Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals
Abstract
:1. Pseudomonas syringae Is a Type III Secretion System-Producing Plant Pathogen
1.1. The P. syringae Type III Secretion System (T3SS) and Its Role in Virulence
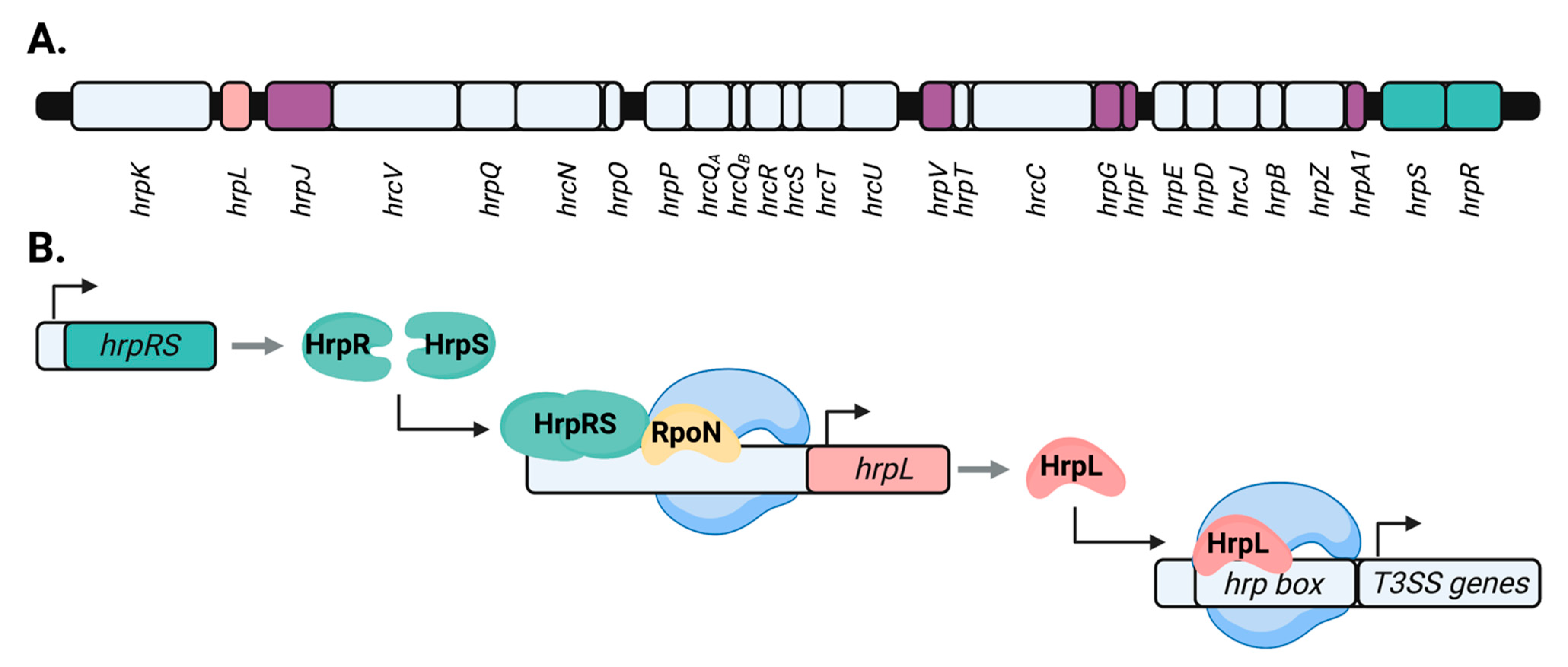
1.2. Transcriptional Control of P. syringae T3SS Genes by hrp/hrc Regulators
1.3. Post-Translational Control of the P. syringae T3SS by hrp/hrc Regulators
2. Environmental Regulation of the P. syringae T3SS
2.1. Induction of the P. syringae T3SS in Response to Synthetic Media Conditions
2.2. Induction of the P. syringae T3SS by Specific Plant-Derived Signals
2.3. Negative Regulation of T3SS Gene Expression by Plant-Derived Compounds
2.4. Dynamics of T3SS Deployment within the Host Plant Environment
3. Molecular Mechanisms of T3SS Deployment in Response to Environmental Stimuli
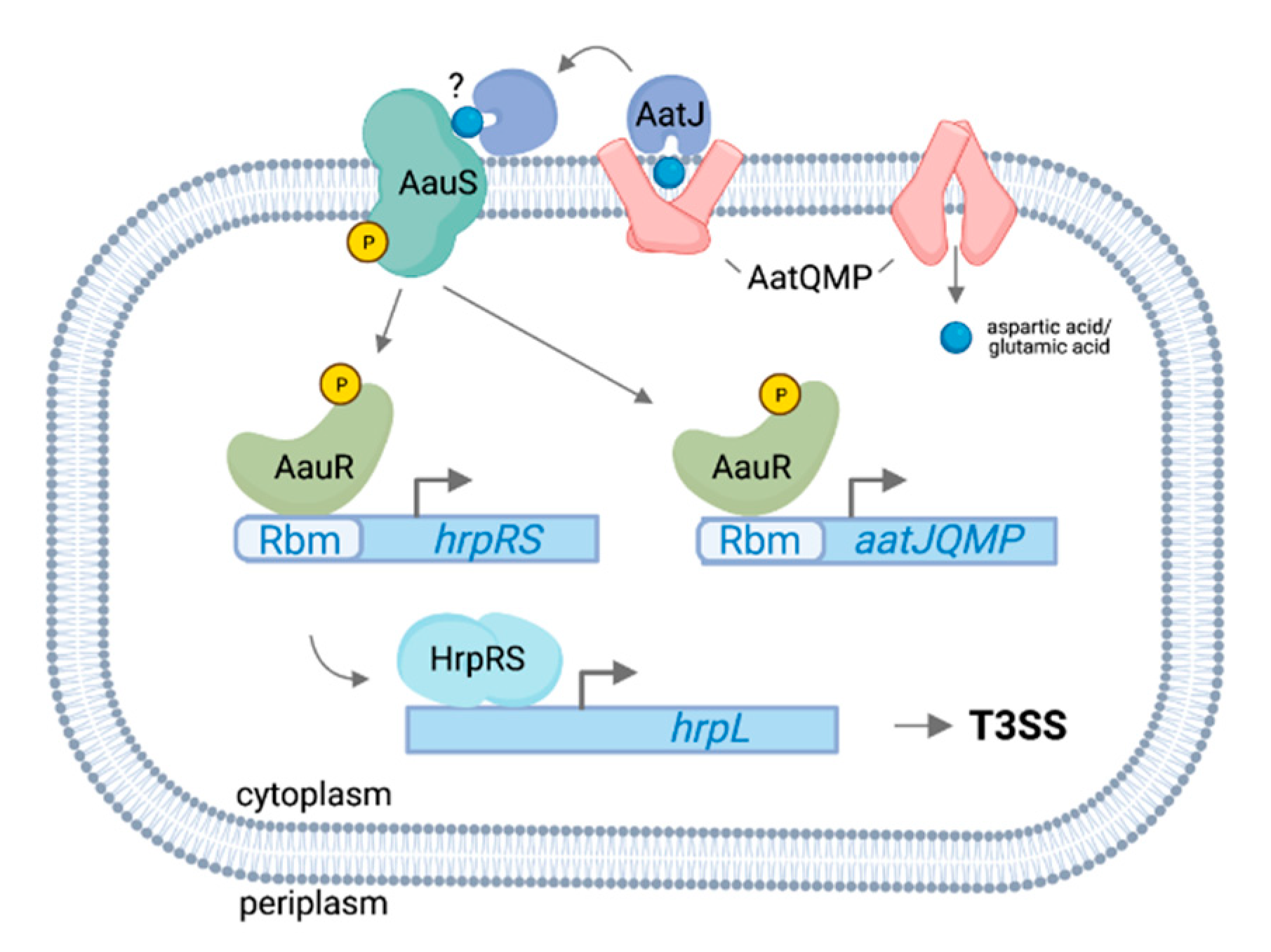
3.1. AauS-AauR Directly Regulates hrpRS Expression in P. syringae in Response to Specific Host-Derived Amino Acids
3.2. The DeoR-Type Regulator SetA Is Necessary for hrpL Expression in Response to Sugar Signals
3.3. Negative Regulation of the T3SS by the GacSA Global Regulatory System
3.4. Negative Regulation of the T3SS by the RhpSR Two-Component System
3.5. Regulation of the T3SS by the CvsSR Two-Component System
4. Additional Global Regulators of the T3SS in P. syringae
4.1. Dual Positive and Negative Regulation of the T3SS by Bifunctional Lon Protease
4.2. Regulation of the T3SS by Alginate Master Regulator AlgU
4.3. Regulation of the T3SS by AHL Quorum Sensing Regulator AefR
4.4. Modulation of T3SS Gene Expression by RsmA RNA-Binding Proteins
4.5. Nucleotide Second Messengers in T3SS Regulation
5. Conclusions and Future Directions
- P. syringae rapidly deploys the T3SS during the initial hours of plant host infection, and primarily relies on the T3SS to establish growth within the host apoplast. T3SS deployment may be negatively regulated in later stages of infection, possibly as a means to maximize fitness in the host environment.
