Temporal Trend of ST131 Clone among Urinary Escherichia coli Isolates in the Community: A Taiwan National Surveillance from 2002 to 2016
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. Antibiotic Susceptibility Testing (AST) and Data Analysis
2.3. ESBL and AmpC β-Lactamase Gene Detection
2.4. Multiplex PCR for Sequence Type (ST) 131, 69, 73, and 95 Determination
2.5. Pulsed-Field Gel Electrophoresis Analysis (PFGE)
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foxman, B.; Brown, P. Epidemiology of urinary tract infections: Transmission and risk factors, incidence, and costs. Infect. Dis. Clin. N. Am. 2003, 17, 227–241. [Google Scholar] [CrossRef]
- Swami, S.K.; Liesinger, J.T.; Shah, N.; Baddour, L.M.; Banerjee, R. Incidence of Antibiotic-Resistant Escherichia coli Bacteriuria According to Age and Location of Onset: A Population-Based Study from Olmsted County, Minnesota. Mayo Clin. Proc. 2012, 87, 753–759. [Google Scholar] [CrossRef]
- Asensio, A.; Alvarez-Espejo, T.; Fernandez-Crehuet, J.; Ramos, A.; Vaque-Rafart, J.; Bishopberger, C.; Navarrete, M.J.H.; Calbo-Torrecillas, F.; Campayo, J.; Cantón, R.; et al. Trends in yearly prevalence of third-generation cephalosporin and fluoroquinolone resistant Enterobacteriaceae infections and antimicrobial use in Spanish hospitals, Spain, 1999 to 2010. Eurosurveillance 2011, 16, 19983. [Google Scholar] [CrossRef]
- Wang, J.-T.; Chang, S.-C.; Chang, F.-Y.; Fung, C.-P.; Chuang, Y.-C.; Chen, Y.-S.; Shiau, Y.-R.; Tan, M.-C.; Wang, H.-Y.; Lai, J.-F.; et al. Antimicrobial Non-Susceptibility of Escherichia coli from Outpatients and Patients Visiting Emergency Rooms in Taiwan. PLoS ONE 2015, 10, e0144103. [Google Scholar] [CrossRef]
- Collignon, P. Resistant Escherichia coli—We are what we eat. Clin. Infect. Dis. 2009, 49, 202–204. [Google Scholar] [CrossRef]
