocatalyst for the synthesis of levan and fructooligosaccharides. Biotechnol. Progr. 2013, 29, 1405. [Google Scholar] [CrossRef]
- Ni, D.; Xu, W.; Bai, Y.; Zhang, W.; Zhang, T.; Mu, W. Biosynthesis of levan from sucrose using a thermostable levansucrase from Lactobacillus reuteri LTH5448. Int. J. Biol. Macromol. 2018, 113, 29–37. [Google Scholar] [CrossRef]
- Okuyama, M.; Serizawa, R.; Tanuma, M.; Kikuchi, A.; Sadahiro, J.; Tagami, T.; Lang, W.; Kimura, A. Molecular insight into regioselectivity of transfructosylation catalyzed by GH68 levansucrase and β-fructofuranosidase. J. Biol. Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Oyewusi, H.A.; Wahab, R.A.; Edbeib, M.F.; Mohamad, M.A.N.; Hamid, A.A.A.; Kaya, Y.; Huyop, F. Functional profiling of bacterial communities in Lake Tuz using 16S rRNA gene sequences. Biotechol. Biotechnol. Equip. 2021, 35, 1–10. [Google Scholar] [CrossRef]
- Ni, D.; Xu, W.; Zhu, Y.; Zhang, W.; Guang, C.; Mu, W. Inulin and its enzymatic production by inulosucrases: Characteristics, structural features, molecular modifications and applications. Biotech. Adv. 2019, 37, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Kralj, S.; Leeflang, C.; Sierra, E.I.; Kempinski, B.; Alkan, V. Synthesis of fructooligosaccharides (FosA) and inulin (InuO) by GH68 fructosyltransferases from Bacillus agaradhaerens strain WDG185. Carb. Pol. 2018, 179, 350–359. [Google Scholar] [CrossRef]
- Wada, T.; Ohguchi, M.; Iwai, Y. A novel enzyme of Bacillus sp. 217c-11 that produces inulin from sucrose. Biosci. Biotech. Biochem. 2003, 67, 1327–1334. [Google Scholar] [CrossRef] [Green Version]
- Charoenwongpaiboon, T.R.; Pichyangkura, R.; Field, A.; Prousoontorn, M.H. Preparation of Cross-Linked Enzyme Aggregates (CLEAs) of an inulosucrase mutant for the enzymatic synthesis of inulin-type fructooligosaccharides. Catalysts 2019, 9, 641. [Google Scholar] [CrossRef] [Green Version]
- Mostafa, F.A.; Wahab, W.A.A.; Salah, H.A.; Nawwar, G.A.M.; Esawy, M.A. Kinetic and thermodynamic characteristic of Aspergillus awamori EM66 levansucrase. Int. J. Biol. Macromol. 2018, 119, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Porras-Domínguez, J.R.; Ávila-Fernández, Á.; Miranda-Molina, A.; Rodríguez-Alegría, M.E.; Munguía, A.L. Bacillus subtilis 168 levansucrase (SacB) activity affects average levan molecular weight. Carbohydr. Polym. 2015, 132, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Visnapuu, T.; Mardo, K.; Alamaë, T. Levansucrases of a Pseudomoans syringae pathovar as catalysts for the synthesis of potentially prebiotic oligo-and polysaccharides. New Biotechnol. 2015, 32, 597–605. [Google Scholar] [CrossRef]
- Ozimek, L.K.; Euverink, G.J.; Van Der Maarel, M.J.E.C.; Dijkhuizen, L. Mutational analysis of the role of calcium ions in the Lactobacillus reuteri strain 121 fructosyltransferase (levansucrase and inulosucrase) enzymes. FEBS Lett. 2005, 579, 1124–1128. [Google Scholar] [CrossRef] [Green Version]
- Olivares-Illana, V.; Lopez-Munguia, A.; Olvera, C. Molecular Characterization of Inulosucrase from Leuconostoc citreum: A Fructosyltransferase within a Glucosyltransferase. J. Bacteriol. 2003, 185, 3606–3612. [Google Scholar] [CrossRef] [Green Version]
