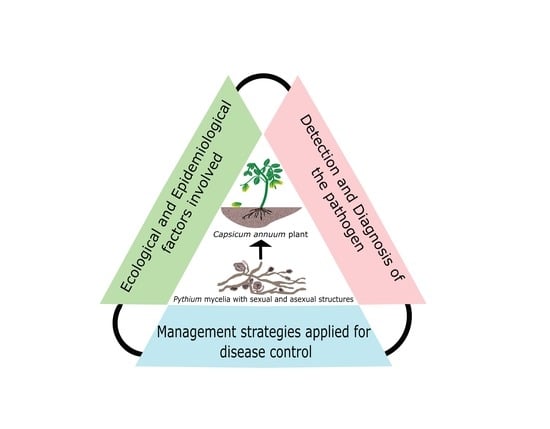
Pythium Damping-Off and Root Rot of Capsicum annuum L.: Impacts, Diagnosis, and Management
Abstract
1. Introduction
2. Pythium Species: Causal Agent of Damping-Off and Root Rot
2.1. Ecology
2.2. Epidemiology
3. Detection and Diagnosis of Pythium Species
4. Control Measures
4.1. Cultural Control
4.2. Chemical Control
4.3. Biological Control
5. Virulence Mechanism of Pythium spp. and Challenges in Resistant Breeding against Pythium spp.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, B.S.; Chahwala, F.D.; Rathod, S.; Singh, A.K. Identification and molecular characterization of a new recombinant begomovirus and associated betasatellite DNA infecting Capsicum annuum in India. Arch. Virol. 2016, 161, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Martins, K.C.; Pereira, T.N.S.; Souza, S.A.M.; Rodrigues, R.; Amaral Junior, A.T. do Crossability and evaluation of incompatibility barriers in crosses between Capsicum species. Crop Breed. Appl. Biotechnol. 2015, 15, 139–145. [Google Scholar] [CrossRef]
- García, C.C.; Barfuss, M.H.J.; Sehr, E.M.; Barboza, G.E.; Samuel, R.; Moscone, E.A.; Ehrendorfer, F. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 2016, 118, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Reyes-Escogido, M.D.L.; Gonzalez-mondragon, E.G.; Vazquez-tzompantzi, E. Chemical and Pharmacological Aspects of Capsaicin. Molecules 2011, 16, 1253. [Google Scholar] [CrossRef] [PubMed]
- Naves, E.R.; Silva, L.D.Á.; Sulpice, R.; Araújo, W.L.; Nunes-nesi, A.; Peres, L.E.P.; Zsögön, A. Capsaicinoids: Pungency beyond Capsicum. Trends Plant Sci. 2019, 24, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Uarrota, V.G.; Maraschin, M.; De Bairros, Â.d.F.M.; Pedreschi, R. Factors affecting the capsaicinoid profile of hot peppers and biological activity of their non-pungent analogs (Capsinoids) present in sweet peppers. Crit. Rev. Food Sci. Nutr. 2021, 61, 649–665. [Google Scholar] [CrossRef]
- Loizzo, M.R.; Pugliese, A.; Bonesi, M.; Menichini, F.; Tundis, R. Evaluation of chemical profile and antioxidant activity of twenty cultivars from Capsicum annuum, Capsicum baccatum, Capsicum chacoense and Capsicum chinense: A comparison between fresh and processed peppers. LWT Food Sci. Technol. 2015, 64, 623–631. [Google Scholar] [CrossRef]
- Arimboor, R.; Natarajan, R.B.; Menon, K.R.; Chandrasekhar, L.P.; Moorkoth, V. Red pepper (Capsicum annuum) carotenoids as a source of natural food colors: Analysis and stability—A review. J. Food Sci. Technol. 2015, 52, 1258–1271. [Google Scholar] [CrossRef]
- Hernández-Pérez, T.; Gómez-García, M.d.R.; Valverde, M.E.; Paredes-López, O. Capsicum annuum (hot pepper): An ancient Latin-American crop with outstanding bioactive compounds and nutraceutical potential. A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2972–2993. [Google Scholar] [CrossRef]
- Olatunji, T.L.; Afolayan, A.J. The suitability of chili pepper (Capsicum annuum L.) for alleviating human micronutrient dietary deficiencies: A review. Food Sci. Nutr. 2018, 6, 2239–2251. [Google Scholar] [CrossRef]
- Saleh, B.K.; Omer, A.; Teweldemedhin, B. Medicinal uses and health benefits of chili pepper (Capsicum spp.): A review. MOJ Food Process. Technol. 2018, 6, 325–328. [Google Scholar] [CrossRef]
- Sarafi, E.; Siomos, A.; Tsouvaltzis, P.; Chatzissavvidis, C.; Therios, I. Boron and maturity effects on biochemical parameters and antioxidant activity of pepper ( Capsicum annuum L.) cultivars. Turk. J. Agric. For. 2018, 42, 237–247. [Google Scholar] [CrossRef]
- Srinivasan, K. Biological Activities of Red Pepper (Capsicum annuum) and Its Pungent Principle Capsaicin: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Nicolaï, M.; Cantet, M.; Lefebvre, V.; Sage-Palloix, A.-M.; Palloix, A. Genotyping a large collection of pepper (Capsicum spp.) with SSR loci brings new evidence for the wild origin of cultivated C. annuum and the structuring of genetic diversity by human selection of cultivar types. Genet. Resour. Crop Evol. 2013, 60, 2375–2390. [Google Scholar] [CrossRef]
- Kamvorn, W.; Techawongstien, S.; Techawongstien, S. Compatibility of inter-specific crosses between Capsicum chinense Jacq. and Capsicum baccatum L. at different fertilization stages. Sci. Hortic. 2014, 179, 9–15. [Google Scholar] [CrossRef]
- Saxena, A.; Raghuwanshi, R.; Gupta, V.K.; Singh, H.B. Chilli Anthracnose: The Epidemiology and Management. Front. Microbiol. 2016, 7, 1527. [Google Scholar] [CrossRef]
