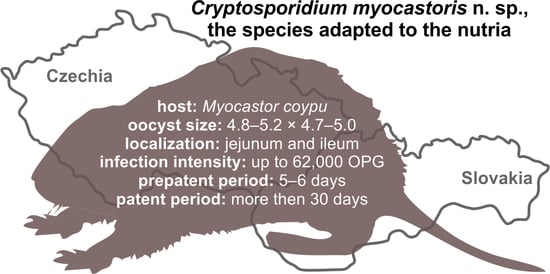
Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Area and Specimens Studied
2.2. Molecular Study
2.3. Source of Oocysts of Cryptosporidium myocastoris n. sp.
2.4. Animals for Transmission Studies
2.5. Animal Care
2.6. Design of Transmission Studies
2.7. Morphological Evaluation
2.8. Clinical and Pathomorphological Examinations
2.9. Statistical Analysis
3. Results
- Taxonomic summary:
- Family Cryptosporidiidae Léger, 1911
- Genus Cryptosporidium Tyzzer, 1907
- Cryptosporidium myocastoris n. sp.
- Type-host:Myocastor coypus (Molina, 1782), nutria.
- Other natural hosts: No other natural hosts are known.
- Type-locality: Dunajská Streda (N 47°55.90470′, E 17°28.42662′), Slovakia.
- Other localities: Lanžhot (N 48°43.41558′, E 16°58.30782′), Czech Republic; Šaľa (N 48°9.08273′, E 17°52.49152′), Slovakia.
- Type-material: Faecal smear slides with oocysts stained by ACMV and ZN staining (nos. MV1/31132 and ZN2/31132); scanning electron microscopy specimens of infected jejunum (no. SEM23/2017) and ileum (no. SEM27/2017); histological sections of infected jejunum (no. H23/2017) and ileum (no. H27/2017); gDNA isolated from faecal samples of naturally (isolate 31132) and experimentally (isolate 32235) infected nutrias; gDNA isolated from jejunum and ileum of experimentally infected nutrias (isolates 32235 and 32236). All specimens are deposited at the Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Czech Republic.
- Prepatent period:Myocastor coypus 5–6 days (present study).
- Patent period: At least 30 days in experimentally infected nutrias (Myocastor coypus; present study).
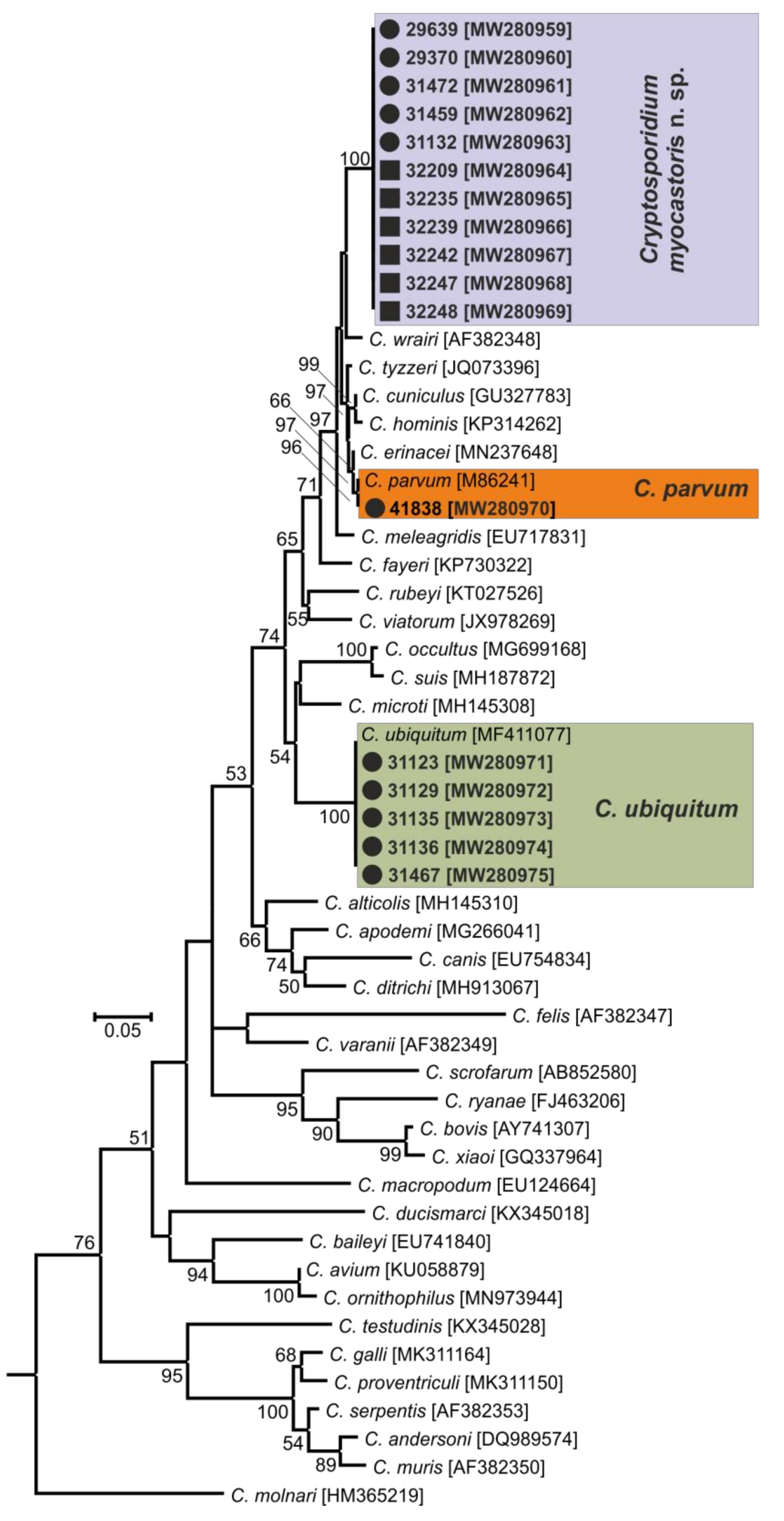
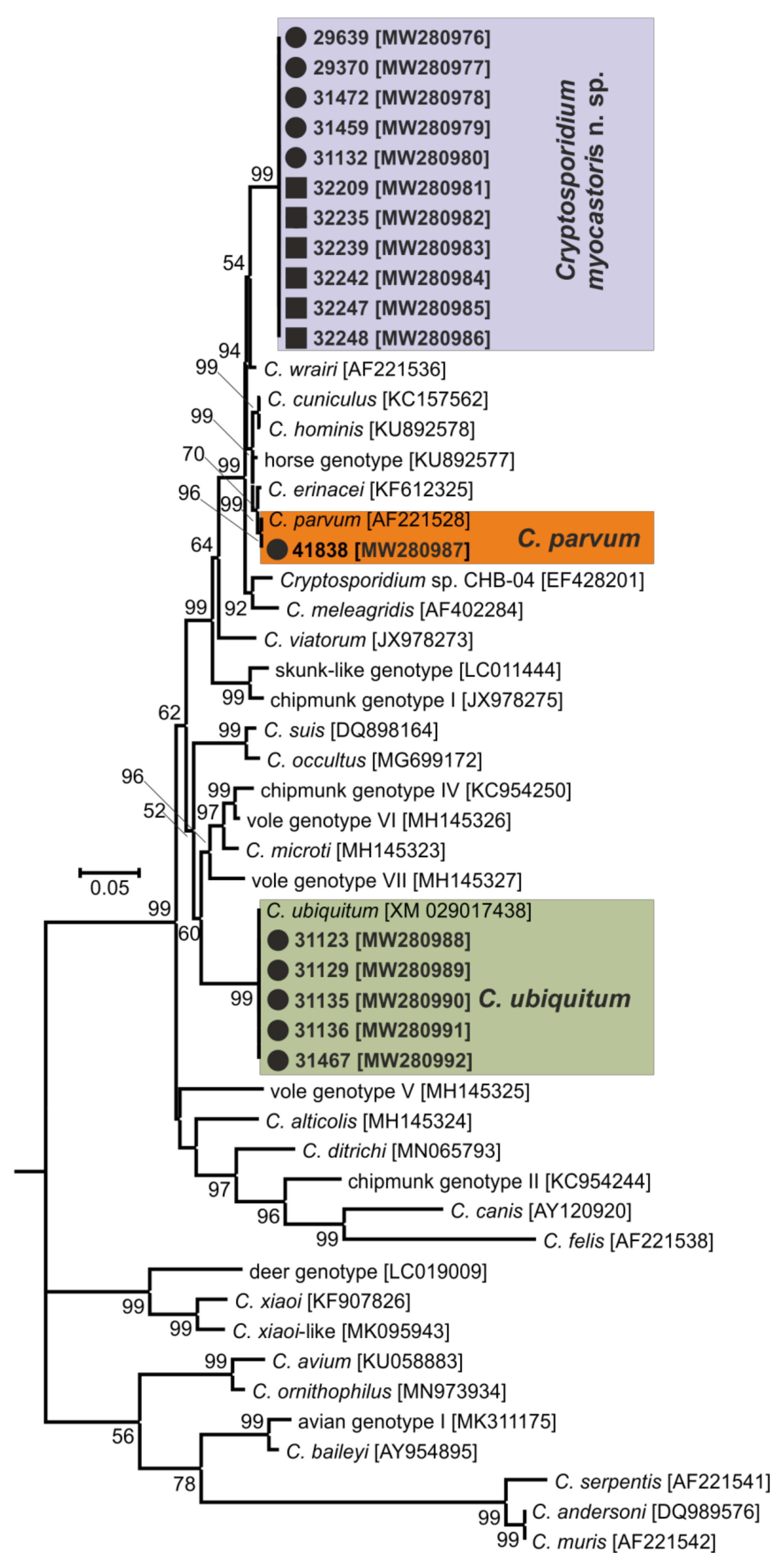
- Representative DNA sequences: Representative nucleotide sequences of SSU [MW274649], actin [MW280963], HSP70 [MW280980] and gp60 [MW280997 and MW280994] genes were saved in the GenBank database.
- ZooBank registration: To comply with the regulations set out in Article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [40], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:FCAD0ED3-2DD0-4A79-93DD-D0C206EC6ACF. The LSID for the new name Cryptosporidium myocastoris n. sp. is urn:lsid:zoobank.org:act:E447F777-5495-4613-8447-D015339F6B32.
- Etymology: The species name myocastoris is derived from the Latin noun myocastor, meaning nutria.
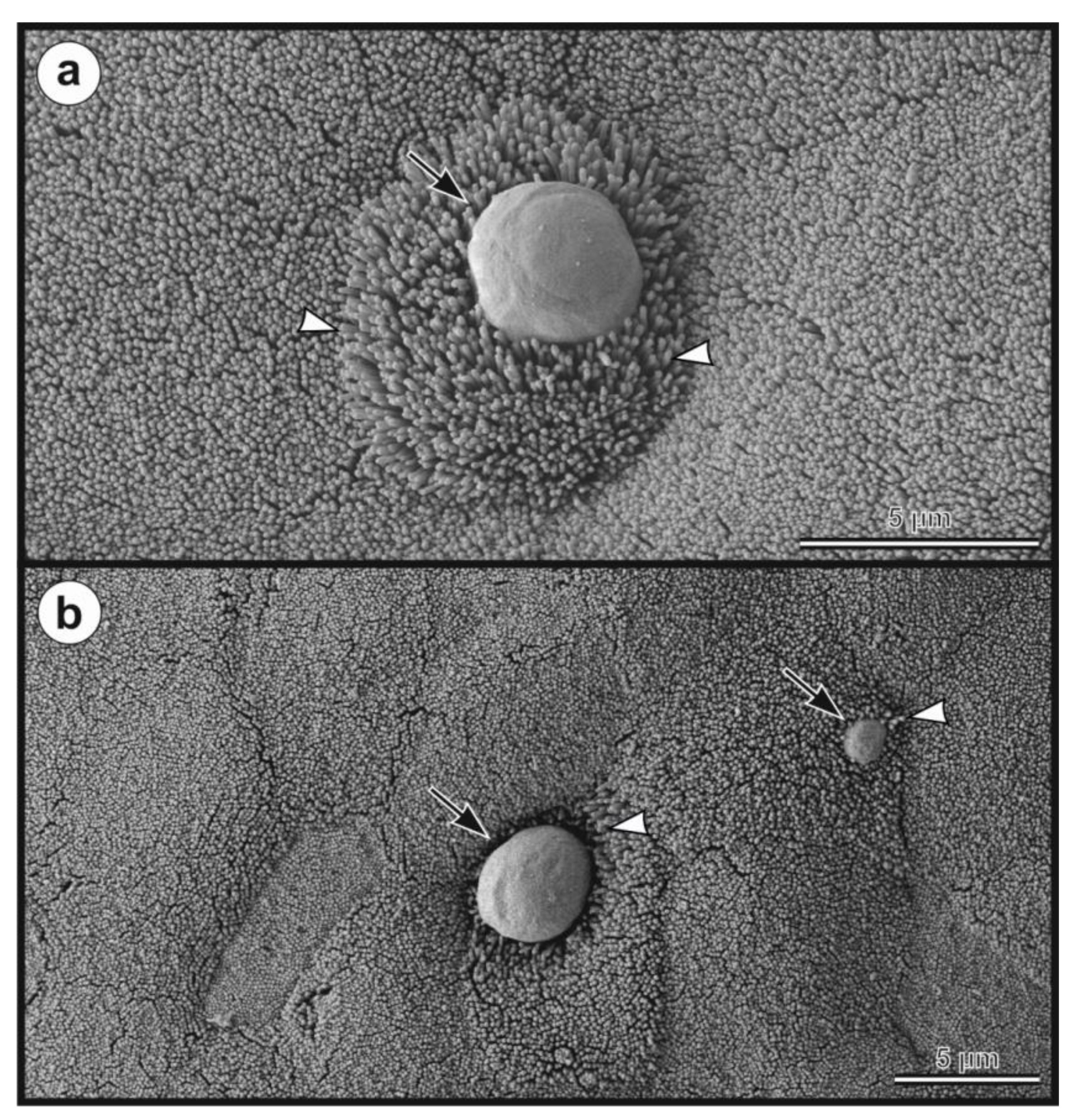
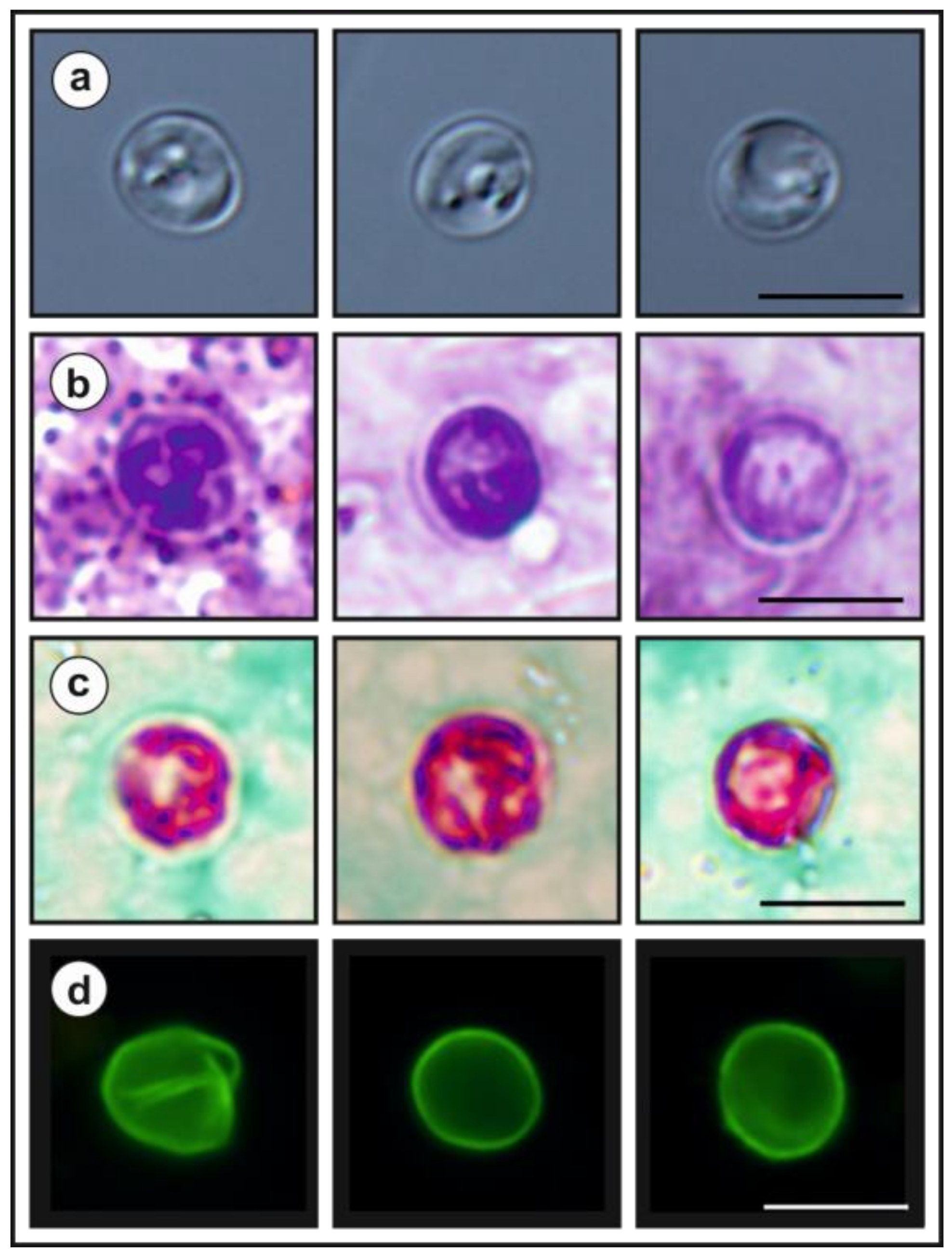
- Description: Oocysts of C. myocastoris n. sp. (isolate 31132) are spherical, measuring 4.8–5.2 × 4.7–5.0 (5.02 ± 0.13 × 4.85 ± 0.10) with a length-to-width ratio of 1.00–1.08 (1.04 ± 0.02) (Figure 9). The oocyst wall is smooth and colourless (Figure 9a). The oocyst residuum is composed of numerous small granules and one spherical globule is clearly visible; a suture is not noticeable. Four sporozoites are clearly visible within oocysts. The morphology and morphometry of other developmental stages is unknown.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Species | Oocyst Size (μm) | Length/Width Ratio | Reference |
|---|---|---|---|
| Cryptosporidium myocastoris | 4.8–5.2 × 4.7–5.0 | 1.00–1.08 | This study |
| Cryptosporidium abrahamseni | 3.82 ± 0.22 × 3.16 ± 0.18 | 1.22 | [13] |
| Cryptosporidiumalticolis | 4.9–5.7 × 4.6–5.2 | 1.00–1.20 | [6] |
| Cryptosporidiumandersoni | 6.0–8.1 × 5.0–6.5 | 1.07–1.50 | [90] |
| Cryptosporidiumapodemi | 3.9–4.7 × 3.8–4.4 | 1.00–1.06 | [76] |
| Cryptosporidiumavium | 5.3–6.9 × 4.3–5.5 | 1.14–1.47 | [91] |
| Cryptosporidiumbaileyi | 6.0–7.5 × 4.8–5.7 | 1.05–1.79 | [92,93] |
| Cryptosporidium bollandi | 2.82–3.11 | unknown | [94] |
| Cryptosporidiumbovis | 4.76–5.35 × 4.17–4.76 | 1.06 | [64] |
| Cryptosporidiumcanis | 3.68–5.88 × 3.68–5.88 | 1.04–1.06 | [95] |
| Cryptosporidiumcichlidis | 4.0–4.7 × 2.5–3.5 | unknown | [96] |
| Cryptosporidiumcuniculus | 5.55–6.40 × 5.02–5.92 | 1.11 | [97] |
| Cryptosporidiumditrichi | 4.5–5.2 × 4.0–4.6 | 1.0–1.2 | [76] |
| Cryptosporidiumducismarci | 4.4–5.4 × 4.3–5.3 | 1.1 ± 0.03 | [98] |
| Cryptosporidiumerinacei | 4.5–5.8 × 4.0–4.8 | 1.02–1.35 | [73] |
| Cryptosporidiumfayeri | 4.5–5.1 × 3.8–5.0 | 1.02–1.18 | [99] |
| Cryptosporidium felis | 5.0 × 4.5 | unknown | [100] |
| Cryptosporidiumfragile | 5.5–7.0 × 5.0–6.5 | 1.0–1.3 | [101] |
| Cryptosporidiumgalli | 8.0–8.5 × 6.2–6.4 | 1.3 | [102] |
| Cryptosporidiumhominis | 4.4–5.4 × 4.4–5.9 | 1.01–1.09 | [103] |
| Cryptosporidiumhuwi | 4.4–4.9 × 4.0–4.8 | 0.92–1.35 | [9] |
| Cryptosporidiummacropodum | 4.5–6.0 × 5.0–6.0 | 1.1 | [67] |
