The Role of Bifidobacteria in Predictive and Preventive Medicine: A Focus on Eczema and Hypercholesterolemia
Abstract
1. Introduction
2. Gut Microbiota
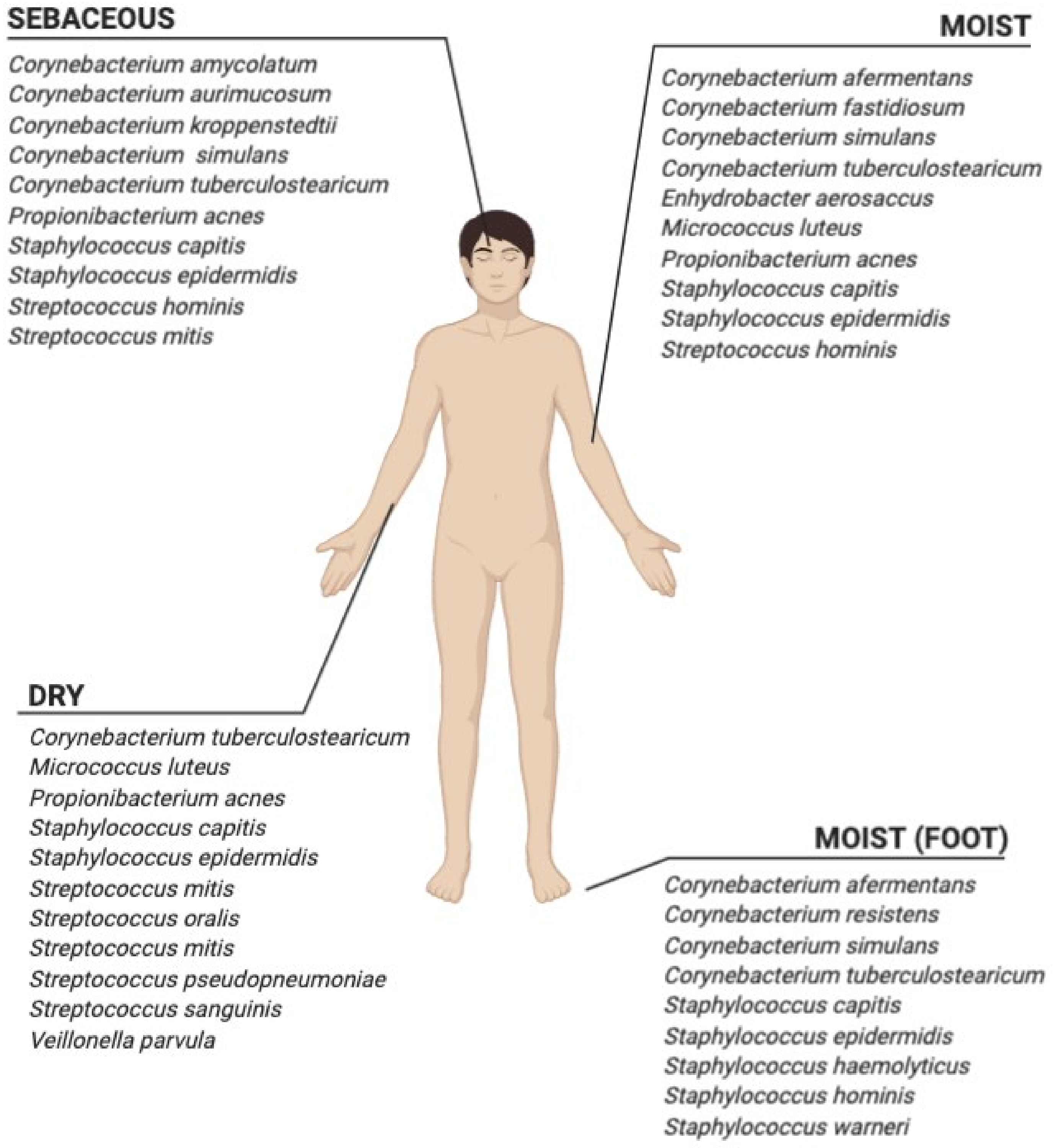
3. Skin Microbiota
4. Hypercholesterolemia and Gut Microbiota
5. Atopic Eczema/Dermatitis Syndrome and Skin Microbiota
6. Bifidobacteria and Their Role in Hypercholesterolemia
7. Bifidobacteria and Their Therapeutic Potential in Eczema
| Strain | Dosage/Dye | Study Design/Subjects | Principal Results | References |
|---|---|---|---|---|
| Bifidobacterium animalis subsp. lactis BB-12 | 1 × 109 CFU/g 1 | Infants | ↓ Neutral lipids in plasma ↑ Phospholipids | Kankaanpaa et al. (2002) [130] |
| Bifidobacterium longum BB536 | 3 × 107 CFU/mL 3 | Rat model | ↓ Total cholesterol ↓ Liver lipids deposition ↓ Adipocyte size | Al-Sheraji et al. (2015) [111] |
| Bifidobacterium infantis 35624 | 1 × 1010 CFU/mL 3 | Human | ↓ C-reactive protein level ↓ TNF-alfa | Groeger et al. (2013) [131] |
| Bifidobacterium breve B-3 | 2 × 109 CFU/mouse 2 | Mouse model | ↓ IL-1beta ↓ Claudin-1 ↓ skin damage ↑ TJs integrity | Satoh et al. (2015) [128] |
| Bifidobacterium adolescentis Ad1-6 | 1 × 109 CFU/0.2 mL | Mouse model 4 | ↓ IgE ↓ IL-4 ↓ AD-simptoms | Fang et al. (2019) [131] |
| Bifidobacterium breve BR03 Lactobacillus salivarius LS01 | 1x109 CFU/g 5 | Human | ↓ SCORAD | Iemoli et al. (2012) [132] |
| Bifidobacterium longum CECT 7347 Bifidobacterium lactis CECT 8145 Lactobacillus casei CECT 9104 | 1 × 109 CFU/mL 6 | Human 7 | ↓ AD SCORAD | Navarro-López et al. (2018) [133] |
| Bifidobacterium breve Bbi99 Lactobacillus rhamnosus GG Lactobacillus rhamnosus LC705 Propionibacterium freudenreichii spp. shermanii JD | 2 × 108 CFU/mL 5 × 109 CFU/mL 5 × 109 CFU/mL 2 × 109 CFU/mL 8 | Infants | ↓ AD rates | Kukkonen et al. (2007) [127] |