- Del Moral, S.; Olvera, C.; Rodriguez, M.; Munguia, A. Functional role of the additional domains in inulosucrase (IslA) from Leuconostoc citreum CW28. BMC Biochem. 2008, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.F.; Jeong, H.; Lee, J.-S.; Choi, S.-H.; Ha, M.; Hur, C.-G.; Kim, J.-S.; Lee, S.; Park, H.-S.; Park, Y.-H.; et al. Complete Genome Seqence of Leuconostoc citreum KM20. J. Bacteriol. 2008, 190, 3093–3094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, S.; Koga, T.; Inoue, M. Isolation and some properties of extracellular D-glucosyltransferases and D-fructosyltransferases from Streptococcus mutans serotypes c, e and f. Carbohydr. Res. 1984, 134, 293–304. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Belanger, M.; Rodrigues, P.H.; Simpson-Haidaris, P.J.; Akin, D.; Burne, R. A Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol. Immunol. 2009, 24, 141–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abranches, J.; Miller, J.H.; Martinez, A.R.; Simpson-Haidaris, P.J.; Burne, R.A.; Lemos, J.A. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect. Immun. 2011, 79, 2277–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Abbeele, P.; Marzorati, M.; Derde, M.; De Weirdt, R.; Vermeiren, J.; Possemiers, S.; Van de Wiele, T. Arabinoxylans, inulin and Lactobacillus reuteri 1063 repress the adherent-invasive Escherichia coli from mucus in a mucose-comprizing gut model. Biofilms Microbiomes 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Ates, O. Systems Biology of Microbial Exopolysaccharides Production. Front. Bioeng. Biotech. 2015, 3, 200. [Google Scholar] [CrossRef] [Green Version]
- Mancinelli, R.; Boyyi, A.; Bruni, F.; Ricci, M.A.; Soper, A.K. Hydration of Sodium, Potassium, and Chloride Ions in Solution and the Concept of Structure Maker/Braker. J. Phys. Chem. B 2007, 111, 13570–13577. [Google Scholar] [CrossRef]
- Benigar, E.; Dogsa, I.; Stopar, D.; Jamnik, A.; Kralj Cigic, I.; Tomsic, M. Structure and Dynamics of a Polysaccharide Matrix: Aqueous Solutions of Bacterial Levan. Langmuir 2014, 30, 4172–4182. [Google Scholar] [CrossRef]
- Hong, B.H.; Joe, M.M.; Delvakumar, G.; Kim, K.Y.; Choi, J.H.; Sa, T.M. Influence of salinity variations on exocellular polysaccharide production, biofilm formation and flocculation in halotolerant bacteria. J. Env. Biol. 2017, 38, 657–664. [Google Scholar] [CrossRef]
- Yuan, S.; Le Roy, K.; Venken, T.; Lammens, W.; Van den Ende, W.; de Maeyer, M. pKa Modulation of the Acid/Base Catalyst within GH32 and GH68: A Role in Substrate/Inhibitor Specificity? PLoS ONE 2012, 5, e37453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Fleites, C.; Ortiz-Lombardia, M.; Pons, T.; Tarbouriech, N.; Taylor, E.J.; Arrieta, J.G.; Hernández, L.; Davies, G.J. Crystal structure of levansucrase from the Gram-negative bacterium Gluconacetobacter diazotrophicus. Biochem. J. 2005, 390, 19–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, G.; Futterer, K. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 2003, 10, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Tamir, A.; Eichler, J. N-Glycosylation is important for proper Haloferax volcanii S-layer stability and function. Appl. Environ. Microbiol. 2017, 83, e03152-16. [Google Scholar] [CrossRef] [Green Version]