| Cryptosporidiummeleagridis | 4.5–6.0 × 4.2–5.3 | 1.00–1.33 | [93,104] |
| Cryptosporidiummicroti | 3.9–4.7 × 3.8–4.4 | 1.00–1.06 | [6] |
| Cryptosporidiummolnari | 3.23–5.45 × 3.02–5.04 | 1.00–1.17 | [105] |
| Cryptosporidiummuris | 6.6–7.9 × 5.3–6.5 | 1.1–1.5 | [106] |
| Cryptosporidiumnasoris | 3.6 | unknown | [107] |
| Cryptosporidiumoccultus | 4.66–5.53 × 4.47–5.44 | 1.00–1.17 | [63] |
| Cryptosporidiumornithophilus | 5.24–6.77 × 4.68–5.50 | 1.06–1.36 | [81] |
| Cryptosporidiumparvum | 4.5–5.4 × 4.5–5.4 | 1.0–1.3 | [108] |
| Cryptosporidiumproliferans | 6.8–8.8 × 4.8–6.2 | 1.48 | [109] |
| Cryptosporidiumproventriculi | 6.70–8.40 × 5.10–6.3 | 1.08–1.41 | [11] |
| Cryptosporidium ratti | 4.4–5.4 × 4.3–5.1 | 1.0–1.1 | [12] |
| Cryptosporidiumreichenbachklinkei | 2.4–3.18 × 2.4–3.0 | unknown | [96] |
| Cryptosporidiumrubeyi | 4.4–5.0 × 4.0–5.0 | 1.08 | [110] |
| Cryptosporidiumryanae | 2.94–4.41 × 2.94–3.68 | 1.18 | [111] |
| Cryptosporidiumscophthalmi | 3.7–5.03 × 3.03–4.69 | 1.05–1.34 | [112] |
| Cryptosporidiumscrofarum | 4.81–5.96 × 4.23–5.29 | 1.07 ± 0.06 | [66] |
| Cryptosporidiumserpentis | 6.3 × 5.5 | 1.14 ± 0.11 | [113] |
| Cryptosporidiumsuis | 6.0–6.8 × 5.3–5.7 | 1.14 | [65] |
| Cryptosporidiumtestudinis | 5.8–6.9 × 5.3–6.5 | 1.1 ± 0.05 | [98] |
| Cryptosporidium tyzzeri | 4.64 ± 0.05 × 4.19 ± 0.06 | 1.11 ± 0.06 | [71] |
| Cryptosporidiumubiquitum | 4.71–5.32 × 4.33–4.98 | 1.08 | [79] |
| Cryptosporidiumvaranii | 4.8–5.1 × 4.4–4.8 | 1.03 ± 0.03 | [114] |
| Cryptosporidiumviatorum | 4.87–5.87 × 4.15–5.20 | 1.03–1.32 | [115] |
| Cryptosporidiumwrairi | 4.0–5.0 × 4.8–5.6 | unknown | [61] |
| Cryptosporidiumxiaoi | 2.94–4.41 × 2.94–4.41 | 1.15 | [116] |
| Species/Genotype | Gene Locus | ||
|---|---|---|---|
| SSU | Actin | HSP70 | |
| C. andersoni | 0.090 | 0.231 | 0.268 |
| C. avium | 0.045 | 0.175 | 0.215 |
| C. baileyi | 0.051 | 0.181 | 0.202 |
| C. bovis | 0.042 | 0.202 | 0.180 |
| C. canis | 0.043 | 0.167 | 0.213 |
| C. cuniculi | 0.018 | 0.039 | 0.031 |
| C. felis | 0.054 | 0.222 | 0.212 |
| C. galli | 0.094 | 0.206 | 0.252 |
| C. hominis | 0.023 | 0.044 | 0.031 |
| C. muris | 0.090 | 0.209 | 0.268 |
| C. occultus | 0.021 | 0.122 | 0.031 |
| C. parvum | 0.014 | 0.039 | 0.035 |
| C. rubeyi | 0.033 | 0.098 | 0.085 |
| C. ryanae | 0.045 | 0.222 | 0.186 |
| C. suis | 0.023 | 0.118 | 0.095 |
| C. ubiquitum | 0.035 | 0.108 | 0.109 |
| C. xiaoi | 0.048 | 0.205 | 0.201 |
| C. wrairi | 0.018 | 0.036 | 0.028 |
References
- Fayer, R. Taxonomy and species delimitation in Cryptosporidium. Exp. Parasitol. 2010, 124, 90–97. [Google Scholar] [CrossRef]
- Ryan, U.; Xiao, L. Taxonomy and Molecular Taxonomy. In Cryptosporidium: Parasite and Disease, 1st ed.; Cacciò, S.M., Widmer, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–42. [Google Scholar]
- Nakamura, A.A.; Meireles, M.V. Cryptosporidium infections in birds—A review. Rev. Bras. Parasitol. Vet. 2015, 24, 253–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zintl, A.; Neville, D.; Maguire, D.; Fanning, S.; Mulcahy, G.; Smith, H.V.; De Waal, T. Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitology 2007, 134, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Čondlová, S.; Horčičková, M.; Havrdová, N.; Sak, B.; Hlásková, L.; Perec-Matysiak, A.; Kicia, M.; McEvoy, J.; Kváč, M. Diversity of Cryptosporidium spp. in Apodemus spp. in Europe. Eur. J. Protistol. 2019, 69, 1–13. [Google Scholar] [CrossRef]
- Horčičková, M.; Čondlová, S.; Holubová, N.; Sak, B.; Květoňová, D.; Hlásková, L.; Konečný, R.; Sedláček, F.; Clark, M.; Giddings, C.; et al. Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. n. and Cryptosporidium microti sp. n. (Apicomplexa: Cryptosporidiidae). Parasitology 2019, 146, 220–233. [Google Scholar] [CrossRef]
- Stenger, B.L.S.; Horčičková, M.; Clark, M.E.; Kváč, M.; Čondlová, S.; Khan, E.; Widmer, G.; Xiao, L.; Giddings, C.W.; Pennil, C.; et al. Cryptosporidium infecting wild cricetid rodents from the subfamilies Arvicolinae and Neotominae. Parasitology 2017, 145, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Liu, Q.; Zhao, W.; Jiang, X.; Zhang, Y.; Zhao, A.; Jing, B.; Lu, G.; Qi, M. Prevalence and diversity of Cryptosporidium spp. in bamboo rats (Rhizomys sinensis) in South Central China. Int. J. Parasitol. Parasites Wildl. 2019, 9, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Paparini, A.; Tong, K.S.; Yang, R.C.; Gibson-Kueh, S.; O’Hara, A.; Lymbery, A.; Xiao, L.H. Cryptosporidium huwi n. sp (Apicomplexa: Eimeriidae) from the guppy (Poecilia reticulata). Exp. Parasitol. 2015, 150, 31–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahedi, A.; Durmic, Z.; Gofton, A.W.; Kueh, S.; Austen, J.; Lawson, M.; Callahan, L.; Jardine, J.; Ryan, U. Cryptosporidium homai n. sp. (Apicomplexa: Cryptosporidiiae) from the guinea pig (Cavia porcellus). Vet. Parasitol. 2017, 245, 92–101. [Google Scholar] [CrossRef] [Green Version]