- Pagoch, K.; Srivastava, J.N.; Singh, A.K. Damping-Off Disease of Seedlings in Solanaceous Vegetables: Current Status and Disease Management. In Recent Advances in the Diagnosis and Management of Plant Diseases; Springer: New Delhi, India, 2015; pp. 35–46. [Google Scholar]
- Muthukumar, A.; Udhayakumar, R.; Naveenkumar, R. Eco friendly management of damping-off of solanaceous crops caused by Pythium species. In Current Trends in Plant Disease Diagnostics and Management Practices; Kumar, P., Gupta, V., Tiwari, A., Kamle, M., Eds.; Springer: Cham, Switzerland, 2016; pp. 49–90. [Google Scholar] [CrossRef]
- Hyder, S.; Inam-ul-Haq, M.; Ashfaq, M.; Ahmad, A.; Gondal, A.S.; Iqbal, M. First report of Pythium myriotylum D., causing damping off and root rot in chili pepper (Capsicum annum L.) from Punjab, Pakistan. Plant Dis. 2018, 102, 687. [Google Scholar] [CrossRef]
- Dubey, M.K.; Zehra, A.; Aamir, M.; Yadav, M.; Samal, S.; Upadhyay, R.S. Isolation, identification, carbon utilization profile and control of Pythium graminicola, the causal agent of chilli damping-off. J. Phytopathol. 2020, 168, 88–102. [Google Scholar] [CrossRef]
- Ali, M.; Shahid, A.A.; Subhani, M.N. Maping and monitoring for the valuation of soil fungi and chili damping off. J. Anim. Plant Sci. 2019, 29, 737–745. [Google Scholar]
- Nawaz, K.; Shahid, A.A.; Subhani, M.N.; Anwar, W.; Aslam, M. First Report of Pythium spinosum Causing Root Rot of Chili (Capsicum annuum) in Pakistan. Plant Dis. 2016, 100, 526. [Google Scholar] [CrossRef]
- Middleton, J.T. The taxonomy, host range and geographic distribution of the genus Pythium. Mem. Torrey Bot. Club 1943, 20, 1–171. [Google Scholar]
- Nema, K.G.; Mahmud, K.A. Damping-off of chilli seedlings in Madhya Pradesh due to Pythium aphanidermatum. Mag. Nagpuragric. Coll 1951, 25, 10–12. [Google Scholar]
- Muthukumar, A.; Eswaran, A.; Sangeetha, G. Occurrence, virulence and pathogenicity of species of Pythium inciting damping-off disease in chilli. J. Mycol. Plant Pathol. 2010, 40, 67. [Google Scholar]
- Jadhav, V.T.; Ambadkar, C. V Effect of Trichoderma spp. on seedling emergence and seedling mortality of tomato, chilli and brinjal. J. Plant Dis. Sci 2007, 2, 190–192. [Google Scholar]
- Zagade, S.N.; Deshpande, G.D.; Gawade, D.B.; Atnoorkar, A.A.; Pawar, S.V. Biocontrol agents and fungicides for management of damping off in chilli. World J. Agric. Sci. 2012, 8, 590–597. [Google Scholar]
- Dar, G.H.; Mir, G.H.; Rashid, H.; Dar, W.A.; Majeed, M. Evaluation of microbial antagonists for the management of wilt/root rot and damping-off diseases in chilli (Capsicum annuum). Vegetos 2015, 28, 102–110. [Google Scholar] [CrossRef]
- Majeed, M.; Hassan Mir, G.; Hassan, M.; Mohuiddin, F.A. Biological management of damping-off disease of Chilli (Capsicum annuum L.). Ecol. Environ. Conserv. 2019, 25, 353–356. [Google Scholar]
- Kavitha, K.; Meenakumari, K.S.; Sivaprasad, P. Effect of dual inoculation of native arbuscular mycorrhizal fungi and Azospirillum on suppression of damping off in chilli. Indian Phytopathol. 2003, 56, 112–113. [Google Scholar]
- Shahid, A.A.; Ali, M.; Ali, S. First report of Pythium debaryanum causing chili damping off in Pakistan. Plant Dis. 2017, 101, 391. [Google Scholar] [CrossRef]
- Mushtaq, M.; Hashmi, M.H. Fungi associated with wilt disease of Capsicum in Sindh, Pakistan. Pak. J. Bot. 1997, 29, 217–222. [Google Scholar]
- Back, M.A.; Haydock, P.P.J.; Jenkinson, P. Disease complexes involving plant parasitic nematodes and soilborne pathogens. Plant Pathol. 2002, 51, 683–697. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Dürr, C.; Schwanck, A.A.; Robin, M.-H.; Sarthou, J.-P.; Cellier, V.; Messéan, A.; Aubertot, J.-N. Integrated management of damping-off diseases. A review. Agron. Sustain. Dev. 2017, 37, 10. [Google Scholar] [CrossRef]
- Mostowfizadeh-Ghalamfarsa, R.; Salmaninezhad, F. Taxonomic Challenges in the Genus Pythium. In Pythium; CRC Press: Boca Raton, FL, USA, 2020; pp. 179–199. [Google Scholar]
- Kageyama, K. Molecular taxonomy and its application to ecological studies of Pythium species. J. Gen. Plant Pathol. 2014, 80, 314–326. [Google Scholar] [CrossRef]
- Schroeder, K.L.; Martin, F.N.; De Cock, A.W.A.M.; Lévesque, C.A.; Spies, C.F.J.; Okubara, P.A.; Paulitz, T.C. Molecular detection and quantification of Pythium species: Evolving taxonomy, new tools, and challenges. Plant Dis. 2013, 97, 4–20. [Google Scholar] [CrossRef]
- Nzungize, J.R.; Lyumugabe, F.; Busogoro, J.-P.; Baudoin, J.-P. Pythium root rot of common bean: Biology and control methods. A review. Biotechnol. Agron. Soc. Environ. 2012, 16, 405–413. [Google Scholar]