| Bifidobacterium animalis subspecies lactis CUL34 Bifidobacterium bifidum CUL20 Lactobacillus salivarius CUL61 Lactobacillus paracasei CUL08 | 1 × 1010 CFU/mL 9 | Infants | ↑ AD prevention | Allen et al. 2014 [134] |
| Bifidobacterium lactis HN019 Lactobacillus acidophilus NCFM Lactobacillus rhamnosus HN001 Lactobacillus paracasei LPC37 | 1 × 1010 CFU/mL 9 | Infants 10 | ↓ SCORAD ↓ BSA ↓ FDLQI | Lise et al. (1992) [96] |
| Bifidobacterium breve Bbi99 Lactobacillus rhamnosus GG Lactobacillus rhamnosus LC705 Propionibacterium freudenreichii spp. shermanii JD | 2 × 108 CFU/mL 5 × 109 CFU/mL 5 × 109 CFU/mL 2 × 1010 CFU/mL 8 | Children 11 | ↑ AD prevention | Kuitunen et al. (2009) [135] |
| Bifidobacterium animalis subsp. lactis BB-12 Lactobacillus rhamnosus GG | 1 × 1010 CFU/mL | Human adults 12 | ↑ AD prevention | Rautava et al. 2006 [136] |
| Bifidobacterium longum BL999 Lactobacillus rhamnosus LPR | 1 × 1010 CFU/mL | Human adults 12 | ↑ AD prevention | Rautava et al. 2012 [137] |
| Bifidobacterium lactis BB-13 Lactobacillus rhamnosus GG | 1 × 1010 CFU/mL | Human adults 12 | ↑ AD prevention | Huurre et al. (2008) [138] |
| Bifidobacterium lactis Lactobacillus rhamnosus | 2 × 1010 CFU/g 6 | Children 13 | ↓ SCORAD | Sistek et al. (2006) [139] |
| Bifidobacterium breve YIT 12272 Lactococcus lactis YIT 2027 Streptococcus thermophilus YIT 2021 | 5–6 × 1010 CFU/100 mL 14 | Human adults 12 | ↑ Cathepsin L-like activity ↑ Hydration level ↓ Serum phenol ↓ Urine phenol | Kano et al. (2013) [129] |
8. Conclusions
Funding
Conflicts of Interest
References and Note
- Proctor, L.; Ravel, J.; Turnbaugh, P. The Human microbiome. Report from Am. Acad. Microbiol. www.asm.org, 2014.