- Holubová, N.; Zikmundová, V.; Limpouchová, Z.; Sak, B.; Konečný, R.; Hlásková, L.; Rajský, D.; Kopacz, Z.; McEvoy, J.; Kváč, M. Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur. J. Protistol. 2019, 69, 70–87. [Google Scholar] [CrossRef]
- Ježková, J.; Prediger, J.; Holubová, N.; Sak, B.; Konečný, R.; Feng, Y.; Xiao, L.; Rost, M.; McEvoy, J.; Kváč, M. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology 2021, 148, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, A.; Bolland, S.J.; Oskam, C.L.; Ryan, U. Cryptosporidium abrahamseni n. sp. (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exp. Parasitol. 2021, 223, 108089. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Xiao, L. Cryptosporidium and Cryptosporidiosis; CRC Press: New York, NY, USA, 2007; p. 560. [Google Scholar]
- Martino, P.E.; Radman, N.; Parrado, E.; Bautista, E.; Cisterna, C.; Silvestrini, M.P.; Corba, S. Note on the occurrence of parasites of the wild nutria (Myocastor coypus, Molina, 1782). Helminthologia 2012, 49, 164–168. [Google Scholar] [CrossRef] [Green Version]
- Tsiamis, K.; Gervasini, E.; Deriu, I.; D’Amico, F.; Nunes, A.L.; Addamo, A.M.; Cardoso, A.C. Baseline Distribution of Invasive Alien Species of Union Concern; Commision, E., Ed.; JRC Science for Poplicy Report: Luxembourg, 2017; p. 96. [Google Scholar] [CrossRef]
- Martino, P.E.; Radman, N.E.; Gamboa, M.I.; Samartino, L.E.; Parrado, E.J. Ectoparasites from some Myocastor coypus (Molina, 1782) populations (Coypus or Nutria) in Argentina. Rev. Bras. Parasitol. Vet. 2018, 27, 254–257. [Google Scholar] [CrossRef]
- Kellnerová, K.; Holubová, N.; Jandová, A.; Vejčík, A.; McEvoy, J.; Sak, B.; Kváč, M. First description of Cryptosporidium ubiquitum XIIa subtype family in farmed fur animals. Eur. J. Protistol. 2017, 59, 108–113. [Google Scholar] [CrossRef]
- Babero, B.B.; Lee, J.W. Studies on the helminths of nutria, Myocastor coypus (Molina), in Louisiana with check-list of other worm parasites from this host. J. Parasitol. 1961, 47, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Pavlásek, I.; Kozakiewicz, B. Coypus (Myocastor coypus) as a new host of Cryptosporidium parvum (Apicomplexa: Cryptosporidiidae). Folia Parasitol. (Praha) 1991, 38, 90. [Google Scholar]
- Zanzani, S.A.; Di Cerbo, A.; Gazzonis, A.L.; Epis, S.; Invernizzi, A.; Tagliabue, S.; Manfredi, M.T. Parasitic and Bacterial Infections of Myocastor coypus in a Metropolitan Area of Northwestern Italy. J. Wildl. Dis. 2016, 52, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Miláček, P.; Vítovec, J. Differential staining of cryptosporidia by aniline-carbol-methyl violet and tartrazine in smears from feces and scrapings of intestinal mucosa. Folia Parasitol. 1985, 32, 50. [Google Scholar]
- Kváč, M.; Ondráčková, Z.; Květoňová, D.; Sak, B.; Vítovec, J. Infectivity and pathogenicity of Cryptosporidium andersoni to a novel host, southern multimammate mouse (Mastomys coucha). Vet. Parasitol. 2007, 143, 229–233. [Google Scholar] [CrossRef]
- Jiang, J.; Alderisio, K.A.; Xiao, L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 2005, 71, 4446–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Morgan, U.M.; Limor, J.; Escalante, A.; Arrowood, M.; Shulaw, W.; Thompson, R.C.; Fayer, R.; Lal, A.A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999, 65, 3386–3391. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, I.M.; Lal, A.A.; Xiao, L.H. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 2002, 88, 388–394. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Morgan, U.M.; Thompson, R.C.; Lal, A.A.; Xiao, L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 2000, 66, 2385–2391. [Google Scholar] [CrossRef] [Green Version]
- Alves, M.; Xiao, L.H.; Sulaiman, I.; Lal, A.A.; Matos, O.; Antunes, F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 2003, 41, 2744–2747. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Xiao, L.; Alderisio, K.; Elwin, K.; Cebelinski, E.; Chalmers, R.; Santin, M.; Fayer, R.; Kváč, M.; Ryan, U.; et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014, 20, 217–224. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Arrowood, M.J.; Donaldson, K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J. Eukaryot. Microbiol. 1996, 43, 89S. [Google Scholar] [CrossRef]