- Doumith, M.; Day, M.; Ciesielczuk, H.; Hope, R.; Underwood, A.; Reynolds, R.; Wain, J.; Livermore, D.M.; Woodford, N. Rapid Identification of Major Escherichia coli Sequence Types Causing Urinary Tract and Bloodstream Infections. J. Clin. Microbiol. 2015, 53, 160–166. [Google Scholar] [CrossRef]
- Coque, T.M.; Novais, A.; Carattoli, A.; Poirel, L.; Pitout, J.; Peixe, L.; Baquero, F.; Cantón, R.; Nordmann, P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 2008, 14, 195–200. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.-J.; Lavigne, J.-P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2007, 61, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The Epidemic of Extended-Spectrum-β-Lactamase-Producing Escherichia coli ST131 Is Driven by a Single Highly Pathogenic Subclone, H30-Rx. mBio 2013, 4, e00377-13. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Johnson, J.R. A New Clone Sweeps Clean: The Enigmatic Emergence of Escherichia coli Sequence Type 131. Antimicrob. Agents Chemother. 2014, 58, 4997–5004. [Google Scholar] [CrossRef] [PubMed]
- Izdebski, R.; Baraniak, A.; Fiett, J.; Adler, A.; Kazma, M.; Salomon, J.; Lawrence, C.; Rossini, A.; Salvia, A.; Vidal Samso, J.; et al. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 2013, 57, 309–316. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Bertrand, X.; Madec, J.-Y. Escherichia coli ST131, an Intriguing Clonal Group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Day, M.J.; Doumith, M.; Abernethy, J.; Hope, R.; Reynolds, R.; Wain, J.; Livermore, D.M.; Woodford, N. Population structure of Escherichia coli causing bacteraemia in the UK and Ireland between 2001 and 2010. J. Antimicrob. Chemother. 2016, 71, 2139–2142. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kallonen, T.; Brodrick, H.J.; Harris, S.R.; Corander, J.; Brown, N.M.; Martin, V.; Peacock, S.J.; Parkhill, J. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017, 27, 1437–1449. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Fluoroquinolone-Resistant Escherichia coli Sequence Type 131 Isolates Causing Bloodstream Infections in a Canadian Region with a Centralized Laboratory System: Rapid Emergence of the H30-Rx Sublineage. Antimicrob. Agents Chemother. 2014, 58, 2699–2703. [Google Scholar] [CrossRef]
- Kuo, S.C.; Huang, W.C.; Wang, H.Y.; Shiau, Y.R.; Cheng, M.F.; Lauderdale, T.L. Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J. Antimicrob. Chemother. 2016, 71, 2327–2329. [Google Scholar] [CrossRef]
- Wu, C.-J.; Lai, J.-F.; Huang, I.-W.; Hsieh, L.-Y.; Wang, H.-Y.; Shiau, Y.-R.; Lauderdale, T.-L. Multiclonal emergence of levofloxacin-resistant group B Streptococcus, Taiwan. J. Antimicrob. Chemother. 2017, 72, 3263–3271. [Google Scholar] [CrossRef] [PubMed]
- CLSI. CLSI supplement M100. In Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. CLSI supplement M100. In Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- O’Brien, T.F.; Stelling, J. Integrated multilevel surveillance of the world’s infecting microbes and their resistance to antimicrobial agents. Clin. Microbiol. Rev. 2011, 24, 281–295. [Google Scholar] [CrossRef][Green Version]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Hunter, S.B.; Vauterin, P.; Lambert-Fair, M.A.; van Duyne, M.S.; Kubota, K.; Graves, L.; Wrigley, D.; Barrett, T.; Ribot, E. Establishment of a Universal Size Standard Strain for Use with the PulseNet Standardized Pulsed-Field Gel Electrophoresis Protocols: Converting the National Databases to the New Size Standard. J. Clin. Microbiol. 2005, 43, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.F.; Chen, W.L.; Hung, W.Y.; Huang, I.F.; Chiou, Y.H.; Chen, Y.S.; Lee, S.S.; Hung, C.H.; Wang, J.L. Emergence of extended spectrum-β-lactamase-producing Escherichia coli O25b-ST131: A major community-acquired uropathogen in infants. Pediatr. Infect. Dis. J. 2015, 34, 469–475. [Google Scholar] [CrossRef]