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.; Gonzalez, A.; Ackermann, G.; Wendel, D.; Vázquez-Baeza, Y.; Jansson, J.K.; Gordon, J.I.; Knight, R. Meta-analyses of studies of the human microbiota. Genome Res. 2013, 23, 1704–1714. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Jeon, Y.; Chin, J. Current status and future promise of human micro-biome pediatric gastroenterol. Hepatol. Nutr. 2013, 16, 71–79. [Google Scholar]
- Marcy, Y.; Ouverney, C.; Bik, E.M.; Lösekann, T.; Ivanova, N.; Martin, H.G.; Szeto, E.; Platt, D.; Hugenholtz, P.; Relman, D.A.; et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 2007, 104, 11889–11894. [Google Scholar] [CrossRef]
- Inturri, R.; Stivala, A.; Blandino, G. Microbiological characteristics of the probiotic strain B. longum BB536 and L. rhamnosus HN001 used in combination. Minerva Gastroenterol. Dietol. 2015, 61, 191–197. [Google Scholar]
- Duranti, S.; Gaiani, F.; Mancabelli, L.; Milani, C.; Grandi, A.; Bolchi, A.; Santoni, A.; Lugli, G.A.; Ferrario, C.; Mangifesta, M.; et al. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. FEMS Microbiol. Ecol. 2016, 92, fiw191. [Google Scholar] [CrossRef] [PubMed]
- Inturri, R.; Stivala, A.; Furneri, P.M.; Blandino, G. Growth and adhesion to HT-29 cells inhibition of Gram-negatives by Bifidobacterium longum BB536 e Lactobacillus rhamnosus HN001 alone and in combination. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4943–4949. [Google Scholar]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Inturri, R.; Trovato, L.; Volti, G.L.; Oliveri, S.; Blandino, G. In vitro inhibitory activity of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 alone or in combination against bacterial and Candida reference strains and clinical isolates. Heliyon 2019, 5, e02891. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Kennedy, E.A.; Connolly, J.; Hourihane, J.O.; Fallon, P.G.; McLean, W.H.I.; Murray, D.; Jo, J.H.; Segre, J.A.; Kong, H.H.; Irvine, A.D. Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J. Allergy Clin. Immunol. 2017, 139, 166–172. [Google Scholar] [CrossRef]
- Schneider, A.M.; Nelson, A.M. Skin microbiota: Friend or foe in pediatric skin health and skin disease. Pediatr. Dermatol. 2019, 36, 815–822. [Google Scholar] [CrossRef]
- Sanders, M.E.; Guarner, F.; Guerrant, R.; Holt, P.R.; Quigley, E.M.; Sartor, R.B.; Sherman, P.M.; Mayer, E.A. An update on the use and investigation of probiotics in health and disease. Gut 2013, 62, 787–796. [Google Scholar] [CrossRef]
- Golubnitschaja, O.; Costigliola, V.; EPMA. General report & recommendations in predictive, preventive and personalised medicine 2012: White paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012, 3, 14. [Google Scholar] [PubMed]
- Al-Sharea, A.; Murphy, A.J.; Huggins, L.A.; Hu, Y.; Goldberg, I.J.; Nagareddy, P.R. SGLT2 inhibition reduces atherosclerosis by enhancing lipoprotein clearance in Ldlr-/- type 1 diabetic mice. Atherosclerosis 2018, 271, 166–176. [Google Scholar] [CrossRef]
- Kumar, S.; Jeong, Y.; Ashraf, M.U.; Bae, Y.S. Dendritic Cell-Mediated Th2 immunity and immune disorders. Int. J. Mol. Sci. 2019, 20, 2159. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Mandal, S. Bifidobacteria-Insight into clinical outcomes and mechanisms of its probiotic action. Microbiol. Res. 2016, 192, 159–171. [Google Scholar] [CrossRef]
- Kuugbee, E.D.; Shang, X.; Gamallat, Y.; Bamba, D.; Awadasseid, A.; Suliman, M.A.; Zang, S.; Ma, Y.; Chiwala, G.; Xin, Y.; et al. Structural change in microbiota by a probiotic cocktail enhances the gut barrier and reduces cancer via TLR2 signaling in a rat model of colon cancer. Dig. Dis. Sci. 2016, 61, 2908–2920. [Google Scholar] [CrossRef]
- Asadollahi, P.; Ghanavati, R.; Rohani, M.; Razavi, S.; Esghaei, M.; Talebi, M. Anti-cancer effects of Bifidobacterium species in colon cancer cells and a mouse model of carcinogenesis. PLoS ONE 2020, 15, e0232930. [Google Scholar] [CrossRef]
- Blandino, G.; Inturri, R.; Lazzara, F.; di Rosa, M.; Malaguarnera, L. Impact of gut microbiota on diabetes mellitus. Diabetes Metab. 2016, 42, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Belviso, S.; Giordano, M.; Dolci, P.; Zeppa, G. In vitro cholesterol-lowering activity of Lactobacillus plantarum and Lactobacillus paracasei strains isolated from the Italian Castelmagno PDO cheese. Dairy Sci. Technol. 2009, 89, 169–176. [Google Scholar] [CrossRef]
- Lin, M.; Chen, T.W. Reduction of cholesterol by Lactobacillus acidophilus in culture broth. J. Food Drug Anal. 2000, 8, 97–102. [Google Scholar]
- Tahri, K.; Grill, J.P.; Schneider, F. Bifidobacteria strain behavior toward cholesterol: Coprecipitation with bile salts and assimilation. Curr. Microbiol. 1996, 33, 187–193. [Google Scholar] [CrossRef]
- Albano, C.; Morandi, S.; Silvetti, T.; Casiraghi, M.C.; Manini, F.; Brasca, M. Lactic acid bacteria with cholesterol-lowering properties for dairy applications: In vitro and in situ activity. J. Dairy Sci. 2018, 101, 10807–10818. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Grover, S.; Batish, V.K. Hypocholesterolaemic effect of dietary inclusion of two putative probiotic bile salt hydrolase-producing Lactobacillus plantarum strains in Sprague-Dawley rats. Br. J. Nutr. 2011, 105, 561–573. [Google Scholar] [CrossRef]