- Arrowood, M.J.; Sterling, C.R. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J. Parasitol. 1987, 73, 314–319. [Google Scholar] [CrossRef]
- Henriksen, S.A.; Pohlenz, J.F. Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Vet. Scand. 1981, 22, 594–596. [Google Scholar] [CrossRef]
- Kváč, M.; Vítovec, J. Prevalence and pathogenicity of Cryptosporidium andersoni in one herd of beef cattle. J. Vet. Med. B 2003, 50, 451–457. [Google Scholar] [CrossRef]
- Nordhausen, K.; Sirkia, S.; Oja, H.; Tyler, D. CSNP: Tools for Multivariate Nonparametrics. R package version 1.1-1. Available online: https://CRAN.R-project.org/package=ICSNP (accessed on 20 February 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Kváč, M.; Horčičková, M.; Holubová, N.; Květoňová, D.; Hlásková, L.; McEvoy, J.; Rajský, D.; Sak, B. Cryptosporidium ubiquitum and Cryptosporidium coypu genotype in wild coypu (Myocastor coypus). In Proceedings of the 14th Interantional Congress of Parasitology, Daegu, Korea, 19–24 August 2010. [Google Scholar]
- ICZN. International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the international code of zoologicalnomenclature to expand and refine methods of publication. Bull. Zool. Nomencl. 2012, 69, 161–169. [Google Scholar] [CrossRef]
- Meireles, M.V.; Soares, R.M.; Bonello, F.; Gennari, S.M. Natural infection with zoonotic subtype of Cryptosporidium parvum in Capybara (Hydrochoerus hydrochaeris) from Brazil. Vet. Parasitol. 2007, 147, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Fayer, R.; Trout, J.M.; Ryan, U.M.; Schaefer, F.W., 3rd; Xiao, L. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl. Environ. Microbiol. 2004, 70, 7574–7577. [Google Scholar] [CrossRef] [Green Version]
- Paziewska, A.; Bednarska, M.; Nieweglowski, H.; Karbowiak, G.; Bajer, A. Distribution of Cryptosporidium and Giardia spp. in selected species of protected and game mammals from north-eastern Poland. Ann. Agric. Environ. Med. 2007, 14, 265–270. [Google Scholar] [PubMed]
- Nasser, A.M. Removal of Cryptosporidium by wastewater treatment processes: A review. J. Water Health 2016, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, K.A.; Waso, M.; Reyneke, B.; Saeidi, N.; Levine, A.; Lalancette, C.; Besner, M.C.; Khan, W.; Ahmed, W. Cryptosporidium and Giardia in Wastewater and Surface Water Environments. J. Environ. Qual. 2018, 47, 1006–1023. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.V.; Nichols, R.A.B. Cryptosporidium: Detection in water and food. Exp. Parasitol. 2010, 124, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y. Cryptosporidium in wild placental mammals. Exp. Parasitol. 2010, 124, 128–137. [Google Scholar] [CrossRef]
- Perz, J.F.; Le Blancq, S.M. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 2001, 67, 1154–1162. [Google Scholar] [CrossRef] [Green Version]
- Hattori, K.; Donomoto, T.; Manchanayake, T.; Shibahara, T.; Sasai, K.; Matsubayashi, M. First surveillance and molecular identification of the Cryptosporidium skunk genotype and Cryptosporidium parvum in wild raccoons (Procyon lotor) in Osaka, Japan. Parasitol. Res. 2018, 117, 3669–3674. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhang, Z.J.; Zhang, Y.Q.; Yang, Y.; Zhao, J.F.; Wang, R.J.; Jian, F.C.; Ning, C.S.; Zhang, W.Y.; Zhang, L.X. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasit. Vectors 2018, 11, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kváč, M.; McEvoy, J.; Stenger, B.; Clark, M. Cryptosporidiosis in other vertebrates. In Cryptosporidium: Parasite and Disease, 1st ed.; Cacciò, S.M., Widmer, G., Eds.; Springer: Vienna, Austria, 2014; pp. 237–326. [Google Scholar]
- Tzanidakis, N.; Sotiraki, S.; Claerebout, E.; Ehsan, A.; Voutzourakis, N.; Kostopoulou, D.; Stijn, C.; Vercruysse, J.; Geurden, T. Occurrence and molecular characterization of Giardia duodenalis and Cryptosporidium spp. in sheep and goats reared under dairy husbandry systems in Greece. Parasite 2014, 21, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, R.; Wang, X.; Huang, Y.; Zhou, P.; Liu, Y.; Chen, Y.; Chen, J.; Zhu, W.; Chen, Z. Prevalence and molecular characterization of Cryptosporidium in goats across four provincial level areas in China. PLoS ONE 2014, 9, e111164. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, G.; Cui, B.; Huang, J.; Cui, Z.; Zhang, S.; Dong, H.; Yue, D.; Zhang, L.; Ning, C.; et al. Prevalence, molecular characterization and zoonotic potential of Cryptosporidium spp. in goats in Henan and Chongqing, China. Exp. Parasitol. 2014, 142, 11–16. [Google Scholar] [CrossRef]