- T3SS expression by P. syringae is induced by specific organic acids and amino acids that are abundant in the plant environment. These T3SS-inducing plant metabolites require the presence of a simple sugar such as fructose for maximal bioactivity, suggesting P. syringae coordinates T3SS deployment by sensing multiple distinct host signals.
- The abundance of T3SS-inducing metabolites in the host environment is genetically regulated by the plant host and significantly impacts the progression of P. syringae disease, as evidenced by the enhanced disease resistance phenotypes of the mkp1 mutant of Arabidopsis.
- AauSR, a two-component system in P. syringae associated with the uptake of acidic amino acids by the ABC transporter AatQMP, directly regulates the expression of T3SS genes in response to host-derived aspartic acid and glutamic acid signals. Additional proteins including RhpSR, CvsSR and SetA may also function as sensors for host signals leading to T3SS regulation. However, the specific signal(s) these proteins detect and the means by which these various response pathways interact to synergistically regulate the T3SS are unknown.
- Do all P. syringae detect and respond to the same T3SS-inducing metabolites? Molecular studies of T3SS induction have so far been limited to a small number of P. syringae strains. Broadening future analyses to include additional strains that represent the diversity of the P. syringae species complex will be necessary to fully evaluate the conservation of host-perception mechanisms.
- How many distinct host signals does a single strain of P. syringae respond to? Mutants lacking putative host signal receptors (e.g., AauSR, SetA) are only partially attenuated in virulence, suggesting that multiple input signals may additively or synergistically contribute to T3SS induction in P. syringae.
- Multiple regulatory systems in P. syringae are known to influence T3SS dynamics—how do these distinct pathways intersect and converge on T3SS regulation? A newly published analysis of gene regulatory networks in P. syringae (termed PSRnet) indicates that complex crosstalk occurs between multiple known virulence regulators [198].
- At what point(s) during P. syringae infection is T3SS deployment repressed, and why? While evidence suggests that T3SS expression may be downregulated in P. syringae on the leaf surface and/or during later stages of apoplast infection, the mechanisms responsible for T3SS repression during host infection are not fully understood. Moreover, the role T3SS repression may play in P. syringae virulence and life cycle remains ambiguous. Further elucidation of how the T3SS is co-regulated with other virulence-related processes in P. syringae, as well as the impact of T3SS deployment on bacterial cell homeostasis and growth, may help to elucidate the potential benefits of T3SS repression.
- How is deployment of the T3SS spatially regulated within the host environment? Additionally, how is T3SS deployment regulated on the population level during P. syringae infection? Experiments to date suggest that only a subpopulation of P. syringae cells may deploy their T3SS during infection. Is this heterogeneity in part due to variation in the abundance of inducing signals within plant tissues? Continued development of transcriptomic methods for profiling gene expression by P. syringae within the host environment, such as single cell RNA-seq, as well as fluorescence-based reporters for in planta detection of T3SS expression, will be necessary to fully address these questions.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Young, J.M. Taxonomy of Pseudomonas syringae. J. Plant Pathol. 2010, 92, S1.5–S1.14. [Google Scholar] [CrossRef]
- Jun, S.R.; Wassenaar, T.M.; Nookaew, I.; Hauser, L.; Wanchai, V.; Land, M.; Timm, C.M.; Lu, T.Y.; Schadt, C.W.; Doktycz, M.J.; et al. Diversity of Pseudomonas genomes, including populus-associated isolates, as revealed by comparative genome analysis. Appl. Environ. Microbiol. 2016, 82, 375–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamichhane, J.R.; Varvaro, L.; Parisi, L.; Audergon, J.-M.; Morris, C.E. Chapter Four—Disease and frost damage of woody plants caused by Pseudomonas syringae: Seeing the forest for the trees. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 126, pp. 235–295. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Messéan, A.; Morris, C.E. insights into epidemiology and control of diseases of annual plants caused by the Pseudomonas syringae species complex. J. Gen. Plant Pathol. 2015, 81, 331–350. [Google Scholar] [CrossRef]
- Sarkar, S.F.; Guttman, D.S. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 2004, 70, 1999–2012. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, H.E.; Thakur, S.; Guttman, D.S. Evolution of plant pathogenesis in Pseudomonas syringae: A genomics perspective. Annu. Rev. Phytopathol. 2011, 49, 269–289. [Google Scholar] [CrossRef]
- Morris, C.E.; Sands, D.C.; Vinatzer, B.A.; Glaux, C.; Guilbaud, C.; Buffière, A.; Yan, S.; Dominguez, H.; Thompson, B.M. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008, 2, 321–334. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.A.X.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [Green Version]
- Hirano, S.S.; Upper, C.D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—A pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000, 64, 624–653. [Google Scholar] [CrossRef] [Green Version]
- Hirano, S.S.; Upper, C.D. Population biology and epidemiology of Pseudomonas syringae. Annu. Rev. Phytopathol. 1990, 28, 155–177. [Google Scholar] [CrossRef]
- Xin, X.-F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef]
- Lindgren, P.B.; Peet, R.C.; Panopoulos, N.J. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J. Bacteriol. 1986, 168, 512–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.H.; Desveaux, D.; Creason, A.L. The ABCs and 123s of bacterial secretion systems in plant pathogenesis. Annu. Rev. Phytopathol. 2014, 52, 317–345. [Google Scholar] [CrossRef] [PubMed]
- Roine, E.; Wei, W.; Yuan, J.; Nurmiaho-Lassila, E.L.; Kalkkinen, N.; Romantschuk, M.; He, S.Y. Hrp pilus: An hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 1997, 94, 3459–3464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubori, T.; Matsushima, Y.; Nakamura, D.; Uralil, J.; Lara-Tejero, M.; Sukhan, A.; Galán, J.E.; Aizawa, S.I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 1998, 280, 602–605. [Google Scholar] [CrossRef] [Green Version]
- Büttner, D.; He, S.Y. Type III Protein secretion in plant pathogenic bacteria. Plant Physiol. 2009, 150, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Petnicki-Ocwieja, T.; Schneider, D.J.; Tam, V.C.; Chancey, S.T.; Shan, L.; Jamir, Y.; Schechter, L.M.; Janes, M.D.; Buell, C.R.; Tang, X.; et al. Genomewide identification of proteins secreted by the hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2002, 99, 7652–7657. [Google Scholar] [CrossRef] [Green Version]