- Gilliland, S.E.; Nelson, C.R.; Maxwell, C. Assimilation of cholesterol by Lactobacillus acidophilus. Appl. Environ. Microbiol. 1985, 49, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhang, S.; Lu, J.; Zhang, C.; Pang, X.; Lv, J. Screening for cholesterol-lowering probiotics from lactic acid bacteria isolated from corn silage based on three hypothesized pathways. Int. J. Mol. Sci. 2019, 26, 2073. [Google Scholar] [CrossRef]
- Bubnov, R.V.; Spivak, M.Y.; Lazarenko, L.M.; Bomba, A.; Boyko, N.V. Probiotics and immunity: Provisional role for personalized diets and disease prevention. EPMA J. 2015, 6, 14. [Google Scholar] [CrossRef]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef]
- Satsangi, J. Gene discovery in IBD: A decade of progress. J. Pediatr. Gastroenterol. Nutr. 2008, 46, E1–E2. [Google Scholar] [PubMed]
- Schultz, M.; Lindström, A.L. Rationale for probiotic treatment strategies in inflammatory bowel disease. Expert Rev. Gastroenterol. Hepatol. 2008, 2, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, U.C.; Shukla, R.; Ghoshal, U.; Gwee, K.A.; Ng, S.C.; Quigley, E.M. The gut microbiota and irritable bowel syndrome: Friend or foe? Int. J. Inflamm. 2012, 2012, 151085. [Google Scholar] [CrossRef] [PubMed]
- Bubnov, R.V.; Babenko, L.P.; Lazarenko, L.M.; Mokrozub, V.V.; Spivak, M.Y. Specific properties of probiotic strains: Relevance and benefits for the host. EPMA J. 2018, 9, 205–223. [Google Scholar] [CrossRef]
- Butler, É.; Lundqvist, C.; Axelsson, J. Lactobacillus reuteri DSM 17938 as a Novel Topical Cosmetic Ingredient: A Proof of Concept Clinical Study in Adults with Atopic Dermatitis. Microorganisms 2020, 8, 1026. [Google Scholar] [CrossRef]
- Makrgeorgou, A.; Leonardi-Bee, J.; Bath-Hextall, F.J.; Murrell, D.F.; Tang, M.L.K.; Roberts, A.; Boyle, R.J. Probiotics for treating eczema. Cochrane Database Syst. Rev. 2018, 11, CD006135. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, H.S. Microbiome of the Skin and Gut in Atopic Dermatitis (AD): Understanding the Pathophysiology and Finding Novel Management Strategies. J. Clin. Med. 2019, 8, 444. [Google Scholar] [CrossRef]
- FAO/WHO, 2007. FAO Therapeutical Meeting on Prebiotics. Food Quality and Standard Service September 15–16, 2007. Available online: www.fao.org/ag/agn/agns/index.en.stm (accessed on 13 April 2021).
- FAO/WHO. Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation; Paper N. 85; FAO Food and Nutrition: Rome, Italy, 2006; ISBN 92-5-105513-0. [Google Scholar]
- Gibson, G.R.; Probert, H.M.; Loo, J.V.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Abrahamsson, T.; Bailey, M.; Bindels, L.B.; Bubnov, R.; Ganguli, K.; Martoni, C.; O’Neill, C.; Savignac, H.M.; Stanton, C.; et al. How do probiotics and prebiotics function at distant sites? Benef. Microbes 2017, 8, 521–533. [Google Scholar] [CrossRef]
- Belizário, J.E.; Napolitano, M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 2015, 6, 1050. [Google Scholar] [CrossRef]
- NIH Human Microbiome Project. Available online: http://hmpdacc.org (accessed on 13 April 2021).
- Metagenomics of the Human Intestinal Tract. Available online: http://www.metahit.eu (accessed on 13 April 2021).
- Turroni, F.; Duranti, S.; Bottacini, F.; Guglielmetti, S.; Van Sinderen, D.; Ventura, M. Bifidobacterium bifidum a sane example of a specialized human gut commensal. Front. Microbiol. 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef]
- Rodríguez, J.M. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef]
- Cheng, M.; Ning, K. Stereotypes about enterotype: The old and new ideas. Genom. Proteom. Bioinform. 2019, 17, 4–12. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ogata, K.; Matsuda, K.; Tsuji, H.; Kubota, H.; Takada, T.; Kado, Y.; Asahara, T.; Takahashi, T.; Nomoto, K. Diversity of Intestinal Clostridium coccoides Group in the Japanese Population, as Demonstrated by Reverse Transcription-Quantitative PCR. PLoS ONE 2015, 10, e0126226. [Google Scholar] [CrossRef]
- Petschow, B.; Doré, J.; Hibberd, P.; Dinan, T.; Reid, G.; Blaser, M.; Cani, P.D.; Degnan, F.H.; Foster, J.; Gibson, G.; et al. Probiotics, prebiotics, and the host microbiome: The science of translation. Ann. N. Y. Acad. Sci. 2013, 1306, 1–17. [Google Scholar] [CrossRef]
- Turroni, F.; Ventura, M.; Buttó, L.F.; Duranti, S.; O’Toole, P.W.; Motherway, M.O.; van Sinderen, D. Molecular dialogue between the human gut microbiota and the host: A Lactobacillus and Bifidobacterium perspective. Cell Mol. Life Sci. 2014, 71, 183–203. [Google Scholar] [CrossRef]
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef]
- Frank, D.N.; Pace, N.R. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 2008, 24, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Major, G.; Spiller, R. Irritable bowel syndrome, inflammatory bowel disease and the microbiome. Curr. Opin. Endocrinol. Diabetes Obes. 2014, 21, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Asar, T.O.; Kumar, V.; Al-Abbasi, F.A.; Alhayyani, S.; Kamal, M.A.; Anwar, F. A cross-talk between gut microbiome, salt and hypertension. Biomed. Pharmacother. 2021, 134, 111156. [Google Scholar] [CrossRef]