- Li, P.; Cai, J.; Cai, M.; Wu, W.; Li, C.; Lei, M.; Xu, H.; Feng, L.; Ma, J.; Feng, Y.; et al. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 2016, 215, 58–62. [Google Scholar] [CrossRef]
- Kotková, M.; Němejc, K.; Sak, B.; Hanzal, V.; Květoňová, D.; Hlásková, L.; Čondlová, S.; McEvoy, J.; Kváč, M. Cryptosporidium ubiquitum, C. muris and Cryptosporidium deer genotype in wild cervids and caprines in the Czech Republic. Folia Parasitol. 2016, 63, 003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, M.; Luo, N.; Wang, H.; Yu, F.; Wang, R.; Huang, J.; Zhang, L. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol. Int. 2015, 64, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Kubota, R.; Matsubara, K.; Tamukai, K.; Ike, K.; Tokiwa, T. Molecular and histopathological features of Cryptosporidium ubiquitum infection in imported chinchillas Chinchilla lanigera in Japan. Parasitol. Int. 2019, 68, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, S.H.; Jiang, W.; Zhao, J.G.; Li, N.; Guo, Y.Q.; Liao, C.H.; Han, Q.; Feng, Y.Y.; Xiao, L.H. Cryptosporidium parvum and Cryptosporidium hominis subtypes in crab-eating macaques. Parasit. Vectors 2019, 12, 350. [Google Scholar] [CrossRef] [Green Version]
- Prediger, J.; Horčičková, M.; Hofmannová, L.; Sak, B.; Ferrari, N.; Mazzamuto, M.V.; Romeo, C.; Wauters, L.A.; McEvoy, J.; Kváč, M. Native and introduced squirrels in Italy host different Cryptosporidium spp. Eur. J. Protistol. 2017, 61, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Vetterling, J.M.; Jervis, H.R.; Merrill, T.G.; Sprinz, H. Cryptosporidium wrairi sp. n. from the guinea pig Cavia porcellus, with an emendation of the genus. J. Protozool. 1971, 18, 243–247. [Google Scholar] [CrossRef]
- Robertson, L.J.; Björkman, C.; Axén, C.; Fayer, R. Cryptosporidiosis in Farmed Animals. In Cryptosporidium: Parasite and Disease; Cacciò, S.M., Widmer, G., Eds.; Springer: Vienna, Austria, 2014; pp. 149–236. [Google Scholar]
- Kváč, M.; Vlnatá, G.; Ježková, J.; Horčičková, M.; Konečný, R.; Hlásková, L.; McEvoy, J.; Sak, B. Cryptosporidium occultus sp. n. (Apicomplexa: Cryptosporidiidae) in rats. Eur. J. Protistol. 2018, 63, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santín, M.; Xiao, L. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J. Parasitol. 2005, 91, 624–629. [Google Scholar] [CrossRef]
- Ryan, U.M.; Monis, P.; Enemark, H.L.; Sulaiman, I.; Samarasinghe, B.; Read, C.; Buddle, R.; Robertson, I.; Zhou, L.; Thompson, R.C.A.; et al. Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa). J. Parasitol. 2004, 90, 769–773. [Google Scholar] [CrossRef] [Green Version]
- Kváč, M.; Kestřánová, M.; Pinková, M.; Květoňová, D.; Kalinová, J.; Wagnerová, P.; Kotková, M.; Vítovec, J.; Ditrich, O.; McEvoy, J.; et al. Cryptosporidium scrofarum n. sp. (Apicomplexa: Cryptosporidiidae) in domestic pigs (Sus scrofa). Vet. Parasitol. 2013, 191, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Power, M.L.; Ryan, U.M. A new species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) from eastern grey kangaroos (Macropus giganteus). J. Parasitol. 2008, 94, 1114–1117. [Google Scholar] [CrossRef]
- Kváč, M.; Květoňová, D.; Sak, B.; Ditrich, O. Cryptosporidium pig genotype II in immunocompetent man. Emerg. Infect. Dis. 2009, 15, 982–983. [Google Scholar] [CrossRef]
- Khan, S.M.; Debnath, C.; Pramanik, A.K.; Xiao, L.; Nozaki, T.; Ganguly, S. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Vet. Parasitol. 2010, 171, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Navarr-I-Martinez, L.; da Silva, A.J.; Bornay-Llinares, F.J.; Moura, I.N.S.; del Aguila, C.; Oleaga, A.; Pieniazek, N.J. Detection and molecular characterization of Cryptosporidium bovis-like isolate from a newborn lamb in Spain. J. Parasitol. 2007, 93, 1536–1538. [Google Scholar] [CrossRef]
- Ren, X.; Zhao, J.; Zhang, L.; Ning, C.; Jian, F.; Wang, R.; Lv, C.; Wang, Q.; Arrowood, M.J.; Xiao, L. Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus). Exp. Parasitol. 2012, 130, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Tzipori, S.; Smith, M.; Halpin, C.; Angus, K.W.; Sherwood, D.; Campbell, I. Experimental Cryptosporidiosis in Calves—Clinical Manifestations and Pathological Findings. Vet. Rec. 1983, 112, 116–120. [Google Scholar] [CrossRef]
- Kváč, M.; Hofmannová, L.; Hlásková, L.; Květoňová, D.; Vítovec, J.; McEvoy, J.; Sak, B. Cryptosporidium erinacei n. sp. (Apicomplexa: Cryptosporidiidae) in hedgehogs. Vet. Parasitol. 2014, 201, 9–17. [Google Scholar] [CrossRef]