- Le, D.P.; Smith, M.; Hudler, G.W.; Aitken, E. Pythium soft rot of ginger: Detection and identification of the causal pathogens, and their control. Crop Prot. 2014, 65, 153–167. [Google Scholar] [CrossRef]
- Okubara, P.A.; Dickman, M.B.; Blechl, A.E. Molecular and genetic aspects of controlling the soilborne necrotrophic pathogens Rhizoctonia and Pythium. Plant Sci. 2014, 228, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Gaastra, W.; Lipman, L.J.A.; De Cock, A.W.A.M.; Exel, T.K.; Pegge, R.B.G.; Scheurwater, J.; Vilela, R.; Mendoza, L. Pythium insidiosum: An overview. Vet. Microbiol. 2010, 146, 1–16. [Google Scholar] [CrossRef]
- Qiu, L.; Mao, Y.; Tang, L.; Tang, X.; Mo, Z. Characterization of Pythium chondricola associated with red rot disease of Pyropia yezoensis (Ueda) (Bangiales, Rhodophyta) from Lianyungang, China. J. Oceanol. Limnol. 2019, 37, 1102–1112. [Google Scholar] [CrossRef]
- Gerbore, J.; Benhamou, N.; Vallance, J.; Le Floch, G.; Grizard, D.; Regnault-Roger, C.; Rey, P. Biological control of plant pathogens: Advantages and limitations seen through the case study of Pythium oligandrum. Environ. Sci. Pollut. Res. 2014, 21, 4847–4860. [Google Scholar] [CrossRef]
- Sharma, P.; Jambhulkar, P.P.; Raja, M.; Javeria, S. Pythium spp. on Vegetable Crops: Research Progress and Major Challenges. In Pythium; CRC Press: Boca Raton, FL, USA, 2020; pp. 136–161. [Google Scholar]
- Weiland, J.E.; Scagel, C.F.; Grünwald, N.J.; Davis, E.A.; Beck, B.R.; Foster, Z.S.L.; Fieland, V.J. Soilborne Phytophthora and Pythium Diversity From Rhododendron in Propagation, Container, and Field Production Systems of the Pacific Northwest. Plant Dis. 2020, 104, 1841–1850. [Google Scholar] [CrossRef]
- Redekar, N.R.; Eberhart, J.L.; Parke, J.L. Diversity of Phytophthora, Pythium, and Phytopythium species in recycled irrigation water in a container nursery. Phytobiomes J. 2019, 3, 31–45. [Google Scholar] [CrossRef]
- Kavitha, K.; Mathiyazhagan, S.; Senthilvel, V.; Nakkeeran, S.; Chandrasekar, G. Development of bioformulations of antagonistic bacteria for the management of damping off of Chilli (Capsicum annuum L). Arch. Phytopathol. Plant Prot. 2005, 38, 19–30. [Google Scholar] [CrossRef]
- Lodhi, A.M.; Khanzada, M.A.; Shahzad, S.; Ghaffar, A. Prevalence of Pythium aphanidermatum in agro-ecosystem of Sindh province of Pakistan. Pak. J. Bot. 2013, 45, 635–642. [Google Scholar]
- Nawaz, K.; Shahid, A.A.; Anwar, W.; Iftikhar, S.; Nasir, M. Genetic Diversity of the Pythium spp. associated with Root Rot of Chili (Capsicum annum L.) in Pakistan and its Management through Plant extracts. In Proceedings of the 5th International Conference on Food, Agricultural, Biological and Medical Science (FABMS-2017), Bangkok, Thailand, 6–7 February 2017; pp. 84–89. [Google Scholar]
- Muthukumar, A.; Nakkeeran, S.; Eswaran, A.; Sangeetha, G. In vitro efficacy of bacterial endophytes against the chilli damping-off pathogen Pythium aphanidermatum. Phytopathol. Mediterr. 2010, 49, 179–186. [Google Scholar]
- Muthukumar, A.; Eswaran, A.; Sangeetha, G. Induction of systemic resistance by mixtures of fungal and endophytic bacterial isolates against Pythium aphanidermatum. Acta Physiol. Plant. 2011, 33, 1933–1944. [Google Scholar] [CrossRef]
- Hussain, F.; Shaukat, S.S.; Abid, M.; Usman, F.; Akbar, M. Pathogenicity of some important root rot fungi to the chilli crop and their biological control. Int. J. Biol. Biotechnol. 2013, 10, 101–108. [Google Scholar]
- Ho, H.H. The Taxonomy and Biology of Phytophthora and Pythium. J. Bacteriol. Mycol. Open Access 2018, 6, 174. [Google Scholar] [CrossRef]
- Martin, F.N.; Loper, J.E. Soilborne Plant Diseases Caused by Pythium spp.: Ecology, Epidemiology, and Prospects for Biological Control. Crit. Rev. Plant Sci. 1999, 18, 111–181. [Google Scholar] [CrossRef]
- Serrano, M.; Robertson, A.E. The Effect of Cold Stress on Damping-Off of Soybean Caused by Pythium sylvaticum. Plant Dis. 2018, 102, 2194–2200. [Google Scholar] [CrossRef]
- Guo, L.Y.; Ko, W.H. Distribution of mating types and the nature of survival of Pythium splendens in soil. Soil Biol. Biochem. 1993, 25, 839–842. [Google Scholar] [CrossRef]
- Stanghellini, M.E. The Sporangium of Pythium ultimum as a Survival Structure in Soil. Phytopathology 1971, 61, 157. [Google Scholar] [CrossRef]
- Hendrix, F.F.; Campbell, W.A. Pythiums as Plant Pathogens. Annu. Rev. Phytopathol. 1973, 11, 77–98. [Google Scholar] [CrossRef]