- Allayee, H.; Hazen, S.L. Contribution of Gut Bacteria to Lipid Levels: Another Metabolic Role for Microbes? Circ. Res. 2015, 117, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Cantabrana, C.; López, P.; Gueimonde, M.; de Los Reyes-Gavilán, C.G.; Suárez, A.; Margolles, A.; Ruas-Madiedo, P. Immune modulation capability of exopolysaccharides synthesised by lactic acid bacteria and bifidobacteria. Probiotics Antimicrob. Proteins 2012, 4, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Inturri, R.; Mangano, K.; Santagati, M.; Intrieri, M.; di Marco, R.; Blandino, G. Immunomodulatory Effects of Bifidobacterium longum W11 produced exopolysaccharide on cytokine production. Curr. Pharm. Biotechnol. 2017, 18, 883–889. [Google Scholar] [CrossRef]
- Inturri, R.; Molinaro, A.; di Lorenzo, F.; Blandino, G.; Tomasello, B.; Hidalgo-Cantabrana, C.; de Castro, C.; Ruas-Madiedo, P. Chemical and biological properties of the novel exopolysaccharide produced by a probiotic strain of Bifidobacterium longum. Carbohydr. Polym. 2017, 174, 1172–1180. [Google Scholar] [CrossRef]
- Szöllősi, A.G.; Gueniche, A.; Jammayrac, O.; Szabó-Papp, J.; Blanchard, C.; Vasas, N.; Andrási, M.; Juhász, I.; Breton, L.; Bíró, T. Bifidobacterium longum extract exerts pro-differentiating effects on human epidermal keratinocytes, in vitro. Exp. Dermatol. 2017, 26, 92–94. [Google Scholar] [CrossRef]
- Odamaki, T.; Bottacini, F.; Kato, K.; Mitsuyama, E.; Yoshida, K.; Horigome, A.; Xiao, J.Z.; van Sinderen, D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018, 8, 85. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Huys, G.; Daube, G. Probiotics an update. J. Pediatr. 2015, 91, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Domingues, C.P.F.; Rebelo, J.S.; Dionisio, F.; Botelho, A.; Nogueira, T. The social distancing imposed to contain COVID-19 can affect our microbiome: A double-edged sword in human health. mSphere 2020, 5, e00716-20. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.I.; Conlan, S.; Belkaid, Y.; Segre, J.A.; Kong, H.H. NISC Comparative Sequencing Program Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Nakamura, Y.; Núñez, G. Role of the microbiota in skin immunity and atopic dermatitis. Allergol. Int. 2017, 66, 539–544. [Google Scholar] [CrossRef]
- Egert, M.; Simmering, R.; Riedel, C.U. The Association of the Skin Microbiota with Health, Immunity, and Disease. Clin. Pharmacol. Ther. 2017, 102, 62–69. [Google Scholar] [CrossRef]
- Loomis, K.H.; Wu, S.K.; Ernlund, A.; Zudock, K.; Reno, A.; Blount, K.; Karig, D.K. A mixed community of skin microbiome representatives influences cutaneous processes more than individual members. Microbiome 2021, 9, 22. [Google Scholar] [CrossRef]
- Meisel, J.S.; Hannigan, G.D.; Tyldsley, A.S.; SanMiguel, A.J.; Hodkinson, B.P.; Zheng, Q.; Grice, E.A. Skin microbiome surveys are strongly influenced by experimental design. J. Investig. Dermatol. 2016, 136, 947–956. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chiang, H.I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 2013, 4, 1431. [Google Scholar] [CrossRef]
- Atarashi, K.; Nishimura, J.; Shima, T.; Umesaki, Y.; Yamamoto, M.; Onoue, M.; Yagita, H.; Ishii, N.; Evans, R.; Honda, K.; et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 2008, 455, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Bouladoux, N.; Sun, C.M.; Wohlfert, E.A.; Blank, R.B.; Zhu, Q.; Grigg, M.E.; Berzofsky, J.A.; Belkaid, Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 2008, 29, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef]
- Yuki, T.; Yoshida, H.; Akazawa, Y.; Komiya, A.; Sugiyama, Y.; Inoue, S. Activation of TLR2 enhances tight junction barrier in epidermal keratinocytes. J. Immunol. 2011, 187, 3230–3237. [Google Scholar] [CrossRef] [PubMed]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Götz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef]
- Doz, E.; Rose, S.; Nigou, J.; Gilleron, M.; Puzo, G.; Erard, F.; Ryffel, B.; Quesniaux, V.F. Acylation determines the toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor-, and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J. Biol. Chem. 2007, 282, 26014–26025. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.L.; Nielsen, T.B.; Pantapalangkoor, P.; Zhao, F.; Lee, J.C.; Montgomery, C.P.; Luna, B.; Spellberg, B.; Daum, R.S. Evaluation of serotypes 5 and 8 capsular polysaccharides in protection against Staphylococcus aureus in murine models of infection. Hum. Vaccin. Immunother. 2017, 13, 1609–1614. [Google Scholar] [CrossRef]
- Zimmermann, M.; Fischbach, M.A. A family of pyrazinone natural products from a conserved nonribosomal peptide synthetase in Staphylococcus aureus. Chem. Biol. 2010, 17, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Busnelli, M.; Manzini, S.; Sirtori, C.R.; Chiesa, G.; Parolini, C. Effects of Vegetable Proteins on Hypercholesterolemia and Gut Microbiota Modulation. Nutrients 2018, 10, 1249. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: https://www.who.int/ (accessed on 13 April 2021).