- Deming, C.; Greiner, E.; Uhl, E.W. Prevalence of Cryptosporidium infection and characteristics of oocyst shedding in a breeding colony of leopard geckos (Eublepharis macularius). J. Zoo Wildl. Med. 2008, 39, 600–607. [Google Scholar] [CrossRef]
- Tzipori, S.; Smith, M.; Makin, T.; Halpin, C. Enterocolitis in piglets caused by Cryptosporidium sp. purified from calf faeces. Vet. Parasitol. 1982, 11, 121–126. [Google Scholar] [CrossRef]
- Čondlová, S.; Horčičková, M.; Sak, B.; Květoňová, D.; Hlásková, L.; Konečný, R.; Stanko, M.; McEvoy, J.; Kváč, M. Cryptosporidium apodemi sp. n. and Cryptosporidium ditrichi sp. n. (Apicomplexa: Cryptosporidiidae) in Apodemus spp. Eur. J. Protistol. 2018, 63, 1–12. [Google Scholar] [CrossRef]
- Umemiya, R.; Fukuda, M.; Fujisaki, K.; Matsui, T. Electron microscopic observation of the invasion process of Cryptosporidium parvum in severe combined immunodeficiency mice. J. Parasitol. 2005, 91, 1034–1039. [Google Scholar] [CrossRef]
- Borowski, H.; Thompson, R.C.; Armstrong, T.; Clode, P.L. Morphological characterization of Cryptosporidium parvum life-cycle stages in an in vitro model system. Parasitology 2010, 137, 13–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fayer, R.; Santín, M.; Macarisin, D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet. Parasitol. 2010, 172, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Hofmannová, L.; Hauptman, K.; Huclová, K.; Květoňová, D.; Sak, B.; Kváč, M. Cryptosporidium erinacei and C. parvum in a group of overwintering hedgehogs. Eur. J. Protistol. 2016, 56, 15–20. [Google Scholar] [CrossRef]
- Holubová, N.; Tůmová, L.; Sak, B.; Hejzlerová, A.; Konečný, R.; McEvoy, J.; Kváč, M. Description of Cryptosporidium ornithophilus sp. n. (Apicomplexa: Cryptosporidiidae) as a new species and diversity in farmed ostriches. Parasit. Vectors 2020, 13, 340. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Osorio, S.; Chaparro-Gutierrez, J.J.; Gomez-Osorio, L.M. Overview of Poultry Eimeria Life Cycle and Host-Parasite Interactions. Front. Vet. Sci. 2020, 7, 384. [Google Scholar] [CrossRef] [PubMed]
- Koehler, A.V.; Šlapeta, J. Use of Markers to Determine Cryptosporidium Genotypes for Epidemiology Tracking and Detection. In Cryptosporidium Methods and Protocols; Methods in Molecular Biology; Volume 2052, Mead, J.R., Arrowood, M.J., Eds.; Springer Science+Business Media, LLC: New York, NY, USA, 2019; pp. 117–127. [Google Scholar]
- Qari, S.H.; Goldman, I.F.; Pieniazek, N.J.; Collins, W.E.; Lal, A.A. Blood and sporozoite stage-specific small subunit ribosomal RNA-encoding genes of the human malaria parasite Plasmodium vivax. Gene 1994, 150, 43–49. [Google Scholar] [CrossRef]
- Tang, Y.; Li, N.; Song, M.; Roellig, D.M.; Feng, Y.; Xiao, L. Development of a multilocus sequence typing tool for high-resolution subtyping and genetic structure characterization of Cryptosporidium ubiquitum. Infect. Genet. Evol. 2016, 45, 256–261. [Google Scholar] [CrossRef] [Green Version]
- El-Sherry, S.; Ogedengbe, M.E.; Hafeez, M.A.; Barta, J.R. Divergent nuclear 18S rDNA paralogs in a turkey coccidium, Eimeria meleagrimitis, complicate molecular systematics and identification. Int. J. Parasitol. 2013, 43, 679–685. [Google Scholar] [CrossRef]
- Le Blancq, S.M.; Khramtsov, N.V.; Zamani, F.; Upton, S.J.; Wu, T.W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 1997, 90, 463–478. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Arisue, N.; Kawai, S.; Escalante, A.A.; Horii, T.; Tanabe, K.; Hashimoto, T. Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus Plasmodium. Mol. Phylogenet. Evol. 2008, 47, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Stenger, B.L.; Clark, M.E.; Kváč, M.; Khan, E.; Giddings, C.W.; Dyer, N.W.; Schultz, J.L.; McEvoy, J.M. Highly divergent 18S rRNA gene paralogs in a Cryptosporidium genotype from eastern chipmunks (Tamias striatus). Infect. Genet. Evol. 2015, 32, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, D.S.; Upton, S.J.; Owens, D.S.; Morgan, U.M.; Mead, J.R.; Blagburn, B.L. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from cattle, Bos taurus. J. Eukaryot. Microbiol. 2000, 47, 91–95. [Google Scholar] [CrossRef]
- Holubová, N.; Sak, B.; Horčičková, M.; Hlásková, L.; Květoňová, D.; Menchaca, S.; McEvoy, J.; Kváč, M. Cryptosporidium avium n. sp. (Apicomplexa: Cryptosporidiidae) in birds. Parasitol. Res. 2016, 115, 2243–2251. [Google Scholar] [CrossRef] [PubMed]