- Kilany, M.; Ibrahim, E.H.; Al Amry, S.; Al Roman, S.; Siddiqi, S. Microbial Suppressiveness of Pythium Damping-Off Diseases. In Organic Amendments and Soil Suppressiveness in Plant Disease Management; Meghvansi, M.K., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 187–206. [Google Scholar]
- Sutton, J.C.; Sopher, C.R.; Owen-Going, T.N.; Liu, W.; Grodzinski, B.; Hall, J.C.; Benchimol, R.L. Etiology and epidemiology of Pythium root rot in hydroponic crops: Current knowledge and perspectives. Summa Phytopathol. 2006, 32, 307–321. [Google Scholar] [CrossRef]
- Hyder, S.; Naseem, S.; Azhar, S.; Ashfaq, M.; Ali, Z.; Khalid, A.; Inam-Ul-Haq, M. Disease incidence and severity of Pythium spp. And Phytophthora spp. affecting chili pepper and tomato crops in punjab, Pakistan. Philipp. Agric. Sci. 2018, 101, 136–147. [Google Scholar]
- Manjunath, M.; Prasanna, R.; Nain, L.; Dureja, P.; Singh, R.; Kumar, A.; Jaggi, S.; Kaushik, B.D. Biocontrol potential of cyanobacterial metabolites against damping off disease caused by Pythium aphanidermatum in solanaceous vegetables. Arch. Phytopathol. Plant Prot. 2010, 43, 666–677. [Google Scholar] [CrossRef]
- Muthukumar, A.; Eswaran, A.; Sanjeevkumar, K. Effect of different soil types on the incidence of chilli damping-off incited by Pythium aphanidermatum. Agric. Sci. Dig. 2009, 29, 215–217. [Google Scholar]
- Hansen, Z.R.; Keinath, A.P. Increased pepper yields following incorporation of biofumigation cover crops and the effects on soilborne pathogen populations and pepper diseases. Appl. Soil Ecol. 2013, 63, 67–77. [Google Scholar] [CrossRef]
- Lévesque, C.A.; De Cock, A.W.A.M. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 2004, 108, 1363–1383. [Google Scholar] [CrossRef]
- McLeod, A.; Botha, W.J.; Meitz, J.C.; Spies, C.F.J.; Tewoldemedhin, Y.T.; Mostert, L. Morphological and phylogenetic analyses of Pythium species in South Africa. Mycol. Res. 2009, 113, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.K.; Pandey, P.K. Differential response of biocontrol agents against soil pathogens on tomato, chilli and brinjal. Indian Phytopathol. 2005, 58, 329–331. [Google Scholar]
- Chakraborty, B.N.; Chakraborty, U. Molecular detection of fungal pathogens and induction of phytoimmunity using bioinoculants. Indian Phytopathol. 2021, 1–16. [Google Scholar] [CrossRef]
- De Cara, M.; Pérez-Hernández, A.; Aguilera-Lirola, A.; Gómez-Vázquez, J. First report of wilting, root rot, and stunting caused by Pythium aphanidermatum on sweet pepper (Capsicum annuum) in southeastern Spain. Plant Dis. 2017, 101, 1059. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guid. Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Wakeham, A.J.; Pettitt, T.R. Diagnostic tests and their application in the management of soil- and water-borne oomycete pathogen species. Ann. Appl. Biol. 2017, 170, 45–67. [Google Scholar] [CrossRef]
- Ray, M.; Dash, S.; Shahbazi, S.; Achary, K.G.; Nayak, S.; Singh, S. Development and validation of ELISA technique for early detection of rhizome rot in golden spice turmeric from different agroclimatic zones. LWT Food Sci. Technol. 2016, 66, 546–552. [Google Scholar] [CrossRef]
- Ray, M.; Dash, S.; Gopinath Achary, K.; Nayak, S.; Singh, S. Development and evaluation of polyclonal antibodies for detection of Pythium aphanidermatum and Fusarium oxysporum in ginger. Food Agric. Immunol. 2018, 29, 204–215. [Google Scholar] [CrossRef]
- Mirmajlessi, S.M.; Loit, E.; Maend, M.; Mansouripour, S.M. Real-time PCR applied to study on plant pathogens: Potential applications in diagnosis-a review. Plant. Prot. Sci. 2015, 51, 177–190. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Li Destri Nicosia, M.G.; Faedda, R.; Cacciola, S.O.; Schena, L. Use of quantitative PCR detection methods to study biocontrol agents and phytopathogenic fungi and oomycetes in environmental samples. J. Phytopathol. 2014, 162, 1–13. [Google Scholar] [CrossRef]
- Li, M.; Senda, M.; Komatsu, T.; Suga, H.; Kageyama, K. Development of real-time PCR technique for the estimation of population density of Pythium intermedium in forest soils. Microbiol. Res. 2010, 165, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ishiguro, Y.; Otsubo, K.; Suzuki, H.; Tsuji, T.; Miyake, N.; Nagai, H.; Suga, H.; Kageyama, K. Monitoring by real-time PCR of three water-borne zoosporic Pythium species in potted flower and tomato greenhouses under hydroponic culture systems. Eur. J. Plant Pathol. 2014, 140, 229–242. [Google Scholar] [CrossRef]