- Villette, R.; Kc, P.; Beliard, S.; Salas Tapia, M.F.; Rainteau, D.; Guerin, M.; Lesnik, P. Unraveling Host-Gut Microbiota Dialogue and Its Impact on Cholesterol Levels. Front. Pharmacol. 2020, 11, 278. [Google Scholar] [CrossRef]
- Ali, N.; Rahman, S.; Islam, S.; Haque, T.; Molla, N.H.; Sumon, A.H.; Kathak, R.R.; Asaduzzaman, M.; Islam, F.; Mohanto, N.C.; et al. The relationship between serum uric acid and lipid profile in Bangladeshi adults. BMC Cardiovasc. Disord. 2019, 19, 42. [Google Scholar] [CrossRef] [PubMed]
- Woldeamlak, B.; Yirdaw, K.; Biadgo, B. Hyperuricemia and its Association with Cardiovascular Disease Risk Factors in Type Two Diabetes Mellitus Patients at the University of Gondar Hospital, Northwest Ethiopia. EJIFCC 2019, 30, 325–339. [Google Scholar]
- Alphonse, P.A.; Jones, P.J. Revisiting Human Cholesterol Synthesis and Absorption: The Reciprocity Paradigm and its Key Regulators. Lipids 2016, 51, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Onaolapo, O.J.; Onaolapo, A.Y.; Olowe, A.O. The neurobehavioral implications of the brain and microbiota interaction. Front. Biosci. 2020, 25, 363–397. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Lise, M.; Mayer, I.; Silveira, M. Use of probiotics in atopic dermatitis. Rev. Assoc. Med. Bras. 2018, 64, 997–1001. [Google Scholar] [CrossRef]
- Chng, K.R.; Tay, A.S.; Li, C.; Ng, A.H.; Wang, J.; Suri, B.K.; Matta, S.A.; McGovern, N.; Janela, B.; Wong, X.F.; et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat. Microbiol. 2016, 1, 16106. [Google Scholar] [CrossRef] [PubMed]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Mäkelä, M.J.; et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. USA 2012, 109, 8334–8339. [Google Scholar] [CrossRef]
- Fyhrquist, N.; Ruokolainen, L.; Suomalainen, A.; Lehtimäki, S.; Veckman, V.; Vendelin, J.; Karisola, P.; Lehto, M.; Savinko, T.; Jarva, H.; et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J. Allergy Clin. Immunol. 2014, 134, 1301–1309.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.J.; Guerrero-Juarez, C.F.; Hata, T.; Bapat, S.P.; Ramos, R.; Plikus, M.V.; Gallo, R.L. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 2015, 347, 67–71. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Two, A.M.; Chun, K.A.; Narala, S.; Geha, R.S.; Hata, T.R.; Gallo, R.L. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J. Investig. Dermatol. 2016, 136, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Sohn, E. Skin microbiota’s community effort. Nature 2018, 563, S91–S93. [Google Scholar] [CrossRef] [PubMed]
- Coates, M.; Lee, M.J.; Norton, D.; MacLeod, A.S. The Skin and Intestinal Microbiota and Their Specific Innate Immune Systems. Front. Immunol. 2019, 10, 2950. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.; Maguire, G. The role of microbiota, and probiotics and prebiotics in skin health. Arch. Dermatol. Res. 2017, 309, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Murzina, E.; Kaliuzhna, L.; Bardova, K.; Yurchyk, Y.; Barynova, M. Human Skin Microbiota in Various Phases of Atopic Dermatitis. Acta Dermatovenerol. Croat. 2019, 27, 245–249. [Google Scholar]
- Guarner, F.; Schaafsma, G.J. Probiotics. Int. J. Food Microbiol. 1998, 39, 237–238. [Google Scholar] [CrossRef]
- Fijan, S. Microorganisms with claimed probiotic properties: An overview of recent literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, F.; Heinken, A.; Thiele, I.; Lindenburg, P.W.; Harms, A.C.; Hankemeier, T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes 2021, 13, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, M.J.; Gao, Q.; Yang, J.F.; Yang, L.; Pang, X.L.; Jiang, X.J. The effects of probiotics on total cholesterol: A meta-analysis of randomized controlled trials. Medicine 2018, 97, e9679. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, L.; Aceto, S.; Agnisola, C.; de Paolo, S.; Dipineto, L.; Stilling, R.M.; Dinan, T.G.; Cryan, J.F.; Menna, L.F.; Fioretti, A. Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish. Sci. Rep. 2016, 6, 30046. [Google Scholar] [CrossRef]