- Guttman, D.S.; Vinatzer, B.A.; Sarkar, S.F.; Ranall, M.V.; Kettler, G.; Greenberg, J.T. A functional screen for the type III (hrp) secretome of the plant pathogen Pseudomonas syringae. Science 2002, 295, 1722–1726. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.H.; Urbach, J.M.; Law, T.F.; Arnold, L.W.; Hu, A.; Gombar, S.; Grant, S.R.; Ausubel, F.M.; Dangl, J.L. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 2005, 102, 2549–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindeberg, M.; Cartinhour, S.; Myers, C.R.; Schechter, L.M.; Schneider, D.J.; Collmer, A. Closing the circle on the discovery of genes encoding hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 2006, 19, 1151–1158. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.; He, P.; Li, J.; Heese, A.; Peck, S.C.; Nürnberger, T.; Martin, G.B.; Sheen, J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 2008, 4, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Göhre, V.; Spallek, T.; Häweker, H.; Mersmann, S.; Mentzel, T.; Boller, T.; de Torres, M.; Mansfield, J.W.; Robatzek, S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 2008, 18, 1824–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, L.; Velásquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012, 69, 92–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J.; et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eschen-Lippold, L.; Jiang, X.; Elmore, J.M.; Mackey, D.; Shan, L.; Coaker, G.; Scheel, D.; Lee, J. Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol. 2016, 171, 2223–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Block, A.; Guo, M.; Li, G.; Elowsky, C.; Clemente, T.E.; Alfano, J.R. The Pseudomonas Syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol. 2010, 12, 318–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Herva, J.J.; González-Melendi, P.; Cuartas-Lanza, R.; Antúnez-Lamas, M.; Río-Alvarez, I.; Li, Z.; López-Torrejón, G.; Díaz, I.; Del Pozo, J.C.; Chakravarthy, S.; et al. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 2012, 14, 669–681. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Bourdais, G.; Yu, G.; Robatzek, S.; Coaker, G. Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 2015, 27, 2042–2056. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.M. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas Syringae effector protein AvrB for stomatal invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Teixeira, P.J.; Biswas, S.; Finkel, O.M.; He, Y.; Salas-Gonzalez, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J.L. Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 2017, 21, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Xin, X.F.; Nomura, K.; Aung, K.; Velásquez, A.C.; Yao, J.; Boutrot, F.; Chang, J.H.; Zipfel, C.; He, S.Y. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 2016, 539, 524–529. [Google Scholar] [CrossRef] [Green Version]
- Collmer, A.; Badel, J.L.; Charkowski, A.O.; Deng, W.L.; Fouts, D.E.; Ramos, A.R.; Rehm, A.H.; Anderson, D.M.; Schneewind, O.; van Dijk, K.; et al. Pseudomonas syringae hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 8770–8777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfano, J.R.; Charkowski, A.O.; Deng, W.L.; Badel, J.L.; Petnicki-Ocwieja, T.; van Dijk, K.; Collmer, A. The Pseudomonas syringae hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in pl. Proc. Natl. Acad. Sci. USA 2000, 97, 4856–4861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hueck, C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998, 62, 379–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston, G.; Deng, W.-L.; Huang, H.-C.; Collmer, A. negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 1998, 180, 4532–4537. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.-L.; Preston, G.; Collmer, A.; Chang, C.-J.; Huang, H.-C. Characterization of the HrpC and HrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of HrpF, HrpG, HrcC, HrpT, and HrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 1998, 180, 4523–4531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-C.; Hsu, S.-T.; Huang, H.-C. Evidence for the interaction between HrpT and HrcC of Pseudomonas syringae pv. syringae 61. Plant Pathol. Bull. 2006, 15, 171–185. [Google Scholar] [CrossRef]
- Clarke, C.R.; Cai, R.; Studholme, D.J.; Guttman, D.S.; Vinatzer, B.A. Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol. Plant Microbe Interact. 2010, 23, 198–210. [Google Scholar] [CrossRef] [Green Version]
- Leach, J.E.; White, F.F. Bacterial avirulence genes. Annu. Rev. Phytopathol. 1996, 34, 153–179. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Keen, N.T. Characterization of the promoter of avirulence gene D from Pseudomonas syringae pv. tomato. J. Bacteriol. 1993, 175, 5916–5924. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Lu, Y.; Heu, S.; Hutcheson, S.W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J. Bacteriol. 1992, 174, 1734–1741. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Hutcheson, S.W. A Single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 1994, 176, 3089–3091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouts, D.E.; Abramovitch, R.B.; Alfano, J.R.; Baldo, A.M.; Buell, C.R.; Cartinhour, S.; Chatterjee, A.K.; D’Ascenzo, M.; Gwinn, M.L.; Lazarowitz, S.G.; et al. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 2002, 99, 2275–2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwiesler-Vollick, J.; Plovanich-Jones, A.E.; Nomura, K.; Bandyopadhyay, S.; Joardar, V.; Kunkel, B.N.; He, S.Y. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 2002, 45, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Losada, L.; Sussan, T.; Pak, K.; Zeyad, S.; Rozenbaum, I.; Hutcheson, S.W. Identification of a novel Pseudomonas syringae Psy61 effector with virulence and avirulence functions by a HrpL-dependent promoter-trap assay. Mol. Plant Microbe Interact. 2004, 17, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Schechter, L.M.; Vencato, M.; Jordan, K.L.; Schneider, S.E.; Schneider, D.J.; Collmer, A. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 2006, 19, 1180–1192. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, A.O.; Myers, C.R.; Gordon, J.S.; Martin, G.B.; Vencato, M.; Collmer, A.; Wehling, M.D.; Alfano, J.R.; Moreno-Hagelsieb, G.; Lamboy, W.F.; et al. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the hrp cis clement, and identifies novel coregulated genes. Mol. Plant Microbe Interact. 2006, 19, 1167–1179. [Google Scholar] [CrossRef] [Green Version]