- Tsai, W.-L.; Hung, C.-H.; Chen, H.-A.; Wang, J.-L.; Huang, I.-F.; Chiou, Y.-H.; Chen, Y.-S.; Lee, S.S.-J.; Hung, W.-Y.; Cheng, M.-F. Extended-spectrum β-lactamase-producing Escherichia coli bacteremia: Comparison of pediatric and adult populations. J. Microbiol. Immunol. Infect. 2018, 51, 723–731. [Google Scholar] [CrossRef]
- Wang, J.-L.; Lee, C.-C.; Lee, C.-H.; Lee, N.-Y.; Hsieh, C.-C.; Hung, Y.-P.; Tang, H.-J.; Ko, W.-C. Clinical Impact of Sequence Type 131 in Adults with Community-Onset Monomicrobial Escherichia coli Bacteremia. J. Clin. Med. 2018, 7, 508. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-H.; Cheng, M.-F.; Lai, C.-H.; Lin, H.-H.; Hung, C.-H.; Wang, J.-L. The role of Sequence Type (ST) 131 in adult community-onset non-ESBL-producing Escherichia coli bacteraemia. BMC Infect. Dis. 2014, 14, 579. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chung, H.-C.; Lai, C.-H.; Lin, J.-N.; Huang, C.-K.; Liang, S.-H.; Chen, W.-F.; Shih, Y.-C.; Lin, H.-H.; Wang, J.-L. Bacteremia Caused by Extended-Spectrum-β-Lactamase-Producing Escherichia coli Sequence Type ST131 and Non-ST131 Clones: Comparison of Demographic Data, Clinical Features, and Mortality. Antimicrob. Agents Chemother. 2011, 56, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Olesen, B.; Frimodt-Møller, J.; Leihof, R.F.; Struve, C.; Johnston, B.; Hansen, D.S.; Scheutz, F.; Krogfelt, K.A.; Kuskowski, M.A.; Clabots, C.; et al. Temporal Trends in Antimicrobial Resistance and Virulence-Associated Traits within the Escherichia coli Sequence Type 131 Clonal Group and Its H30 and H30-Rx Subclones, 1968–2012. Antimicrob. Agents Chemother. 2014, 58, 6886–6895. [Google Scholar] [CrossRef]