- Al-Sheraji, S.H.; Amin, I.; Azlan, A.; Manap, M.Y.; Hassan, F.A. Effects of Bifidobacterium longum BB536 on lipid profile and histopathological changes in hypercholesterolaemic rats. Benef. Microbes 2015, 6, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kang, J.; Choi, S.; Park, H.; Hwang, E.; Kang, Y.; Kim, A.; Holzapfel, W.; Ji, Y. Cholesterol-lowering effect of Lactobacillus rhamnosus BFE5264 and its influence on the gut microbiome and propionate level in a murine model. PLoS ONE 2018, 13, e0203150. [Google Scholar] [CrossRef]
- Mann, G.V. Studies of a surfactant and cholesteremia in the Maasai. Am. J. Clin. Nutr. 1974, 27, 464–469. [Google Scholar] [CrossRef]
- Zhou, X.; Li, J.; Guo, J.; Geng, B.; Ji, W.; Zhao, Q.; Li, J.; Liu, X.; Liu, J.; Guo, Z.; et al. Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome 2018, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Savilahti, E. Probiotics in the treatment and prevention of allergies in children. Biosci. Microflora 2011, 30, 119–128. [Google Scholar] [CrossRef]
- Yang, H.J.; Min, T.K.; Lee, H.W.; Pyun, B.Y. Efficacy of Probiotic Therapy on Atopic Dermatitis in Children: A Randomized, Double-blind, Placebo-controlled Trial. Allergy Asthma Immunol. Res. 2014, 6, 208–215. [Google Scholar] [CrossRef]
- Ji, G.E. Probiotics in primary prevention of atopic dermatitis. Forum Nutr. 2009, 61, 117–128. [Google Scholar]
- Lee, J.; Seto, D.; Bielory, L. Meta-analysis of clinical trials of probiotics for prevention and treatment of pediatric atopic dermatitis. J. Allergy Clin. Immunol. 2008, 121, 116–121.e11. [Google Scholar] [CrossRef]
- Panduru, M.; Panduru, N.M.; Sălăvăstru, C.M.; Tiplica, G.S. Probiotics and primary prevention of atopic dermatitis: A meta-analysis of randomized controlled studies. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 232–242. [Google Scholar] [CrossRef]
- Betsi, G.I.; Papadavid, E.; Falagas, M.E. Probiotics for the treatment or prevention of atopic dermatitis: A review of the evidence from randomized controlled trials. Am. J. Clin. Dermatol. 2008, 9, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Li, L.; Zhao, J.; Zhang, H.; Lee, Y.K.; Lu, W.; Chen, W. Bifidobacteria adolescentis regulated immune responses and gut microbial composition to alleviate DNFB-induced atopic dermatitis in mice. Eur. J. Nutr. 2020, 59, 3069–3081. [Google Scholar] [CrossRef] [PubMed]
- Fleming, S.A.; Mudd, A.T.; Hauser, J.; Yan, J.; Metairon, S.; Steiner, P.; Donovan, S.M.; Dilger, R.N. Human and bovine milk oligosaccharides elicit improved recognition memory concurrent with alterations in regional brain volumes and hippocampal mRNA Expression. Front. Neurosci. 2020, 14, 770. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, R.; Stone, T.W. The Gut-Brain Axis, BDNF, NMDA and CNS Disorders. Neurochem. Res. 2016, 41, 2819–2835. [Google Scholar] [CrossRef]
- Fleming, S.A.; Mudd, A.T.; Hauser, J.; Yan, J.; Metairon, S.; Steiner, P.; Donovan, S.M.; Dilger, R.N. Dietary oligofructose alone or in combination with 2′-fucosyllactose differentially improves recognition memory and hippocampal mRNA Expression. Nutrients 2020, 12, 2131. [Google Scholar] [CrossRef]
- Franco-Robles, E.; López, M.G. Agavins increase neurotrophic factors and decrease oxidative stress in the brains of high-fat diet-induced obese mice. Molecules 2016, 21, 998. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Yang, X.; Zheng, L.; Wang, Z.; Wu, L.; Jiang, J.; Yang, T.; Ma, L.; Fu, Z. Lactobacillus and bifidobacterium improves physiological function and cognitive ability in aged mice by the regulation of gut microbiota. Mol. Nutr. Food Res. 2019, 63, e1900603. [Google Scholar] [CrossRef]
- Kukkonen, K.; Savilahti, E.; Haahtela, T.; Juntunen-Backman, K.; Korpela, R.; Poussa, T.; Tuure, T.; Kuitunen, M. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: A randomized, double-blind, placebo-controlled trial. J. Allergy Clin. Immunol. 2007, 119, 192–198. [Google Scholar] [CrossRef]
- Satoh, T.; Murata, M.; Iwabuchi, N.; Odamaki, T.; Wakabayashi, H.; Yamauchi, K.; Abe, F.; Xiao, J.Z. Effect of Bifidobacterium breve B-3 on skin photoaging induced by chronic UV irradiation in mice. Benef. Microbes 2015, 6, 497–504. [Google Scholar] [CrossRef]
- Kano, M.; Masuoka, N.; Kaga, C.; Sugimoto, S.; Iizuka, R.; Manabe, K.; Sone, T.; Oeda, K.; Nonaka, C.; Miyazaki, K.; et al. Consecutive intake of fermented milk containing Bifidobacterium breve strain yakult and galacto-oligosaccharides benefits skin condition in healthy adult women. Biosci. Microbiota. Food Health 2013, 32, 33–39. [Google Scholar] [CrossRef]