- Lam, H.N.; Chakravarthy, S.; Wei, H.-L.; Buinguyen, H.; Stodghill, P.V.; Collmer, A.; Swingle, B.M.; Cartinhour, S.W. Global analysis of the HrpL regulon in the plant pathogen Pseudomonas syringae pv. tomato DC3000 reveals new regulon members with diverse functions. PLoS ONE 2014, 9, e106115. [Google Scholar] [CrossRef] [Green Version]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; Deboy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar] [CrossRef] [Green Version]
- Feil, H.; Feil, W.S.; Chain, P.; Larimer, F.; Dibartolo, G.; Copeland, A.; Lykidis, A.; Trong, S.; Nolan, M.; Goltsman, E.; et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2005, 102, 11064–11069. [Google Scholar] [CrossRef] [Green Version]
- Joardar, V.; Lindeberg, M.; Jackson, R.W.; Selengut, J.; Dodson, R.; Brinkac, L.M.; Daugherty, S.C.; Deboy, R.; Durkin, A.S.; Giglio, M.G.; et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 2005, 187, 6488–6498. [Google Scholar] [CrossRef] [Green Version]
- Thwaites, R.; Spanu, P.D.; Panopoulos, N.J.; Stevens, C.; Mansfield, J.W. Transcriptional regulation of components of the type III secretion system and effectors in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 2004, 17, 1250–1258. [Google Scholar] [CrossRef]
- Waite, C.; Schumacher, J.; Jovanovic, M.; Bennett, M.; Buck, M. Negative autogenous control of the master type III secretion system regulator HrpL in Pseudomonas syringae. MBio 2017. [Google Scholar] [CrossRef] [Green Version]
- Hendrickson, E.L.; Guevera, P.; Ausubel, F.M. The alternative sigma factor RpoN is required for hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J. Bacteriol. 2000, 182, 3508–3516. [Google Scholar] [CrossRef] [Green Version]
- Hutcheson, S.W.; Bretz, J.; Sussan, T.; Jin, S.; Pak, K. Enhancer-binding proteins HrpR and HrpS Interact to regulate hrp-encoded type III protein secretion in Pseudomonas syringae strains. J. Bacteriol. 2001, 183, 5589–5598. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Martín, I.; Thwaites, R.; Macho, A.P.; Mansfield, J.W.; Beuzón, C.R. Positive regulation of the hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 2010, 23, 665–681. [Google Scholar] [CrossRef] [Green Version]
- Grimm, C.; Aufsatz, W.; Panopoulos, N.J. The HrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol. Microbiol. 1995, 15, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; James, E.H.; Burrows, P.C.; Rego, F.G.M.; Buck, M.; Schumacher, J. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat. Commun. 2011, 2, 177. [Google Scholar] [CrossRef] [Green Version]
- Lan, L.; Deng, X.; Zhou, J.; Tang, X. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by HrpL and HrpRS. Mol. Plant Microbe Interact. 2006, 19, 976–987. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Shao, X.; Zhang, Y.; Zhu, Y.; Yang, P.; Yuan, J.; Wang, T.; Yin, C.; Wang, W.; Chen, S.; et al. HrpS is a global regulator on type III secretion system (T3SS) and non-T3SS genes in Pseudomonas savastanoi pv. phaseolicola. Mol. Plant Microbe Interact. 2018, 31, 1232–1243. [Google Scholar] [CrossRef]
- Wei, W.; Plovanich-Jones, A.; Deng, W.-L.; Jin, Q.-L.; Collmer, A.; Huang, H.-C.; He, S.Y. The gene coding for the hrp pilus structural protein is required for type III secretion of Hrp and Avr Proteins in Pseudomonas syringae pv. tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 2247–2252. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Cui, Y.; Yang, H.; Collmer, A.; Alfano, J.R.; Chatterjee, A.K. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant Microbe Interact. 2003. [Google Scholar] [CrossRef] [Green Version]
- Jovanovic, M.; Lawton, E.; Schumacher, J.; Buck, M. Interplay among Pseudomonas syringae HrpR, HrpS and HrpV proteins for regulation of the type III secretion system. FEMS Microbiol. Lett. 2014, 356, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.-F.; Deng, W.-L.; Huang, H.-C. A Chaperone-like HrpG protein acts as a suppressor of HrpV in regulation of the Pseudomonas syringae pv. syringae type III secretion system. Mol. Microbiol. 2005, 57, 520–536. [Google Scholar] [CrossRef]
- Charova, S.N.; Gazi, A.D.; Mylonas, E.; Pozidis, C.; Sabarit, B.; Anagnostou, D.; Psatha, K.; Aivaliotis, M.; Beuzon, C.R.; Panopoulos, N.J.; et al. Migration of type III secretion system transcriptional regulators links gene expression to secretion. MBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Crabill, E.; Karpisek, A.; Alfano, J.R. The Pseudomonas syringae HrpJ protein controls the secretion of type III translocator proteins and has a virulence role inside plant cells. Mol. Microbiol. 2012, 85, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.Q.; Guo, M.; Alfano, J.R. Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors, and secretion of the HrpZ1 harpin. J. Bacteriol. 2006, 188, 6060–6069. [Google Scholar] [CrossRef] [Green Version]
- Ramos, A.R.; Morello, J.E.; Ravindran, S.; Deng, W.-L.; Huang, H.-C.; Collmer, A. Identification of Pseudomonas syringae pv. syringae 61 type III secretion system hrp proteins that can travel the type III pathway and contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 2007, 189, 5773–5778. [Google Scholar] [CrossRef] [Green Version]
- Haapalainen, M.; Van Gestel, K.; Pirhonen, M.; Taira, S. Soluble plant cell signals induce the expression of the type III secretion system of Pseudomonas syringae and upregulate the production of pilus protein HrpA. Mol. Plant Microbe Interact. 2009, 22, 282–290. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.-C.; Lin, Y.-C.; Wei, C.-F.; Deng, W.-L.; Huang, H.-C. The pathogenicity factor HrpF interacts with HrpA and HrpG to modulate type III secretion system (T3SS) function and T3SS expression in Pseudomonas syringae pv. averrhoi. Mol. Plant Pathol. 2016, 17, 1080–1094. [Google Scholar] [CrossRef]
- Sturm, A.; Heinemann, M.; Arnoldini, M.; Benecke, A.; Ackermann, M.; Benz, M.; Dormann, J.; Hardt, W.-D. The cost of virulence: Retarded growth of Salmonella typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011, 7, e1002143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, T.V.; Dahlbeck, D.; Staskawicz, B.J. Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science 1989, 245, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Arlat, M.; Gough, C.L.; Zischek, C.; Barberis, P.A.; Trigalet, A.; Boucher, C.A. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 1992, 5, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Mindrinos, M.N.; Panopoulos, N.J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 1992, 174, 3499–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dijk, K.; Fouts, D.E.; Rehm, A.H.; Hill, A.R.; Collmer, A.; Alfano, J.R. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the hrp (Type III) protein secretion system in a temperature- and pH-sensitive manner. J. Bacteriol. 1999, 181, 4790–4797. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Martín, I.; Thwaites, R.; Mansfield, J.W.; Beuzón, C.R. Negative regulation of the hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 2010, 23, 682–701. [Google Scholar] [CrossRef] [Green Version]