- Banerjee, R.; Johnston, B.; Lohse, C.; Porter, S.B.; Clabots, C.; Johnson, J.R. Escherichia coli Sequence Type 131 Is a Dominant, Antimicrobial-Resistant Clonal Group Associated with Healthcare and Elderly Hosts. Infect. Control. Hosp. Epidemiol. 2013, 34, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Lynch, T.; Matsumara, Y.; Nobrega, D.; Finn, T.J.; deVinney, R.; Pitout, J.D. Trends in Population Dynamics of Escherichia coli Sequence Type 131, Calgary, Alberta, Canada, 2006–2016. Emerg. Infect. Dis. 2020, 26, 2907–2915. [Google Scholar] [CrossRef]
- Johnson, J.R.; Porter, S.; Thuras, P.; Castanheira, M. Epidemic Emergence in the United States of Escherichia coli Sequence Type 131-H30 (ST131-H30), 2000–2009. Antimicrob. Agents Chemother. 2017, 61, 61. [Google Scholar] [CrossRef]
- Brodrick, H.J.; Raven, K.E.; Kallonen, T.; Jamrozy, D.; Blane, B.; Brown, N.M.; Martin, V.; Török, M.E.; Parkhill, J.; Peacock, S.J. Longitudinal genomic surveillance of multidrug-resistant Escherichia coli carriage in a long-term care facility in the United Kingdom. Genome Med. 2017, 9, 70. [Google Scholar] [CrossRef]
- Ludden, C.; Cormican, M.; Vellinga, A.; Johnson, J.R.; Austin, B.; Morris, D. Colonisation with ESBL-producing and carbapenemase-producing Enterobacteriaceae, vancomycin-resistant enterococci, and meticillin-resistant Staphylococcus aureus in a long-term care facility over one year. BMC Infect. Dis. 2015, 15, 168. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Chu, Y.P.-S.; Lo, W.-U.; Chow, K.-H.; Law, P.Y.; Tse, C.W.-S.; Ng, T.-K.; Cheng, V.C.-C.; Que, T.-L. High prevalence of Escherichia coli sequence type 131 among antimicrobial-resistant E. coli isolates from geriatric patients. J. Med Microbiol. 2015, 64, 243–247. [Google Scholar] [CrossRef][Green Version]
- Burgess, M.J.; Johnson, J.R.; Porter, S.B.; Johnston, B.; Clabots, C.; Lahr, B.D.; Uhl, J.R.; Banerjee, R. Long-Term Care Facilities Are Reservoirs for Antimicrobial-Resistant Sequence Type 131 Escherichia coli. Open Forum Infect. Dis. 2015, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S.-C.; Chen, P.-C.; Shiau, Y.-R.; Wang, H.-Y.; Lai, J.-F.; Huang, W.; Lauderdale, T.-L.Y. Levofloxacin-Resistant Haemophilus influenzae, Taiwan, 2004–2010. Emerg. Infect. Dis. 2014, 20, 1386–1390. [Google Scholar] [CrossRef]