- Van Der Heyden, H.; Wallon, T.; Lévesque, C.A.; Carisse, O. Detection and Quantification of Pythium tracheiphilum in Soil by Multiplex Real-Time qPCR. Plant Dis. 2019, 103, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Senda, M.; Suga, H.; Kageyama, K. Development of Multiplex PCR to Detect Five Pythium Species Related to Turfgrass Diseases. J. Phytopathol. 2010, 158, 609–615. [Google Scholar] [CrossRef]
- Ishiguro, Y.; Asano, T.; Otsubo, K.; Suga, H.; Kageyama, K. Simultaneous detection by multiplex PCR of the high-temperature-growing Pythium species: P. aphanidermatum, P. helicoides and P. myriotylum. J. Gen. Plant Pathol. 2013, 79, 350–358. [Google Scholar] [CrossRef]
- Fukuta, S.; Takahashi, R.; Kuroyanagi, S.; Ishiguro, Y.; Miyake, N.; Nagai, H.; Suzuki, H.; Tsuji, T.; Hashizume, F.; Watanabe, H.; et al. Development of loop-mediated isothermal amplification assay for the detection of Pythium myriotylum. Lett. Appl. Microbiol. 2014, 59, 49–57. [Google Scholar] [CrossRef]
- Takahashi, R.; Fukuta, S.; Kuroyanagi, S.; Miyake, N.; Nagai, H.; Kageyama, K.; Ishiguro, Y. Development and application of a loop-mediated isothermal amplification assay for rapid detection of Pythium helicoides. FEMS Microbiol. Lett. 2014, 355, 28–35. [Google Scholar] [CrossRef]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef]
- Feng, W.; Nukaya, A.; Satou, M.; Fukuta, N.; Ishiguro, Y.; Suga, H.; Kageyama, K. Use of LAMP detection to identify potential contamination sources of plant-pathogenic Pythium species in hydroponic culture systems of tomato and eustoma. Plant Dis. 2018, 102, 1357–1364. [Google Scholar] [CrossRef]
- Shen, D.; Li, Q.; Yu, J.; Zhao, Y.; Zhu, Y.; Xu, H.; Dou, D. Development of a loop-mediated isothermal amplification method for the rapid detection of Pythium ultimum. Australas. Plant Pathol. 2017, 46, 571–576. [Google Scholar] [CrossRef]
- Feng, H.; Chen, J.; Yu, Z.; Li, Z.; Ye, W.; Wang, Y.; Zheng, X. A loop-mediated isothermal amplification assay can rapidly diagnose soybean root-rot and damping-off diseases caused by Pythium spinosum. Australas. Plant Pathol. 2019, 48, 553–562. [Google Scholar] [CrossRef]
- Cao, Y.; Li, Y.; Li, J.; Wang, L.; Cheng, Z.; Wang, H.; Fan, Z.; Li, H. Rapid and quantitative detection of Pythium inflatum by real-time fluorescence loop-mediated isothermal amplification assay. Eur. J. Plant Pathol. 2016, 144, 83–95. [Google Scholar] [CrossRef]
- Kageyama, K.; Ohyama, A.; Hyakumachi, M. Detection of Pythium ultimum using polymerase chain reaction with species-specific primers. Plant Dis. 1997, 81, 1155–1160. [Google Scholar] [CrossRef]
- Wan, P.H.; Chung, C.Y.; Lin, Y.S.; Yeh, Y. Use of polymerase chain reaction to detect the soft rot pathogen, Pythium myriotylum, in infected ginger rhizomes. Lett. Appl. Microbiol. 2003, 36, 116–120. [Google Scholar] [CrossRef]
- Okubara, P.A.; Schroeder, K.L.; Paulitz, T.C. Real-time polymerase chain reaction: Applications to studies on soilborne pathogens. Can. J. Plant Pathol. 2005, 27, 300–313. [Google Scholar] [CrossRef]
- Schroeder, K.L.; Okubara, P.A.; Tambong, J.T.; Lévesque, C.A.; Paulitz, T.C. Identification and Quantification of Pathogenic Pythium spp. from Soils in Eastern Washington Using Real-Time Polymerase Chain Reaction. Phytopathology 2006, 96, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, J.; Tiu, K.R.; Kumar, A.; Kumar Singh, V.; Khan, Z. Impact of soil solarization on some solanaceous vegetables nursery in Plateau Region of Jharkhand, India. Vegetos 2012, 25, 109–114. [Google Scholar]
- Baysal-Gurel, F.; Liyanapathiranage, P.; Addesso, K.M. Effect of Brassica crop-based biofumigation on soilborne disease suppression in woody ornamentals. Can. J. Plant Pathol. 2020, 42, 94–106. [Google Scholar] [CrossRef]
- Smith, B.J.; Kirkegaard, J.A. In vitro inhibition of soil microorganisms by 2-phenylethyl isothiocyanate. Plant Pathol. 2002, 51, 585–593. [Google Scholar] [CrossRef]
- Handiseni, M.; Brown, J.; Zemetra, R.; Mazzola, M. Use of Brassicaceous seed meals to improve seedling emergence of tomato and pepper in Pythium ultimum infested soils. Arch. Phytopathol. Plant Prot. 2012, 45, 1204–1209. [Google Scholar] [CrossRef]
- Chowdhary, K.; Kumar, A.; Sharma, S.; Pathak, R.; Jangir, M. Ocimum sp.: Source of biorational pesticides. Ind. Crops Prod. 2018, 122, 686–701. [Google Scholar] [CrossRef]
- Saha, S.; Rai, A.B.; Pandey, S. Efficacy of seed dressing agents against damping-off disease of chilli (Capsicum frutescens). Indian J. Agric. Sci. 2011, 81, 92. [Google Scholar]