- Salmeron, J.M.; Staskawicz, B.J. Molecular characterization and hrp dependence of the avirulence gene AvrPro from Pseudomonas syringae pv. tomato. Mol. Gen. Genet. 1993, 239, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Mark Goodwin, S.; Xiao, Y.; Sun, Z.; Baker, D.; Tang, X.; Jenks, M.A.; Zhou, J.-M. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004, 23, 2903–2913. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Rogan, C.J.; Anderson, J.C. Development of a Pseudomonas syringae-Arabidopsis suspension cell infection system for investigating host metabolite-dependent regulation of type III secretion and pattern-triggered immunity. Mol. Plant Microbe Interact. 2019, 32, 527–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axelos, M.; Curie, C.; Mazzolini, L.; Bardet, C.; Lescure, B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol. Biochem. 1992, 30, 123–128. [Google Scholar]
- Anderson, J.C.; Wan, Y.; Kim, Y.M.; Pasa-Tolic, L.; Metz, T.O.; Peck, S.C. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 2014, 111, 6846–6851. [Google Scholar] [CrossRef] [Green Version]
- Rico, A.; Preston, G.M. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant Microbe Interact. 2008, 21, 269–282. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Sharma, A.; Kaur, R.; Thukral, A.K.; Bhardwaj, R.; Ahmad, P. Differential distribution of amino acids in plants. Amino Acids 2017, 49, 821–869. [Google Scholar] [CrossRef]
- Mo, Y.Y.; Gross, D.C. Plant signal molecules activate the syrb gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 1991, 173, 5784–5792. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.C.; Bartels, S.; González Besteiro, M.A.; Shahollari, B.; Ulm, R.; Peck, S.C. Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 2011, 67, 258–268. [Google Scholar] [CrossRef]
- McClerklin, S.A.; Lee, S.G.; Harper, C.P.; Nwumeh, R.; Jez, J.M.; Kunkel, B.N. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 2018, 14, e1006811. [Google Scholar] [CrossRef] [Green Version]
- Djami-Tchatchou, A.T.; Harrison, G.A.; Harper, C.P.; Wang, R.; Prigge, M.J.; Estelle, M.; Kunkel, B.N. Dual role of auxin in regulating plant defense and bacterial virulence gene expression during Pseudomonas syringae Pto DC3000 pathogenesis. Mol. Plant Microbe Interact. 2020, 33, 1059–1071. [Google Scholar] [CrossRef]
- Aragón, I.M.; Pérez-Martínez, I.; Moreno-Pérez, A.; Cerezo, M.; Ramos, C. New insights into the role of indole-3-acetic acid in the virulence of Pseudomonas savastanoi pv. savastanoi. FEMS Microbiol. Lett. 2014, 356, 184–192. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Agnew, J.L.; Cohen, J.D.; He, P.; Shan, L.; Sheen, J.; Kunkel, B.N. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 2007, 104, 20131–20136. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, P.J.; Schmelz, E.A.; Moussatche, P.; Lund, S.T.; Jones, J.B.; Klee, H.J. Susceptible to intolerance—A range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 2003, 33, 245–257. [Google Scholar] [CrossRef]
- Schmelz, E.A.; Engelberth, J.; Tumlinson, J.H.; Block, A.; Alborn, H.T. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 2004, 39, 790–808. [Google Scholar] [CrossRef]
- McAtee, P.A.; Brian, L.; Curran, B.; Van Der Linden, O.; Nieuwenhuizen, N.J.; Chen, X.; Henry-Kirk, R.A.; Stroud, E.A.; Nardozza, S.; Jayaraman, J.; et al. Re-programming of Pseudomonas syringae pv. actinidiae gene expression during early stages of infection of kiwifruit. BMC Genomics 2018, 19. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Yang, J.; Zhang, J.; Liu, Y.-X.; Tian, C.; Qu, B.; Gao, C.; Xin, P.; Cheng, S.; Zhang, W.; et al. An Arabidopsis secondary metabolite directly targets expression of the bacterial type III secretion system to inhibit bacterial virulence. Cell Host Microbe 2020, 27, 601–613. [Google Scholar] [CrossRef]
- Vargas, P.; Farias, G.A.; Nogales, J.; Prada, H.; Carvajal, V.; Barón, M.; Rivilla, R.; Martín, M.; Olmedilla, A.; Gallegos, M.-T. Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environ. Microbiol. Rep. 2013, 5, 841–850. [Google Scholar] [CrossRef]
- Lee, J.S.; Ryu, H.R.; Cha, J.Y.; Baik, H.S. The hrp pathogenicity island of Pseudomonas syringae pv. tomato DC3000 is induced by plant phenolic acids. J. Microbiol. 2015, 53, 725–731. [Google Scholar] [CrossRef]
- Kang, J.E.; Jeon, B.J.; Park, M.Y.; Yang, H.J.; Kwon, J.; Lee, D.H.; Kim, B.S. Inhibition of the type III secretion system of Pseudomonas syringae pv. tomato DC3000 by resveratrol oligomers identified in Vitis vinifera L. Pest Manag. Sci. 2020, 76, 2294–2303. [Google Scholar] [CrossRef]
- Puigvert, M.; Solé, M.; López-Garcia, B.; Coll, N.S.; Beattie, K.D.; Davis, R.A.; Elofsson, M.; Valls, M. Type III secretion inhibitors for the management of bacterial plant diseases. Mol. Plant Pathol. 2019, 20, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.-N.; Chen, L.; Si, N.-G.; Jiang, W.-J.; Zhou, Z.-G.; Liu, J.-L.; Zhang, L.-Q. Identification of benzyloxy carbonimidoyl dicyanide derivatives as novel type III secretion system inhibitors via high-throughput screening. Front. Plant Sci. 2019, 10, 1059. [Google Scholar] [CrossRef] [Green Version]
- Sasser, M. Inhibition by antibacterial compounds of the hypersensitive reaction induced by Pseudomonas pisi in tobacco. Phytopathology 1982, 72. [Google Scholar] [CrossRef]
- Nobori, T.; Velásquez, A.C.; Wu, J.; Kvitko, B.H.; Kremer, J.M.; Wang, Y.; He, S.Y.; Tsuda, K. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. USA 2018. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Lund, S.P.; Scott, R.A.; Greenwald, J.W.; Records, A.H.; Nettleton, D.; Lindow, S.E.; Gross, D.C.; Beattie, G.A. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. USA 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Lund, S.P.; Greenwald, J.W.; Records, A.H.; Scott, R.A.; Nettleton, D.; Lindow, S.E.; Gross, D.C.; Beattie, G.A. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. MBio 2014, 5, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Helmann, T.C.; Deutschbauer, A.M.; Lindow, S.E. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. USA 2019, 116, 18900–18910. [Google Scholar] [CrossRef] [Green Version]
- Vinatzer, B.A.; Teitzel, G.M.; Lee, M.-W.; Jelenska, J.; Hotton, S.; Fairfax, K.; Jenrette, J.; Greenberg, J.T. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 2006, 62, 26–44. [Google Scholar] [CrossRef]
- Lee, J.; Teitzel, G.; Munkvold, K.; Pozo, O.; Martin, G.; Michelmore, R.; Greenberg, J. Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 2012, 158, 1803–1818. [Google Scholar] [CrossRef] [Green Version]
- Rufián, J.S.; Sánchez-Romero, M.A.; López-Márquez, D.; Macho, A.P.; Mansfield, J.W.; Arnold, D.L.; Ruiz-Albert, J.; Casadesús, J.; Beuzón, C.R. Pseudomonas syringae differentiates into phenotypically distinct subpopulations during colonization of a plant host. Environ. Microbiol. 2016, 18, 3593–3605. [Google Scholar] [CrossRef]