- Current, W.L.; Upton, S.J.; Haynes, T.B. The life cycle of Cryptosporidium baileyi n. sp. (Apicomplexa, Cryptosporidiidae) infecting chickens. J. Protozool. 1986, 33, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.S.; Blagburn, B.L.; Sundermann, C.A. Morphometric Comparison of the Oocysts of Cryptosporidium meleagridis and Cryptosporidium baileyi from Birds. Proc. Helminthol. Soc. Wash. 1989, 56, 91–92. [Google Scholar]
- Bolland, S.J.; Zahedi, A.; Oskam, C.; Murphy, B.; Ryan, U. Cryptosporidium bollandi n. sp. (Apicomplexa: Cryptosporidiiae) from angelfish (Pterophyllum scalare) and Oscar fish (Astronotus ocellatus). Exp. Parasitol. 2020, 217, 107956. [Google Scholar] [CrossRef]
- Fayer, R.; Trout, J.M.; Xiao, L.; Morgan, U.M.; Lai, A.A.; Dubey, J.P. Cryptosporidium canis n. sp. from domestic dogs. J. Parasitol. 2001, 87, 1415–1422. [Google Scholar] [CrossRef]
- Paperna, I.; Vilenkin, M. Cryptosporidiosis in the gourami Trichogaster leeri: Description of a new species and a proposal for a new genus, Piscicryptosporidium, for species infecting fish. Dis. Aquat. Organ. 1996, 27, 95–101. [Google Scholar] [CrossRef]
- Robinson, G.; Wright, S.; Elwin, K.; Hadfield, S.J.; Katzer, F.; Bartley, P.M.; Hunter, P.R.; Nath, M.; Innes, E.A.; Chalmers, R.M. Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): Morphology, biology and phylogeny. Int. J. Parasitol. 2010, 40, 1539–1548. [Google Scholar] [CrossRef]
- Ježková, J.; Horčičková, M.; Hlásková, L.; Sak, B.; Květoňová, D.; Novák, J.; Hofmannová, L.; McEvoy, J.; Kváč, M. Cryptosporidium testudinis sp. n., Cryptosporidium ducismarci Traversa, 2010 and Cryptosporidium tortoise genotype III (Apicomplexa: Cryptosporidiidae) in tortoises. Folia Parasitol. 2016, 63, 035. [Google Scholar] [CrossRef] [Green Version]
- Ryan, U.M.; Power, M.; Xiao, L. Cryptosporidium fayeri n. sp. (Apicomplexa: Cryptosporidiidae) from the Red Kangaroo (Macropus rufus). J. Eukaryot. Microbiol. 2008, 55, 22–26. [Google Scholar] [CrossRef]
- Iseki, M. Cryptosporidium felis sp. n. (protozoa: Eimeriorina) from the domestic cat. Jap. J. Protozool. 1979, 28, 285–307. [Google Scholar]
- Jirků, M.; Valigurová, A.; Koudela, B.; Křížek, J.; Modrý, D.; Šlapeta, J. New species of Cryptosporidium Tyzzer, 1907 (Apicomplexa) from amphibian host: Morphology, biology and phylogeny. Folia Parasitol. (Praha) 2008, 55, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Ryan, U.M.; Xiao, L.; Read, C.; Sulaiman, I.M.; Monis, P.; Lal, A.A.; Fayer, R.; Pavlásek, I. A redescription of Cryptosporidium galli Pavlásek, 1999 (Apicomplexa: Cryptosporidiidae) from birds. J. Parasitol. 2003, 89, 809–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan-Ryan, U.M.; Fall, A.; Ward, L.A.; Hijjawi, N.; Sulaiman, I.; Fayer, R.; Thompson, R.C.; Olson, M.; Lal, A.; Xiao, L. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 2002, 49, 433–440. [Google Scholar] [CrossRef]
- Slavin, D. Cryptosporidium meleagridis (sp. nov.). J. Comp. Pathol. 1955, 65, 262–266. [Google Scholar] [CrossRef]
- Alvarez-Pellitero, P.; Sitja-Bobadilla, A. Cryptosporidium molnari n. sp. (Apicomplexa: Cryptosporidiidae) infecting two marine fish species, Sparus aurata L. and Dicentrarchus labrax L. Int. J. Parasitol. 2002, 32, 1007–1021. [Google Scholar] [CrossRef]
- Tyzzer, E.E. An extracellular coccidium, Cryptosporidium muris (gen. et sp. nov.) of the gastric glands of the common mouse. J. Med. Res. 1910, 23, 487–509. [Google Scholar]
- Hoover, D.M.; Hoerr, F.J.; Carlton, W.W.; Hinsman, E.J.; Ferguson, H.W. Enteric Cryptosporidiosis in a Naso Tang, Naso-Lituratus Bloch and Schneider. J. Fish Dis. 1981, 4, 425–428. [Google Scholar] [CrossRef]
- Upton, S.J.; Current, W.L. The species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) infecting mammals. J. Parasitol. 1985, 71, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Kváč, M.; Havrdová, N.; Hlásková, L.; Daňková, T.; Kanděra, J.; Ježková, J.; Vítovec, J.; Sak, B.; Ortega, Y.; Xiao, L.; et al. Cryptosporidium proliferans n. sp. (Apicomplexa: Cryptosporidiidae): Molecular and biological evidence of cryptic species within gastric Cryptosporidium of mammals. PLoS ONE 2016, 11, e0147090. [Google Scholar] [CrossRef]