- Kuo, S.-C.; Shih, S.-M.; Hsieh, L.-Y.; Lauderdale, T.-L.Y.; Chen, Y.-C.; Hsiung, C.A.; Chang, S.-C. Antibiotic restriction policy paradoxically increased private drug consumptions outside Taiwan’s National Health Insurance. J. Antimicrob. Chemother. 2017, 72, 1544–1545. [Google Scholar] [CrossRef] [PubMed]
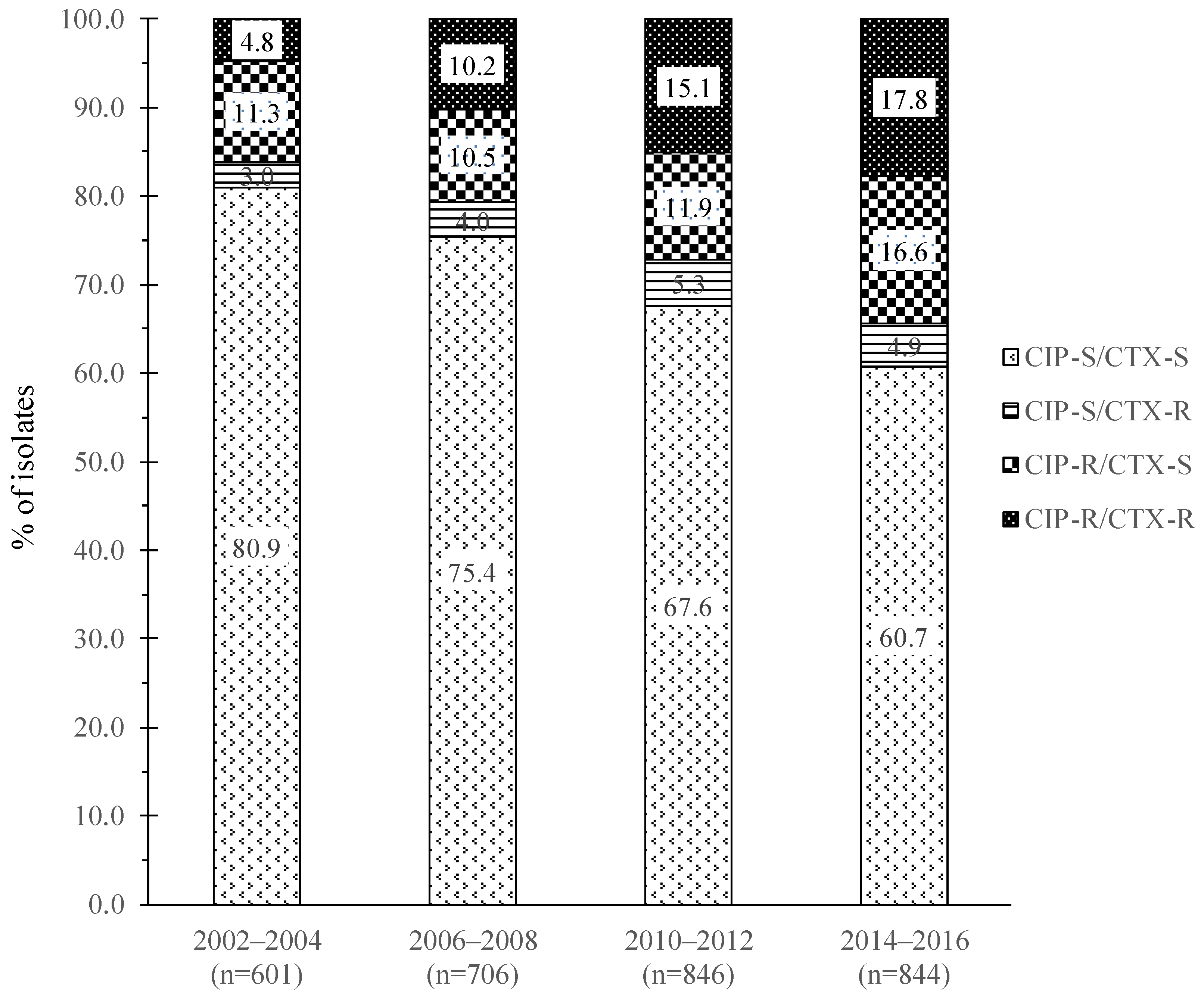
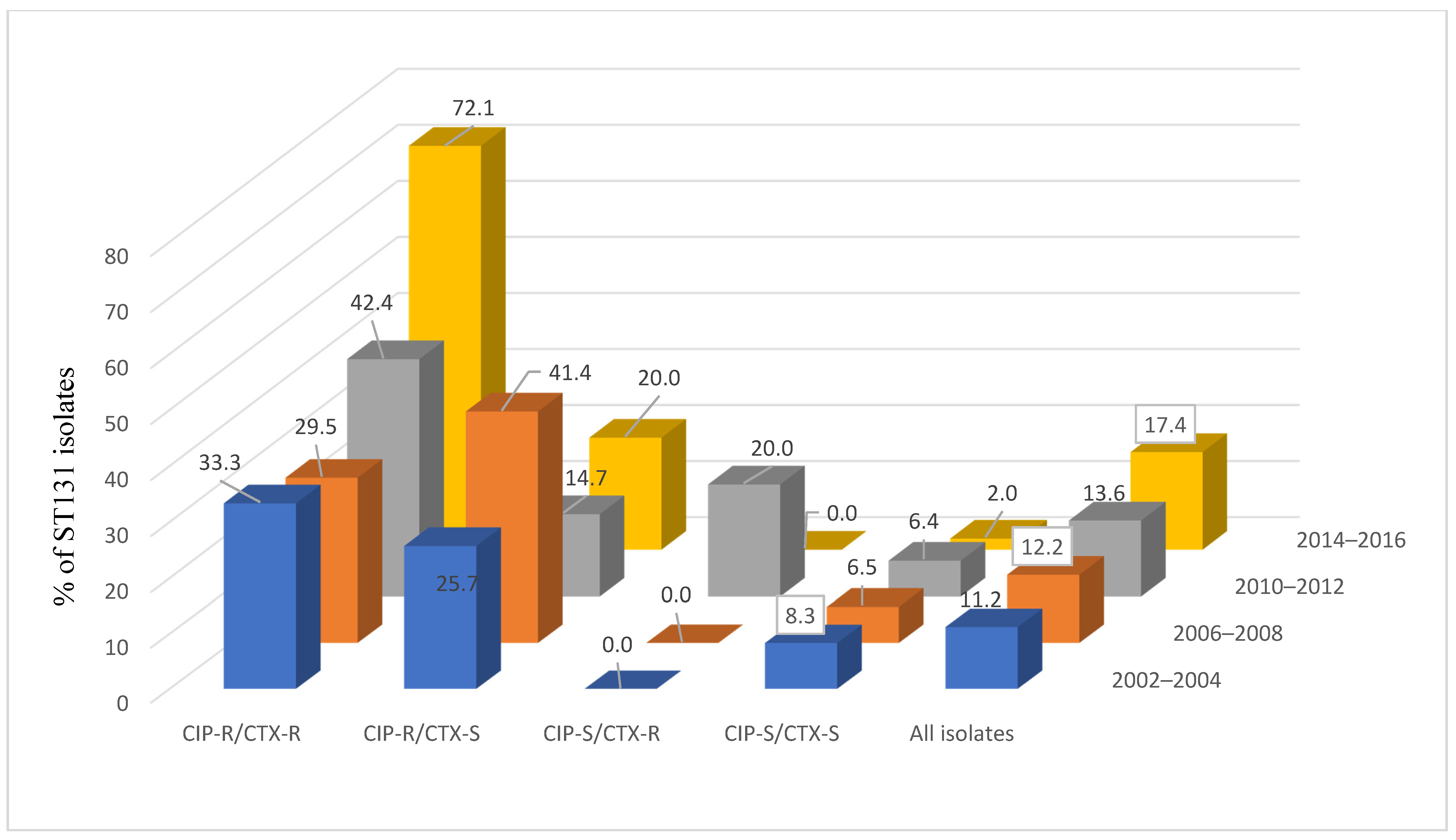
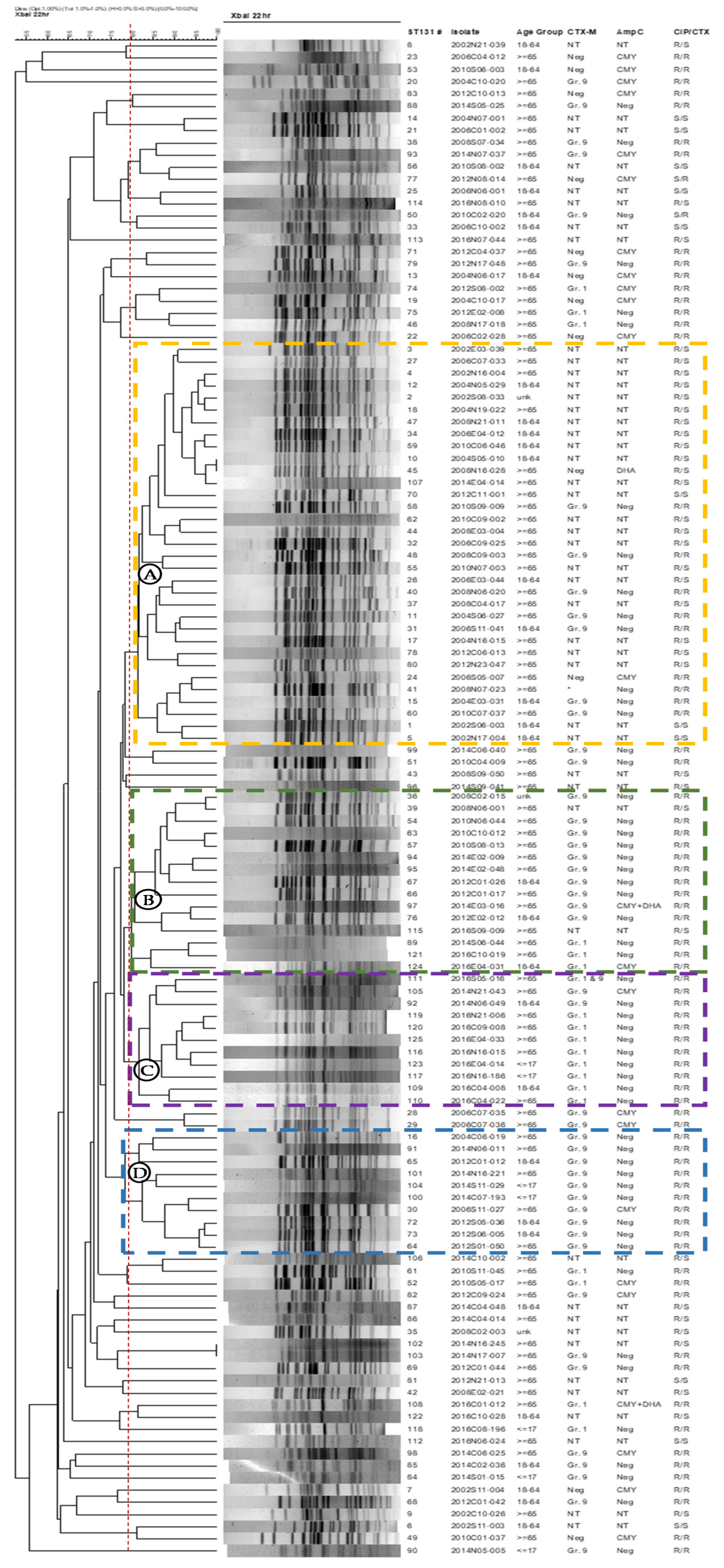
| Antimicrobial Agent b | CIP-R/CTX-R (N = 380) | CIP-R/CTX-S (N = 383) | CIP-S/CTX-R (N = 130) | CIP-S/CTX-S (N = 2104) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| %S | %I | %R | %S | %I | %R | %S | %I | %R | %S | %I | %R | |
| Amikacin | 93.9 | 1.6 | 4.5 | 99.7 | 0.3 | 0 | 96.1 | 0.8 | 3.1 | 100 | 0 | 0 |
| Ampicillin | 0 | 0 | 100 | 14.6 | 0.5 | 84.9 | 0 | 0 | 100 | 39.9 | 0.2 | 59.9 |
| Aztreonam | 16.1 | 22.6 | 61.3 | 99.7 | 0 | 0.3 | 29.2 | 33.1 | 37.7 | 100 | 0 | 0 |
| Cefazolin–UTI | 0.3 | - | 99.7 | 92.3 | - | 7.7 | 2.3 | - | 97.7 | 96.4 | - | 3.6 |
| Cefepime | 39.7 | 15.3 | 45.0 | 100 | 0 | 0 | 64.6 | 10.8 | 24.6 | 100 | 0 | 0 |
| Cefotaxime | 0 | 0.8 | 99.2 | 100 | 0 | 0 | 0 | 3.8 | 96.2 | 100 | 0 | 0 |
| Cefoxitin | 30.3 | 10.8 | 58.9 | 78.6 | 16.4 | 5.0 | 30.0 | 3.8 | 66.2 | 97.4 | 1.9 | 0.7 |
| Ceftazidime | 27.6 | 9.5 | 62.9 | 100 | 0 | 0 | 24.6 | 16.9 | 58.5 | 99.9 | 0.1 | 0 |
| Cefuroxime | 0.5 | 2.1 | 97.4 | 85.1 | 12.3 | 2.6 | 1.5 | 3.9 | 94.6 | 97.3 | 2.3 | 0.4 |
| Ciprofloxacin | 0 | 0 | 100 | 0 | 0 | 100 | 80.8 | 11.5 | 7.7 | 92.5 | 4.8 | 2.7 |
| Ciprofloxacin (2018) | 0 | 1.6 | 98.4 | 0 | 2.3 | 97.7 | 100 | 0 | 0 | 100 | 0 | 0 |
| Gentamicin | 38.9 | 1.1 | 60.0 | 53.3 | 0.8 | 46.0 | 62.3 | 3.1 | 34.6 | 86.4 | 2.0 | 11.6 |
| Imipenem | 98.4 | 1.1 | 0.5 | 100 | 0 | 0 | 99.2 | 0.8 | 0 | 100 | 0 | 0 |