- Rufián, J.S.; Macho, A.P.; Corry, D.S.; Mansfield, J.W.; Ruiz-Albert, J.; Arnold, D.L.; Beuzón, C.R. Confocal microscopy reveals in planta dynamic interactions between pathogenic, avirulent and non-pathogenic Pseudomonas syringae strains. Mol. Plant Pathol. 2018, 19, 537–551. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Laiosa, M.D.; Steeber, D.A.; Biddle, E.M.; Peng, Q.; Yang, C.H. Cell individuality: The bistable gene expression of the type III secretion system in Dickeya dadantii 3937. Mol. Plant Microbe Interact. 2012, 25, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Kussell, E.; Kishony, R.; Balaban, N.Q.; Leibler, S. Bacterial persistence: A model of survival in changing environments. Genetics 2005, 169, 1807–1814. [Google Scholar] [CrossRef] [Green Version]
- Groisman, E.A.; Ochman, H. Pathogenicity islands: Bacterial evolution in quantum leaps. Cell 1996, 87, 791–794. [Google Scholar] [CrossRef] [Green Version]
- Sawada, H.; Suzuki, F.; Matsuda, I.; Saitou, N. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argk and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 1999, 49, 627–644. [Google Scholar] [CrossRef]
- Mohr, T.J.; Liu, H.; Yan, S.; Morris, C.E.; Castillo, J.A.; Jelenska, J.; Vinatzer, B.A. Naturally occurring nonpathogenic isolates of the plant pathogen Pseudomonas syringae lack a type III secretion system and effector gene orthologues. J. Bacteriol. 2008, 190, 2858–2870. [Google Scholar] [CrossRef] [Green Version]
- Grimm, C.; Panopoulos, N.J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several procaryotic regulatory proteins. J. Bacteriol. 1989, 171, 5031–5038. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Xiao, Y.; Zhou, J.-M. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant Microbe Interact. 2006, 19, 1159–1166. [Google Scholar] [CrossRef] [Green Version]
- Beier, D.; Gross, R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006, 9, 143–152. [Google Scholar] [CrossRef] [PubMed]
- West, A.H.; Stock, A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001, 26, 369–376. [Google Scholar] [CrossRef]
- Yan, Q.; Rogan, C.J.; Pang, Y.-Y.; Davis, E.W.; Anderson, J.C. Ancient co-option of an amino acid ABC transporter locus in Pseudomonas syringae for host signal-dependent virulence gene regulation. PLoS Pathog. 2020, 16, e1008680. [Google Scholar] [CrossRef]
- Sonawane, A.M.; Singh, B.; Röhm, K.H. The AauR-AauS two-component system regulates uptake and metabolism of acidic amino acids in Pseudomonas putida. Appl. Environ. Microbiol. 2006, 72, 6569–6577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Röhm, K.H. Characterization of a Pseudomonas putida ABC transporter (AatJMQP) required for acidic amino acid uptake: Biochemical properties and regulation by the Aau two-component system. Microbiology 2008, 154 Pt 3, 797–809. [Google Scholar] [CrossRef] [Green Version]
- Turner, S.E.; Pang, Y.-Y.; O’Malley, M.R.; Weisberg, A.J.; Fraser, V.N.; Yan, Q.; Chang, J.H.; Anderson, J.C. A DeoR-type transcription regulator is required for sugar-induced expression of type III secretion-encoding genes in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2020, 33, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 2001. [Google Scholar] [CrossRef] [Green Version]
- Laville, J.; Voisard, C.; Keelt, C.; Maurhofert, M.; Dtfagot, G.; Haas, D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 1992, 89, 1562–1566. [Google Scholar] [CrossRef] [Green Version]
- Natsch, A.; Keel, C.; Pfirter, H.A.; Haas, D.; Défago, G. Contribution of the global regulator gene GacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 1994, 60, 2553–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rich, J.J.; Kinscherf, T.G.; Kitten, T.; Willis, D.K. Genetic evidence that the GacA gene encodes the cognate response regulator for the LemA sensor in Pseudomonas syringae. J. Bacteriol. 1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latour, X. The evanescent GacS signal. Microorganisms 2020, 8, 1746. [Google Scholar] [CrossRef]
- Willis, D.K.; Hrabak, E.M.; Rich, J.J.; Barta, T.M.; Lindow, S.E.; Panopoulos, N.J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol. Plant Microbe Interact. 1990, 3, 149–156. [Google Scholar] [CrossRef]
- Mizuno, T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997, 4, 161–168. [Google Scholar] [CrossRef] [Green Version]
- Koretke, K.K.; Lupas, A.N.; Warren, P.V.; Rosenberg, M.; Brown, J.R. Evolution of two-component signal transduction. Mol. Biol. Evol. 2000, 17, 1956–1970. [Google Scholar] [CrossRef] [Green Version]
- Pernestig, A.-K.; Melefors, Ö.; Georgellis, D. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 2001, 276, 225–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moolenaar, G.F.; van Sluis, C.A.; Backendorf, C.; de Putte, P.V. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrc structural gene and the region coding for a 24 kd protein. Nucleic Acids Res. 1987, 15, 4273–4289. [Google Scholar] [CrossRef] [Green Version]
- Ferreiro, M.-D.; Nogales, J.; Farias, G.A.; Olmedilla, A.; Sanjuán, J.; Gallegos, M.T. Multiple CsrA proteins control key virulence traits in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2018. [Google Scholar] [CrossRef] [Green Version]
- Marutani, M.; Taguchi, F.; Ogawa, Y.; Hossain, M.M.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Gac two-component system in Pseudomonas syringae pv. tabaci is required for virulence but not for hypersensitive reaction. Mol. Genet. Genomics 2008, 279, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the rsmY and rsmZ regulatory small RNAs. Mol. Microbiol. 2009. [Google Scholar] [CrossRef] [Green Version]
- Valentini, M.; Gonzalez, D.; Mavridou, D.A.; Filloux, A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018, 41, 15–20. [Google Scholar] [CrossRef]
- Brencic, A.; Winans, S.C. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 2005, 69, 155–194. [Google Scholar] [CrossRef] [Green Version]
- Mole, B.M.; Baltrus, D.A.; Dangl, J.L.; Grant, S.R. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 2007, 15, 363–371. [Google Scholar] [CrossRef]
- MacLean, D.; Studholme, D.J. A Boolean model of the Pseudomonas syringae hrp regulon predicts a tightly regulated system. PLoS ONE 2010, 5, e9101. [Google Scholar] [CrossRef]
- O’Malley, M.R.; Chien, C.F.; Peck, S.C.; Lin, N.C.; Anderson, J.C. A Revised model for the role of GacS/GacA in regulating type III secretion by Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2020, 21, 139–144. [Google Scholar] [CrossRef] [Green Version]