- White, D.J.; Chen, W.; Schroeder, K.L. Assessing the contribution of ethaboxam in seed treatment cocktails for the management of metalaxyl-resistant Pythium ultimum var. ultimum in Pacific Northwest spring wheat production. Crop Prot. 2019, 115, 7–12. [Google Scholar] [CrossRef]
- Wang, M.; Van Vleet, S.; McGee, R.; Paulitz, T.C.; Porter, L.D.; Vandemark, G.; Chen, W. Chickpea seed rot and damping-off caused by metalaxyl-resistant Pythium ultimum and its management with ethaboxam. Plant Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rizzati, V.; Briand, O.; Guillou, H.; Gamet-Payrastre, L. Effects of pesticide mixtures in human and animal models: An update of the recent literature. Chem. Biol. Interact. 2016, 254, 231–246. [Google Scholar] [CrossRef]
- Pesticide Control Division, Department of Agriculture Malaysia (DOA). Pesticide Information System (SISMARP). 2021. Available online: http://www.portal.doa.gov.my/sismarp/welcome/detail/14415?jc=perosak&search=pythium (accessed on 20 March 2021).
- Saha, S.K.; McSorley, R.; Wang, K.; McGovern, R.J. Impacts of extreme weather and soil management treatments on disease development of Pythium spp. in field grown pepper. In Proceedings of the 118th Annual Meeting of the Florida State Horticultural Society, Marriott Tampa Westshore, FL, USA, 5–7 June 2005; Volume 118, pp. 146–149. [Google Scholar]
- Kokalis-Burelle, N.; McSorley, R.; Wang, K.H.; Saha, S.K.; McGovern, R.J. Rhizosphere microorganisms affected by soil solarization and cover cropping in Capsicum annuum and Phaseolus lunatus agroecosystems. Appl. Soil Ecol. 2017, 119, 64–71. [Google Scholar] [CrossRef]
- Jørgensen, L.N.; Van den Bosch, F.; Oliver, R.P.; Heick, T.M.; Paveley, N.D. Targeting fungicide inputs according to need. Annu. Rev. Phytopathol. 2017, 55, 181–203. [Google Scholar] [CrossRef]
- Muthukumar, A.; Bhaskaran, R.; Sanjeevkumar, K. Efficacy of endophytic Pseudomonas fluorescens (Trevisan) migula against chilli damping-off. J. Biopestic. 2010, 3, 105. [Google Scholar]
- Amaresan, N.; Jayakumar, V.; Thajuddin, N. Isolation and characterization of endophytic bacteria associated with chilli (Capsicum annuum) grown in coastal agricultural ecosystem. Indian J. Biotechnol. 2014, 13, 247–255. [Google Scholar]
- Mukherjee, P.K.; Raghu, K. Effect of temperature on antagonistic and biocontrol potential of shape Trichoderma sp. on Sclerotium rolfsii. Mycopathologia 1997, 139, 151–155. [Google Scholar] [CrossRef]
- Mehetre, S.T.; Kale, S.P. Comparative efficacy of thermophilic bacterium, Bacillus licheniformis (NR1005) and antagonistic fungi, Trichoderma harzianum to control Pythium aphanidermatum -induced damping off in chilli (Capsicum annuum L.). Arch. Phytopathol. Plant Prot. 2011, 44, 1068–1074. [Google Scholar] [CrossRef]
- Lara-Capistran, L.; Zulueta-Rodriguez, R.; Castellanos-Cervantes, T.; Reyes-Perez, J.J.; Preciado-Rangel, P.; Hernandez-Montiel, L.G. Efficiency of Marine Bacteria and Yeasts on the Biocontrol Activity of Pythium ultimum in Ancho-Type Pepper Seedlings. Agronomy 2020, 10, 408. [Google Scholar] [CrossRef]
- Ali, M.; Shahid, A.A.; Haider, M.S. Isolation and in-vitro screening of potential antagonistic rhizobacteria against Pythium debaryanum. Pak. J. Phytopathol. 2016, 28, 231–240. [Google Scholar]
- Muthukumar, A. Evaluation of introduced Trichoderma species as antagonist of Pythium aphanidermatum causing chilli damping-off. Plant Dis. Res. 2011, 26, 136–138. [Google Scholar]
- Muthukumar, A.; Eswaran, A.; Sanjeevkumas, K. Exploitation of Trichoderma species on the growth of Pythium aphanidermatum in chilli. Braz. J. Microbiol. 2011, 42, 1598–1607. [Google Scholar] [CrossRef]
- Muthukumar, A.; Eswaran, A.; Nakkeeran, S.; Sangeetha, G. Efficacy of plant extracts and biocontrol agents against Pythium aphanidermatum inciting chilli damping-off. Crop. Prot. 2010, 29, 1483–1488. [Google Scholar] [CrossRef]
- Harris, A.R.; Schisler, D.A.; Ryder, M.H.; Adkins, P.G. Bacteria suppress damping-off caused by Pythium ultimum var. Sporangiiferum, and promote growth, in bedding plants. Soil Biol. Biochem. 1994, 26, 1431–1437. [Google Scholar] [CrossRef]
- Zerillo, M.M.; Adhikari, B.N.; Hamilton, J.P.; Buell, C.R.; Lévesque, C.A.; Tisserat, N. Carbohydrate-Active Enzymes in Pythium and Their Role in Plant Cell Wall and Storage Polysaccharide Degradation. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Lévesque, C.A.; Brouwer, H.; Cano, L.; Hamilton, J.P.; Holt, C.; Huitema, E.; Raffaele, S.; Robideau, G.P.; Thines, M.; Win, J.; et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010, 11. [Google Scholar] [CrossRef]