| Trimethoprim/sulfa. | 30.0 | - | 70.0 | 36.6 | 0 | 63.4 | 40.0 | - | 60.0 | 58.3 | - | 41.7 |
| Antimicrobial Agent a | ST131 (N = 125) | ST69 (N = 14) | ST73 (N = 21) | ST95 (N = 58) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| %S | %I | %R | %S | %I | %R | %S | %I | %R | %S | %I | %R | |
| Amikacin | 96.0 | 2.4 | 1.6 | 100 | 0 | 0 | 100 | 0 | 0 | 98.3 | 0 | 1.7 |
| Ampicillin | 3.2 | 0 | 96.8 | 14.3 | 0 | 85.7 | 52.4 | 0 | 47.6 | 36.2 | 0 | 63.8 |
| Aztreonam | 50.4 | 16.0 | 33.6 | 85.7 | 14.3 | 0 | 100 | 0 | 0 | 93.1 | 3.4 | 3.4 |
| Cefazolin–UTI | 33.6 | - | 66.4 | 85.7 | 0 | 14.3 | 100 | - | 0 | 87.9 | - | 12.1 |
| Cefepime | 48.8 | 14.4 | 36.8 | 85.7 | 0 | 14.3 | 100 | 0 | 0 | 94.8 | 0 | 5.2 |
| Cefotaxime | 37.6 | 0 | 62.4 | 85.7 | 0 | 14.3 | 100 | 0 | 0 | 89.7 | 0 | 10.3 |
| Cefoxitin | 65.6 | 10.4 | 24.0 | 92.9 | 7.1 | 0.0 | 100 | 0 | 0 | 93.1 | 1.7 | 5.2 |
| Ceftazidime | 64.0 | 5.6 | 30.4 | 100 | 0 | 0 | 100 | 0 | 0 | 93.1 | 1.7 | 5.2 |
| Cefuroxime | 34.4 | 3.2 | 62.4 | 78.6 | 7.1 | 14.3 | 100 | 0 | 0 | 89.7 | 0 | 10.3 |
| Ciprofloxacin | 9.6 | 0 | 90.4 | 64.3 | 14.3 | 21.4 | 100 | 0 | 0 | 91.4 | 0 | 8.6 |
| Ciprofloxacin (2018) | 10.4 | 0 | 89.6 | 78.6 | 0 | 21.4 | 100 | 0 | 0 | 91.4 | 0 | 8.6 |
| Gentamicin | 48.0 | 0 | 52.0 | 64.3 | 0 | 35.7 | 90.5 | - | 9.5 | 87.9 | 0 | 12.1 |
| Trimethoprim/Sulfa. | 40.8 | - | 59.2 | 28.6 | - | 71.4 | 85.7 | - | 14.3 | 53.4 | - | 46.6 |
| Strata | ST131 | Non-ST131 | pa | OR b | 95%CI b | pb |
|---|---|---|---|---|---|---|
| Number of isolates | 125 | 417 | ||||
| Geographic region | NS | |||||
| Central | 47 (37.6) | 129 (30.9) | ||||
| Eastern | 15 (12.0) | 45 (10.8) | ||||
| Northern | 35 (28.0) | 115 (27.6) | ||||
| Southern | 28 (22.4) | 128 (30.7) | ||||
| Age c | 0.001 | |||||
| 18–64 | 31 (24.8) | 185 (44.4) | Reference | |||
| > = 65 | 84 (51.3) | 194 (46.5) | 1.739 | 1.058–2.858 | 0.029 | |