- O’Malley, M.R.; Weisberg, A.J.; Chang, J.H.; Anderson, J.C. Re-evaluation of a Tn5::gacA mutant of Pseudomonas syringae pv. tomato DC3000 uncovers roles for uvrC and anmK in promoting virulence. PLoS ONE 2019, 14, e0223637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chieda, Y.; Iiyama, K.; Yasunaga-Aoki, C.; Lee, J.M.; Kusakabe, T.; Shimizu, S. Pathogenicity of gacA mutant of Pseudomonas aeruginosa PAO1 in the silkworm, Bombyx mori. FEMS Microbiol. Lett. 2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chieda, Y.; Iiyama, K.; Lee, J.M.; Kusakabe, T.; Yasunaga-aoki, C.; Shimizu, S. The gacA gene of Pseudomonas aeruginosa PAO1 is not required for full virulence in Bombyx mori. J. Insect Biotechnol. Sericol. 2007, 95, 89–95. [Google Scholar] [CrossRef]
- Xiao, Y.; Lan, L.; Yin, C.; Deng, X.; Baker, D.; Zhou, J.M.; Tang, X. Two-component sensor RhpS promotes induction of Pseudomonas syringae type III secretion system by repressing negative regulator RhpR. Mol. Plant Microbe Interact. 2007, 20, 223–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Lan, L.; Xiao, Y.; Kennelly, M.; Zhou, J.-M.; Tang, X. Pseudomonas syringae two-component response regulator RhpR regulates promoters carrying an inverted repeat element. Mol. Plant Microbe Interact. 2010, 23, 927–939. [Google Scholar] [CrossRef]
- Deng, X.; Liang, H.; Chen, K.; He, C.; Lan, L.; Tang, X. Molecular mechanisms of two-component system RhpRS regulating type III secretion system in Pseudomonas syringae. Nucleic Acids Res. 2014, 42, 11472–11486. [Google Scholar] [CrossRef]
- Xie, Y.; Shao, X.; Zhang, Y.; Liu, J.; Wang, T.; Zhang, W.; Hua, C.; Deng, X. Pseudomonas savastanoi two-component system RhpRS switches between virulence and metabolism by tuning phosphorylation state and sensing nutritional conditions. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.; Yin, C.; Zhang, Y.; Shi, H.; Wang, J.; Sun, L.; Shao, X.; Gao, R.; Wang, W.; Deng, X. Lon protease is involved in RhpRS-mediated regulation of type III secretion in Pseudomonas syringae. Mol. Plant Microbe Interact. 2016, 29, 807–814. [Google Scholar] [CrossRef] [Green Version]
- Fishman, M.R.; Zhang, J.; Bronstein, P.A.; Stodghill, P.; Filiatrault, M.J. Ca2+ -induced two-component system CvsSR regulates the type III secretion system and the extracytoplasmic function sigma factor AlgU in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 2018, 200, e00538-17. [Google Scholar] [CrossRef] [Green Version]
- Fishman, M.R.; Filiatrault, M.J. Prevention of surface-associated calcium phosphate by the Pseudomonas syringae two-component system CvsSR. J. Bacteriol. 2019, 201, e00584-18. [Google Scholar] [CrossRef] [Green Version]
- Stael, S.; Wurzinger, B.; Mair, A.; Mehlmer, N.; Vothknecht, U.C.; Teige, M. Plant organellar calcium signalling: An emerging field. J. Exp. Bot. 2012, 63, 1525–1542. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, B.M.; Neale, H.C.; Geilfus, C.-M.; Jackson, R.W.; Arnold, D.L.; Preston, G.M. Early changes in apoplast composition associated with defense and disease in interactions between Phaseolus vulgaris and the halo blight pathogen Pseudomonas syringae pv. phaseolicola. Plant. Cell Environ. 2016, 39, 2172–2184. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 2004, 6, 552–567. [Google Scholar] [CrossRef]
- Tsilibaris, V.; Maenhaut-Michel, G.; Van Melderen, L. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 2006, 157, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Wang, T.; Shao, X.; Xie, Y.; Huang, H.; Liu, J.; Zhang, W.; Zhang, Y.; Ding, Y.; Jiang, L.; et al. Pseudomonas syringae dual-function protein Lon switches between virulence and metabolism by acting as both DNA -binding transcriptional regulator and protease in different environments. Environ. Microbiol. 2020, 22, 2968–2988. [Google Scholar] [CrossRef]
- Bretz, J.; Losada, L.; Lisboa, K.; Hutcheson, S.W. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 2002, 45, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Deng, X.; Xiao, Y.; Zhou, J.-M.; Tang, X. Mutation of Lon protease differentially affects the expression of Pseudomonas syringae type III secretion system genes in rich and minimal media and reduces pathogenicity. Mol. Plant Microbe Interact. 2007, 20, 682–696. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.J.; Lee, J.S.; Cha, J.Y.; Baik, H.S. Negative regulation of pathogenesis in Pseudomonas syringae pv. tabaci 11528 by ATP-dependent Lon protease. Mol. Cells 2011, 32, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losada, L.C.; Hutcheson, S.W. Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 2004, 55, 941–953. [Google Scholar] [CrossRef]
- Taira, S.; Tuimala, J.; Roine, E.; Nurmiaho-Lassila, E.-L.; Savilahti, H.; Romantschuk, M. Mutational analysis of the Pseudomonas syringae pv. tomato HrpA gene encoding hrp pilus subunit. Mol. Microbiol. 1999, 34, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Markel, E.; Stodghill, P.; Bao, Z.; Myers, C.R.; Swingle, B. AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 2016, 198, 2330–2344. [Google Scholar] [CrossRef] [Green Version]
- Ishiga, T.; Ishiga, Y.; Betsuyaku, S.; Nomura, N. AlgU contributes to the virulence of Pseudomonas syringae pv. tomato DC3000 by regulating production of the phytotoxin coronatine. J. Gen. Plant Pathol. 2018, 84, 189–201. [Google Scholar] [CrossRef]
- Schreiber, K.J.; Desveaux, D. AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol. Microbiol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Kidambi, S.P.; Sundin, G.W.; Palmer, D.A.; Chakrabarty, A.M.; Bender, C.L. Copper as a signal for alginate synthesis in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 1995, 61, 2172–2179. [Google Scholar] [CrossRef] [Green Version]
- Keith, L.M.W.; Bender, C.L. AlgT (σ22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J. Bacteriol. 1999, 181, 7176–7184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, A.; Weingart, H.; Ullrich, M.S. The alternative sigma factor AlgT, but not alginate synthesis, promotes in planta multiplication of Pseudomonas syringae pv. glycinea. Microbiology 2008, 154, 413–421. [Google Scholar] [CrossRef] [Green Version]
- Freeman, B.C.; Chen, C.; Yu, X.; Nielsen, L.; Peterson, K.; Beattie, G.A. Physiological and transcriptional responses to osmotic stress of two Pseudomonas syringae strains that differ in epiphytic fitness and osmotolerance. J. Bacteriol. 2013, 195, 4742–4752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fett, W.F.; Dunn, M.F. Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol. 1989, 89, 5–9. [Google Scholar] [CrossRef]
- Rudolph, K.W.E.; Gross, M.; Ebrahim-Nesbat, F.; Nöllenburg, M.; Zomorodian, A.; Wydra, K.; Neugebauer, M.; Hettwer, U.; El-Shouny, W.; Sonnenberg, B.; et al. The role of extracellular polysaccharides as virulence factors for phytopathogenic pseudomonads and xanthomonads. In Molecular Mechanisms of Bacterial Virulence; Springer: Amsterdam, The Netherlands, 1994; pp. 357–378. [Google Scholar] [CrossRef]