- Schulze, S.; Pfeiffer, F.; Garcia, B.A.; Pohlschroder, M. Glycoproteomics of Haloferax volcanii reveals an extensive glycoproteome and occurrence of different N-glycosylation pathways. bioRxiv Preprint 2021. [Google Scholar] [CrossRef]
- Dombkowski, A.A.; Sultana, K.Z.; Craig, D.B. Protein disulfide engineering. FEBS Lett. 2014, 588, 206–212. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Hu, D.; Li, J.; He, Y.; Zhu, T.; Wu, M. Contribution of disulfide bridges to the thermostability of a type A feruloyl esterase from Aspergillus usamii. PLoS ONE 2015, 10, e0126864. [Google Scholar] [CrossRef] [Green Version]
- Raga-Carbajal, E.; Diaz-Vilchis, A.; Rojas-Trejo, S.P.; Rudino-Pinero, E.; Olvera, C. The molecular basis of the nonprocessive elongation mechanism in levansucrases. J. Biol. Chem. 2021, 296, 100178. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.K.; Dey, R.; Thomas, M.; Sherathia, D.; Dalsania, T.; Patel, I.; Savsani, K.; Ghorai, S.; Vanpariya, S.; Sukhadiya, B.; et al. Draft genome sequence of the extremely halophilic Bacillus sp. strain SB49, isolated from a salt crystallizer pond of the Little Rann of Kutch, India. Genome Announc. 2013, 1, e00869-13. [Google Scholar] [CrossRef] [Green Version]






| Compound | Activity at 1 mM (%) | Activity at 5 mM (%) |
|---|---|---|
| None | 100 | 100 |
| SDS | 2.0 ± 0.4 | 1.0 ± 0.2 |
| Triton X-100 | 0.5 ± 0.4 | 1.0 ± 0.5 |
| EDTA | 113 ± 6 | 11.8 ± 12.3 |
| Ba2+ | 108 ± 7 | 91 ± 17 |
| Fe2+ | 96 ± 9 | 69 ± 5 |
| Ca2+ | 32 ± 3 | 0 |
| Cu2+ | 0 | 0 |
| Co2+ | 0 | 0 |
| Ni2+ | 0.5 ± 0.7 | 0 |
| Mg2+ | 52 ± 5 | 68 ± 9 |
| Mn2+ | 61 ± 9 | 24 ± 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abaramak, G.; Porras-Domínguez, J.R.; Janse van Rensburg, H.C.; Lescrinier, E.; Toksoy Öner, E.; Kırtel, O.; Van den Ende, W. Functional and Molecular Characterization of the Halomicrobium sp. IBSBa Inulosucrase. Microorganisms 2021, 9, 749. https://doi.org/10.3390/microorganisms9040749
Abaramak G, Porras-Domínguez JR, Janse van Rensburg HC, Lescrinier E, Toksoy Öner E, Kırtel O, Van den Ende W. Functional and Molecular Characterization of the Halomicrobium sp. IBSBa Inulosucrase. Microorganisms. 2021; 9(4):749. https://doi.org/10.3390/microorganisms9040749
Chicago/Turabian StyleAbaramak, Gülbahar, Jaime Ricardo Porras-Domínguez, Henry Christopher Janse van Rensburg, Eveline Lescrinier, Ebru Toksoy Öner, Onur Kırtel, and Wim Van den Ende. 2021. "Functional and Molecular Characterization of the Halomicrobium sp. IBSBa Inulosucrase" Microorganisms 9, no. 4: 749. https://doi.org/10.3390/microorganisms9040749
APA StyleAbaramak, G., Porras-Domínguez, J. R., Janse van Rensburg, H. C., Lescrinier, E., Toksoy Öner, E., Kırtel, O., & Van den Ende, W. (2021). Functional and Molecular Characterization of the Halomicrobium sp. IBSBa Inulosucrase. Microorganisms, 9(4), 749. https://doi.org/10.3390/microorganisms9040749