- Adhikari, B.N.; Hamilton, J.P.; Zerillo, M.M.; Tisserat, N.; Lévesque, C.A.; Buell, C.R. Comparative Genomics Reveals Insight into Virulence Strategies of Plant Pathogenic Oomycetes. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Ibarra Caballero, J.R.; Tisserat, N.A. Transcriptome and secretome of two Pythium species during infection and saprophytic growth. Physiol. Mol. Plant Pathol. 2017, 99, 41–54. [Google Scholar] [CrossRef]
- Lin, F.; Wani, S.H.; Collins, P.J.; Wen, Z.; Li, W.; Zhang, N.; McCoy, A.G.; Bi, Y.; Tan, R.; Zhang, S.; et al. QTL mapping and GWAS for identification of loci conferring partial resistance to Pythium sylvaticum in soybean (Glycine max (L.) Merr). Mol. Breed. 2020, 40. [Google Scholar] [CrossRef]
- Ai, G.; Yang, K.; Ye, W.; Tian, Y.; Du, Y.; Zhu, H.; Li, T.; Xia, Q.; Shen, D.; Peng, H.; et al. Prediction and characterization of RXLR effectors in Pythium species. Mol. Plant Microbe Interact. 2020, 33, 1046–1058. [Google Scholar] [CrossRef]
- Navarro, F.; Sass, M.E.; Nienhuis, J. Identification and confirmation of quantitative trait loci for root rot resistance in snap bean. Crop Sci. 2008, 48, 962–972. [Google Scholar] [CrossRef]
- Stasko, A.K.; Wickramasinghe, D.; Nauth, B.J.; Acharya, B.; Ellis, M.L.; Taylor, C.G.; McHale, L.K.; Dorrance, A.E. High-density mapping of resistance QTL toward Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Sci. 2016, 56, 2476–2492. [Google Scholar] [CrossRef]
- Urrea, K.; Rupe, J.; Chen, P.; Rothrock, C.S. Characterization of seed rot resistance to Pythium aphanidermatum in soybean. Crop Sci. 2017, 57, 1394–1403. [Google Scholar] [CrossRef]
- Klepadlo, M.; Balk, C.S.; Vuong, T.D.; Dorrance, A.E.; Nguyen, H.T. Molecular characterization of genomic regions for resistance to Pythium ultimum var. ultimum in the soybean cultivar Magellan. Theor. Appl. Genet. 2019, 132, 405–417. [Google Scholar] [CrossRef]



| Country | Species | Identification Criteria | Sequence Used in Molecular Identification | Crops Affected | Reference |
|---|---|---|---|---|---|
| Pakistan | P. myriotylum | Morphological and molecular | ITS sequence | 13.8%–45.4% | [20] |
| India | P. graminicola | Morphological and molecular | ITS sequence | Not available | [21] |
| Pakistan | P. spinosum | Morphological and molecular | ITS sequence | Not available | [23] |
| India | P. aphanidermatum, P. graminicola, P. ultimum, P. diliense, P. heterothallicum | Not available | Not available | Not available | [26] |
| Pakistan | P. debaryanum | Morphological and molecular | ITS sequence | 45% | [32] |
| India | P. aphanidermatum | Morphological | Not available | Not available | [48] |
| Pakistan | P. aphanidermatum | Morphological and molecular | ITS and partial LSU sequence | Not available | [49] |
| Pakistan | P. aphanidermatum P. spinosum P. intermedium | Morphological and molecular | ITS Sequence | Not available | [50] |
| India | P. aphanidermatum | Not available | Not available | Not available | [51] |
| India | P. aphanidermatum | Not available | Not available | Not available | [52] |
| Pakistan | Pythium spp. | Not available | Not available | Not available | [53] |
| Pathogen Species | Microbial Control | Strain Name/Commercial Product | In Vitro Control | In Vivo Control | Mode of Action | Reference |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| P. aphanidermatum | Pseudomonas fluorescens | EBS20 | 76.66% reduction in the growth of mycelia. | Not available | Production of phytopathogen inhibitor phenazine. | [51] |
| P. aphanidermatum | Pseudomonas. fluorescens | Biomonas * | Not available. | 10.46% and 20.28% losses due to pre and post-emergence damping-off, respectively, as opposed to 29.06% and 59.12% in control in nursery fields using seed treatment. | Not available. | [98] |
| P. aphanidermatum | Pseudomonas fluorescens | EBC 5 | 68.88% reduction in the growth of mycelia. | 9.10% and 12.33% incidences of pre and post-emergence damping-off when EBC 5 and EBC 7 were combined in pot culture using seed coating, as opposed to 30.66% and 34% in control. | Production of antifungal metabolites reduced mycelial growth in-vitro. | [106] |
| Pseudomonas fluorescens | EBC 7 | 65.93% reduction in the growth of mycelia. | ||||