- Yu, J.; Penaloza-Vazquez, A.; Chakrabarty, A.M.; Bender, C.L. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 1999, 33, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.N.; Newman, M.-A.; Erbs, G.; Morrissey, K.L.; Chinchilla, D.; Boller, T.; Jensen, T.T.; De Castro, C.; Ierano, T.; Molinaro, A.; et al. bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr. Biol. 2008, 18, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Lovelace, A.H.; Smith, A.; Kvitko, B.H. Pattern-triggered immunity alters the transcriptional regulation of virulence-associated genes and induces the sulfur starvation response in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2018, 31, 750–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Z.; Wei, H.-L.; Ma, X.; Swingle, B. Pseudomonas syringae AlgU downregulates flagellin gene expression, helping evade plant immunity. J. Bacteriol. 2019, 202. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, B.; Pujol, C.J.; Lindow, S.E. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 2004, 17, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Xiao, Y.; Lan, L.; Zhou, J.-M.; Tang, X. Pseudomonas syringae pv. phaseolicola mutants compromised for type III secretion system gene induction. Mol. Plant Microbe Interact. 2009, 22, 964–976. [Google Scholar] [CrossRef]
- Kawakita, Y.; Taguchi, F.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Characterization of each AefR and MexT mutant in Pseudomonas syringae pv. tabaci 6605. Mol. Genet. Genomics 2012, 287, 473–484. [Google Scholar] [CrossRef]
- Quiñones, B.; Dulla, G.; Lindow, S.E. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. MPMI 2005, 18, 682–693. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Cui, Y.; Hasegawa, H.; Chatterjee, A.K. PsrA, the Pseudomonas sigma regulator, controls regulators of epiphytic fitness, quorum-sensing signals, and plant interactions in Pseudomonas syringae pv. tomato strain DC3000. Appl. Environ. Microbiol. 2007, 73, 3684–3694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blumer, C.; Heeb, S.; Pessi, G.; Haas, D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 1999, 96, 14073–14078. [Google Scholar] [CrossRef] [Green Version]
- Heeb, S.; Blumer, C.; Haas, D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 2002. [Google Scholar] [CrossRef] [Green Version]
- Valverde, C.; Haas, D. Small RNAs controlled by two-component systems. Adv. Exp. Med. Biol. 2008, 631, 55–79. [Google Scholar] [CrossRef]
- Lapouge, K.; Schubert, M.; Allain, F.H.T.; Haas, D. Gac/Rsm signal transduction pathway of γ-Proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef]
- Baker, C.S.; Morozov, I.; Suzuki, K.; Romeo, T.; Babitzke, P. CsrA regulates glycogen biosynthesis by preventing translation of GlgC in Escherichia coli. Mol. Microbiol. 2002, 44, 1599–1610. [Google Scholar] [CrossRef]
- Dubey, A.K.; Baker, C.S.; Romeo, T.; Babitzke, P. RNA Sequence and Secondary Structure Participate in High-Affinity CsrA-RNA Interaction. RNA 2005, 11, 1579–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo, T.; Vakulskas, C.A.; Babitzke, P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ. Microbiol. 2013, 15, 313–324. [Google Scholar] [CrossRef]
- Yakhnin, A.V.; Baker, C.S.; Vakulskas, C.A.; Yakhnin, H.; Berezin, I.; Romeo, T.; Babitzke, P. CsrA activates FlhDC expression by protecting FlhDC mRNA from RNase E-mediated cleavage. Mol. Microbiol. 2013, 87, 851–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, M.O.; Farah, C.S.; Wang, N. The post-transcriptional regulator RsmA/CsrA activates T3SS by stabilizing the 5′ UTR of HrpG, the master regulator of Hrp/Hrc genes, in Xanthomonas. PLoS Pathog. 2014, 10, e1003945. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Lee, J.H.; Liu, J.; Yang, H.; Tian, Y.; Hu, B.; Zhao, Y. Homologues of the RNA binding protein RsmA in Pseudomonas syringae pv. tomato DC3000 exhibit distinct binding affinities with non-coding small RNAs and have distinct roles in virulence. Mol. Plant Pathol. 2019, 20, 1217–1236. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yu, M.; Ge, Y.; Tian, Y.; Hu, B.; Zhao, Y. The RsmA RNA-Binding proteins in Pseudomonas syringae exhibit distinct and overlapping roles in modulating virulence and survival under different nutritional conditions. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Ramírez-Zapata, D.; Ramos, C.; Aguilera, S.; Bardaji, L.; Martínez-Gil, M.; Murillo, J. Two homologues of the global regulator Csr/Rsm redundantly control phaseolotoxin biosynthesis and virulence in the plant pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms 2020, 8, 1536. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C.; Valverde, C.; Kay, E.; Haas, D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 2005, 187, 276–285. [Google Scholar] [CrossRef] [Green Version]
- Moll, S.; Schneider, D.J.; Stodghill, P.; Myers, C.R.; Cartinhour, S.W.; Filiatrault, M.J. Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 2010, 7, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Pesavento, C.; Hengge, R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 2009, 12, 170–176. [Google Scholar] [CrossRef]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatnaparat, T.; Li, Z.; Korban, S.S.; Zhao, Y. The bacterial alarmone (p)ppGpp is required for virulence and controls cell size and survival of Pseudomonas syringae on plants. Environ. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Cai, Z.; Shao, X.; Zhang, W.; Xie, Y.; Zhang, Y.; Hua, C.; Schuster, S.C.; Yang, L.; Deng, X. Pleiotropic effects of c-di-GMP content in Pseudomonas syringae. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engl, C.; Waite, C.J.; McKenna, J.F.; Bennett, M.H.; Hamann, T.; Buck, M. Chp8, a diguanylate cyclase from Pseudomonas syringae pv. tomato DC3000, suppresses the pathogen-associated molecular pattern flagellin, increases extracellular polysaccharides, and promotes plant immune evasion. MBio 2014, 5, e01168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.; Tan, M.; Xie, Y.; Yao, C.; Wang, T.; Huang, H.; Zhang, Y.; Ding, Y.; Liu, J.; Han, L.; et al. Integrated regulatory network in Pseudomonas syringae reveals dynamics of virulence. Cell Rep. 2021, 34, 108920. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Malley, M.R.; Anderson, J.C. Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals. Microorganisms 2021, 9, 1227. https://doi.org/10.3390/microorganisms9061227
O’Malley MR, Anderson JC. Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals. Microorganisms. 2021; 9(6):1227. https://doi.org/10.3390/microorganisms9061227
Chicago/Turabian StyleO’Malley, Megan R., and Jeffrey C. Anderson. 2021. "Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals" Microorganisms 9, no. 6: 1227. https://doi.org/10.3390/microorganisms9061227
APA StyleO’Malley, M. R., & Anderson, J. C. (2021). Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals. Microorganisms, 9(6), 1227. https://doi.org/10.3390/microorganisms9061227