- Kankaanpää, P.E.; Yang, B.; Kallio, H.P.; Isolauri, E.; Salminen, S.J. Influence of probiotic supplemented infant formula on composition of plasma lipids in atopic infants. J. Nutr. Biochem. 2002, 13, 364–369. [Google Scholar] [CrossRef]
- Groeger, D.; O’Mahony, L.; Murphy, E.F.; Bourke, J.F.; Dinan, T.G.; Kiely, B.; Shanahan, F.; Quigley, E.M. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes 2013, 4, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Iemoli, E.; Trabattoni, D.; Parisotto, S.; Borgonovo, L.; Toscano, M.; Rizzardini, G.; Clerici, M.; Ricci, E.; Fusi, A.; de Vecchi, E.; et al. Probiotics reduce gut microbial translocation and improve adult atopic dermatitis. J. Clin. Gastroenterol. 2012, 46, S33–S40. [Google Scholar] [CrossRef] [PubMed]
- Navarro-López, V.; Ramírez-Boscá, A.; Ramón-Vidal, D.; Ruzafa-Costas, B.; Genovés-Martínez, S.; Chenoll-Cuadros, E.; Carrión-Gutiérrez, M.; de la Horga Parte, J.; Prieto-Merino, D.; Codoñer-Cortés, F.M. Effect of Oral Administration of a Mixture of Probiotic Strains on SCORAD Index and Use of Topical Steroids in Young Patients with Moderate Atopic Dermatitis: A Randomized Clinical Trial. JAMA Dermatol. 2018, 154, 37–43. [Google Scholar] [CrossRef]
- Allen, S.J.; Jordan, S.; Storey, M.; Thornton, C.A.; Gravenor, M.B.; Garaiova, I.; Plummer, S.F.; Wang, D.; Morgan, G. Probiotics in the prevention of eczema: A randomised controlled trial. Arch. Dis. Child 2014, 99, 1014–1019. [Google Scholar] [CrossRef]
- Kuitunen, M.; Kukkonen, K.; Juntunen-Backman, K.; Korpela, R.; Poussa, T.; Tuure, T.; Haahtela, T.; Savilahti, E. Probiotics prevent IgE-associated allergy until age 5 years in cesarean-delivered children but not in the total cohort. J. Allergy Clin. Immunol. 2009, 123, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Rautava, S.; Arvilommi, H.; Isolauri, E. Specific probiotics in enhancing maturation of IgA responses in formula-fed infants. Pediatr. Res. 2006, 60, 221–224. [Google Scholar] [CrossRef]
- Rautava, S.; Kainonen, E.; Salminen, S.; Isolauri, E. Maternal probiotic supplementation during pregnancy and breast-feeding reduces the risk of eczema in the infant. J. Allergy Clin. Immunol. 2012, 130, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Huurre, A.; Laitinen, K.; Rautava, S.; Korkeamäki, M.; Isolauri, E. Impact of maternal atopy and probiotic supplementation during pregnancy on infant sensitization: A double-blind placebo-controlled study. Clin. Exp. Allergy 2008, 38, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Sistek, D.; Kelly, R.; Wickens, K.; Stanley, T.; Fitzharri, S.P.; Crane, J. Is the effect of probiotics on atopic dermatitis confined to food sensitized children? Clin. Exp. Allergy 2006, 36, 629–633. [Google Scholar] [CrossRef]
- Kulkarni, H.S.; Khoury, C.C. Sepsis associated with Lactobacillus bacteremia in a patient with ischemic colitis. Indian J. Crit. Care Med. 2014, 18, 606–608. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marras, L.; Caputo, M.; Bisicchia, S.; Soato, M.; Bertolino, G.; Vaccaro, S.; Inturri, R. The Role of Bifidobacteria in Predictive and Preventive Medicine: A Focus on Eczema and Hypercholesterolemia. Microorganisms 2021, 9, 836. https://doi.org/10.3390/microorganisms9040836
Marras L, Caputo M, Bisicchia S, Soato M, Bertolino G, Vaccaro S, Inturri R. The Role of Bifidobacteria in Predictive and Preventive Medicine: A Focus on Eczema and Hypercholesterolemia. Microorganisms. 2021; 9(4):836. https://doi.org/10.3390/microorganisms9040836
Chicago/Turabian StyleMarras, Luisa, Michele Caputo, Sonia Bisicchia, Matteo Soato, Giacomo Bertolino, Susanna Vaccaro, and Rosanna Inturri. 2021. "The Role of Bifidobacteria in Predictive and Preventive Medicine: A Focus on Eczema and Hypercholesterolemia" Microorganisms 9, no. 4: 836. https://doi.org/10.3390/microorganisms9040836
APA StyleMarras, L., Caputo, M., Bisicchia, S., Soato, M., Bertolino, G., Vaccaro, S., & Inturri, R. (2021). The Role of Bifidobacteria in Predictive and Preventive Medicine: A Focus on Eczema and Hypercholesterolemia. Microorganisms, 9(4), 836. https://doi.org/10.3390/microorganisms9040836