| < = 17 | 7 (5.6) | 29 (7.0) | 1.139 | 0.368–7.652 | NS | |
| Ciprofloxacin | ||||||
| Susceptible | 13 (10.4) | 214 (51.3) | <0.001 | |||
| Resistant | 112 (89.6) | 203 (48.7) | 26.769 | 3.458–207.191 | 0.002 | |
| Cefotaxime | <0.001 | |||||
| Susceptible | 47 (37.6) | 292 (70.0) | ||||
| Resistant | 78 (62.4) | 125 (30.0) | 1.217 | 0.556–2.664 | 0.622 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.-L.; Ko, W.-C.; Hung, C.-H.; Cheng, M.-F.; Wang, H.-Y.; Shiau, Y.-R.; Lai, J.-F.; Huang, I.-W.; Hsieh, L.-Y.; Lauderdale, T.-L.Y.; et al. Temporal Trend of ST131 Clone among Urinary Escherichia coli Isolates in the Community: A Taiwan National Surveillance from 2002 to 2016. Microorganisms 2021, 9, 963. https://doi.org/10.3390/microorganisms9050963
Wang J-L, Ko W-C, Hung C-H, Cheng M-F, Wang H-Y, Shiau Y-R, Lai J-F, Huang I-W, Hsieh L-Y, Lauderdale T-LY, et al. Temporal Trend of ST131 Clone among Urinary Escherichia coli Isolates in the Community: A Taiwan National Surveillance from 2002 to 2016. Microorganisms. 2021; 9(5):963. https://doi.org/10.3390/microorganisms9050963
Chicago/Turabian StyleWang, Jiun-Ling, Wen-Chien Ko, Chih-Hsin Hung, Ming-Fang Cheng, Hui-Ying Wang, Yih-Ru Shiau, Jui-Fen Lai, I-Wen Huang, Li-Yun Hsieh, Tsai-Ling Yang Lauderdale, and et al. 2021. "Temporal Trend of ST131 Clone among Urinary Escherichia coli Isolates in the Community: A Taiwan National Surveillance from 2002 to 2016" Microorganisms 9, no. 5: 963. https://doi.org/10.3390/microorganisms9050963
APA StyleWang, J.-L., Ko, W.-C., Hung, C.-H., Cheng, M.-F., Wang, H.-Y., Shiau, Y.-R., Lai, J.-F., Huang, I.-W., Hsieh, L.-Y., Lauderdale, T.-L. Y., & on behalf of TSAR Hospitals. (2021). Temporal Trend of ST131 Clone among Urinary Escherichia coli Isolates in the Community: A Taiwan National Surveillance from 2002 to 2016. Microorganisms, 9(5), 963. https://doi.org/10.3390/microorganisms9050963