- Li, X.; Pereira, M.; Larsen, R.; Xiao, C.; Phillips, R.; Striby, K.; McCowan, B.; Atwill, E.R. Cryptosporidium rubeyi n. sp. (Apicomplexa: Cryptosporidiidae) in multiple Spermophilus ground squirrel species. Int. J. Parasitol. Parasites Wildl. 2015, 4, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Fayer, R.; Santín, M.; Trout, J.M. Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). Vet. Parasitol. 2008, 156, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Pellitero, P.; Quiroga, M.I.; Sitja-Bobadilla, A.; Redondo, M.J.; Palenzuela, O.; Padros, F.; Vazquez, S.; Nieto, J.M. Cryptosporidium scophthalmi n. sp (Apicomplexa: Cryptosporidiidae) from cultured turbot Scophthalmus maximus. Light and electron microscope description and histopathological study. Dis. Aquat. Organ. 2004, 62, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Levine, N.D. Some corrections of coccidian (Apicomplexa: Protozoa) nomenclature. J. Parasitol. 1980, 66, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Pavlásek, I.; Lávisková, M.; Horák, P.; Král, J.; Král, B. Cryptosporidium varanii n.sp. (Apicomplexa: Cryptosporidiidae) in Emerald monitor (Varanus prasinus Schlegal, 1893) in captivity in Prague zoo. Gazella 1995, 22, 99–108. [Google Scholar]
- Elwin, K.; Hadfield, S.J.; Robinson, G.; Crouch, N.D.; Chalmers, R.M. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int. J. Parasitol. 2012, 42, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santín, M. Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries). Vet. Parasitol. 2009, 164, 192–200. [Google Scholar] [CrossRef]





| Country | Locality # | No. of Positive/No. of Screened | ID of Positive Animal | Microscopical Positivity (OPG) | Genotyping at the Gene Loci | |||
|---|---|---|---|---|---|---|---|---|
| SSU | Actin | HSP70 | GP60 | |||||
| Czech Republic | Planá nad Lužnicí (1) | 1/11 | 41838 | No | C. parvum | C. parvum | C. parvum | IIa |
| Praha (2) | 0/15 | - | - | - | - | - | - | |
| Jihlava (3) | 0/7 | - | - | - | - | - | - | |
| Třebíč (4) | 0/6 | - | - | - | - | - | - | |
| Břeclav (5) | 0/6 | - | - | - | - | - | - | |
| Týnec (6) | 0/15 | - | - | - | - | - | - | |
| Lanžhot (7) | 2/12 | 29639 | No | C. myocastoris | C. myocastoris | C. myocastoris | XXIIb | |
| 29370 | No | C. myocastoris | C. myocastoris | C. myocastoris | XXIIa | |||
| Slovakia | Nové Zámky (8) | 2/12 | 31467 | No | C. ubiquitum | C. ubiquitum | C. ubiquitum | XIId |
| 31472 | No | C. myocastoris | C. myocastoris | C. myocastoris | XXIIb | |||
| Komárno (9) | 0/3 | - | - | - | - | - | - | |
| Šaľa (10) | 1/5 | 31459 | Yes (25,000) | C. myocastoris | C. myocastoris | C. myocastoris | XXIIa | |
| Dolný Ohaj (11) | 0/7 | - | - | - | - | - | - | |
| Topoľníky (12) | 0/10 | - | - | - | - | - | - | |
| Palárikovo (13) | 1/6 | - | - | - | - | - | - | |
| Nitrianský Hrádok (14) | 0/5 | - | - | - | - | - | - | |
| Dunajská Streda (15) | 5/19 | 31123 | No | C. ubiquitum | C. ubiquitum | C. ubiquitum | XIId | |
| 31129 | Yes (18,000) | C. ubiquitum | C. ubiquitum | C. ubiquitum | XIId | |||
| 31135 | No | C. ubiquitum | C. ubiquitum | C. ubiquitum | XIId | |||
| 31136 | No | C. ubiquitum | C. ubiquitum | C. ubiquitum | XIId | |||
| 31132 | Yes (10,000) | C. myocastoris | C. myocastoris | C. myocastoris | XXIIa | |||
| Vlčany (16) | 0/6 | - | - | - | - | - | - | |
| Diakovce (17) | 0/1 | - | - | - | - | - | - | |
| Lipové (18) | 0/4 | - | - | - | - | - | - | |
| Isolate | Length (μm) Range (Mean ± SD) | Width (μm) Range (Mean ± SD) | Length/Width Ratio Range (Mean ± SD) |
|---|---|---|---|
| Nutria 31132 * | 4.8–5.2 (5.02 ± 0.13) | 4.7–5.0 (4.85 ± 0.10) | 1.00–1.08 (1.04 ± 0.02) |
| Nutria 31459 * | 4.8–5.3 (5.01 ± 0.14) | 4.7–5.0 (4.81 ± 0.10) | 1.00–1.06 (1.04 ± 0.01) |
| Nutria N0# | 4.8–5.2 (5.00 ± 0.12) | 4.7–5.0 (4.79 ± 0.09) | 1.02–1.09 (1.04 ± 0.02) |
| Nutria N1# | 4.8–5.3 (5.02 ± 0.14) | 4.6–5.1 (4.85 ± 0.14) | 1.02–1.07 (1.03 ± 0.01) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ježková, J.; Limpouchová, Z.; Prediger, J.; Holubová, N.; Sak, B.; Konečný, R.; Květoňová, D.; Hlásková, L.; Rost, M.; McEvoy, J.; et al. Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus). Microorganisms 2021, 9, 813. https://doi.org/10.3390/microorganisms9040813
Ježková J, Limpouchová Z, Prediger J, Holubová N, Sak B, Konečný R, Květoňová D, Hlásková L, Rost M, McEvoy J, et al. Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus). Microorganisms. 2021; 9(4):813. https://doi.org/10.3390/microorganisms9040813
Chicago/Turabian StyleJežková, Jana, Zlata Limpouchová, Jitka Prediger, Nikola Holubová, Bohumil Sak, Roman Konečný, Dana Květoňová, Lenka Hlásková, Michael Rost, John McEvoy, and et al. 2021. "Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus)" Microorganisms 9, no. 4: 813. https://doi.org/10.3390/microorganisms9040813
APA StyleJežková, J., Limpouchová, Z., Prediger, J., Holubová, N., Sak, B., Konečný, R., Květoňová, D., Hlásková, L., Rost, M., McEvoy, J., Rajský, D., Feng, Y., & Kváč, M. (2021). Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus). Microorganisms, 9(4), 813. https://doi.org/10.3390/microorganisms9040813