| Pythium spp. | Bacillus megaterium | BECS7 | 45.9% reduction in the growth of mycelia. | 2% incidences of damping-off as opposed to 14.67% in control in field conditions. | Release of hydrolytic enzymes such as lipase, cellulase, amylase, and protease. | [107] |
| P. aphanidermatum | Bacillus licheniformis | NR1005 | 69.96% reduction in the growth of mycelia. | 81.18% reduction in damping-off incidence over control in pot culture using seed treatment. | Not available. | [109] |
| P. ultimum | Stenotrophomonas rhizophila | KM01 | 80% reduction in the growth of mycelia. | 75%–100% reduction in disease index over control in pot culture using root inoculation. | Not available. | [110] |
| Stenotrophomonas rhizophila | KM02 | 76% reduction in the growth of mycelia. | 75%–100% reduction in disease index over control in pot culture using root inoculation. | |||
| Bacillus subtilis | RBM02 | 67%–77% reduction in the growth of mycelia. | 100% reduction in disease index over control in pot culture using root inoculation. | |||
| P. debaryanum | Bacillus subtilis | RB-31 | 91% inhibition of mycelial growth. | Not available. | Not available. | [111] |
| Fungi | ||||||
| Pythium spp. | Trichoderma harzianum | TK8 | 62.8% reduction in the growth of mycelia. | Not available. | Not available. | [29] |
| P. aphanidermatum | Trichoderma harzianum | Not available | 75.34% reduction in the growth of mycelia. | 83.16% reduction in damping-off incidence over control in pot culture using seed treatment. | Not available. | [109] |
| P. ultimum | Cryptococcus laurentii | 2R1CB | 75% reduction in the growth of mycelia. | 75%–100% reduction in disease index over control in pot culture using root inoculation. | Production of β-1,3-glucanase reduced the mycelial growth in vitro. | [110] |
| P. aphanidermatum | Trichoderma viride | Not available | 76.1% reduction in the growth of mycelia. | Not available | Production of antibiotics. | [112] |
| P. aphanidermatum | Trichoderma viride | TVC3 | 88% reduction in the growth of mycelia. | Not available | Volatile and non-volatile antibiotics production and mycoparasitism. | [113] |
| Fungi + Bacteria | ||||||
| P. aphanidermatm | Trichoderma viride + Trichoderma harzianum + Pseudomonas fluorescens + Bacillus subtilis | Not available. | Not available. | 13.33% and 15.36% incidences of pre and post-emergence damping-off, respectively, as opposed to 53.33% and 24.80% in control in pot culture using seed treatment. | Not available. | [30] |
| P. aphanidermatum | Trichoderma viride + Pseudomonas fluorescens | TVA EBL 20-PF | Not available. | Reduction of 84% and 71.5% in pre and post-emergence damping-off incidences, respectively, using seed treatment and soil application in pot culture. | Induced systemic resistance due to increased activities of PAL, PO, PPO, and accumulation of phenolics. | [52] |
| P. aphanidermatum | Trichoderma viride + Pseudomonas fluorescens | Not available. | 82% reduction in mycelial growth over control. | Reduction of 72.2% and 59.2% in pre and post-emergence damping-off incidences, respectively, in pot culture, using seed treatment. | Production of antifungal antibiotic. | [114] |
| Algae | ||||||
| P. aphanidermatum | Calothrix elenkenii | Not available. | Minimum inhibitory concentration of the ethyl acetate extract of culture filtrate was 16.6 ppm. | Seed treatment with ethyl acetate extract of culture filtrate reduced mortality to 10%–20% as opposed to 60%–70% in untreated controls in pot culture. | Not available. | [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, H.; Sharma, A.; Sharma, S.; Haron, F.F.; Gafur, A.; Sayyed, R.Z.; Datta, R. Pythium Damping-Off and Root Rot of Capsicum annuum L.: Impacts, Diagnosis, and Management. Microorganisms 2021, 9, 823. https://doi.org/10.3390/microorganisms9040823
Arora H, Sharma A, Sharma S, Haron FF, Gafur A, Sayyed RZ, Datta R. Pythium Damping-Off and Root Rot of Capsicum annuum L.: Impacts, Diagnosis, and Management. Microorganisms. 2021; 9(4):823. https://doi.org/10.3390/microorganisms9040823
Chicago/Turabian StyleArora, Himanshu, Abhishek Sharma, Satyawati Sharma, Farah Farhanah Haron, Abdul Gafur, R. Z. Sayyed, and Rahul Datta. 2021. "Pythium Damping-Off and Root Rot of Capsicum annuum L.: Impacts, Diagnosis, and Management" Microorganisms 9, no. 4: 823. https://doi.org/10.3390/microorganisms9040823
APA StyleArora, H., Sharma, A., Sharma, S., Haron, F. F., Gafur, A., Sayyed, R. Z., & Datta, R. (2021). Pythium Damping-Off and Root Rot of Capsicum annuum L.: Impacts, Diagnosis, and Management. Microorganisms, 9(4), 823. https://doi.org/10.3390/microorganisms9040823