Behavioral Interactions between Bacterivorous Nematodes and Predatory Bacteria in a Synthetic Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial and Nematode Strains
2.2. Standard Culture Conditions
2.3. Culturing C. elegans
2.4. Culturing M. xanthus
2.5. Culturing Prey Bacteria
2.6. C. elegans’ Binary Choice Assays
2.7. C. elegans Half-Plate Choice Assays
2.8. M. xanthus Swarming Assays
2.9. Statistical Analysis
3. Results
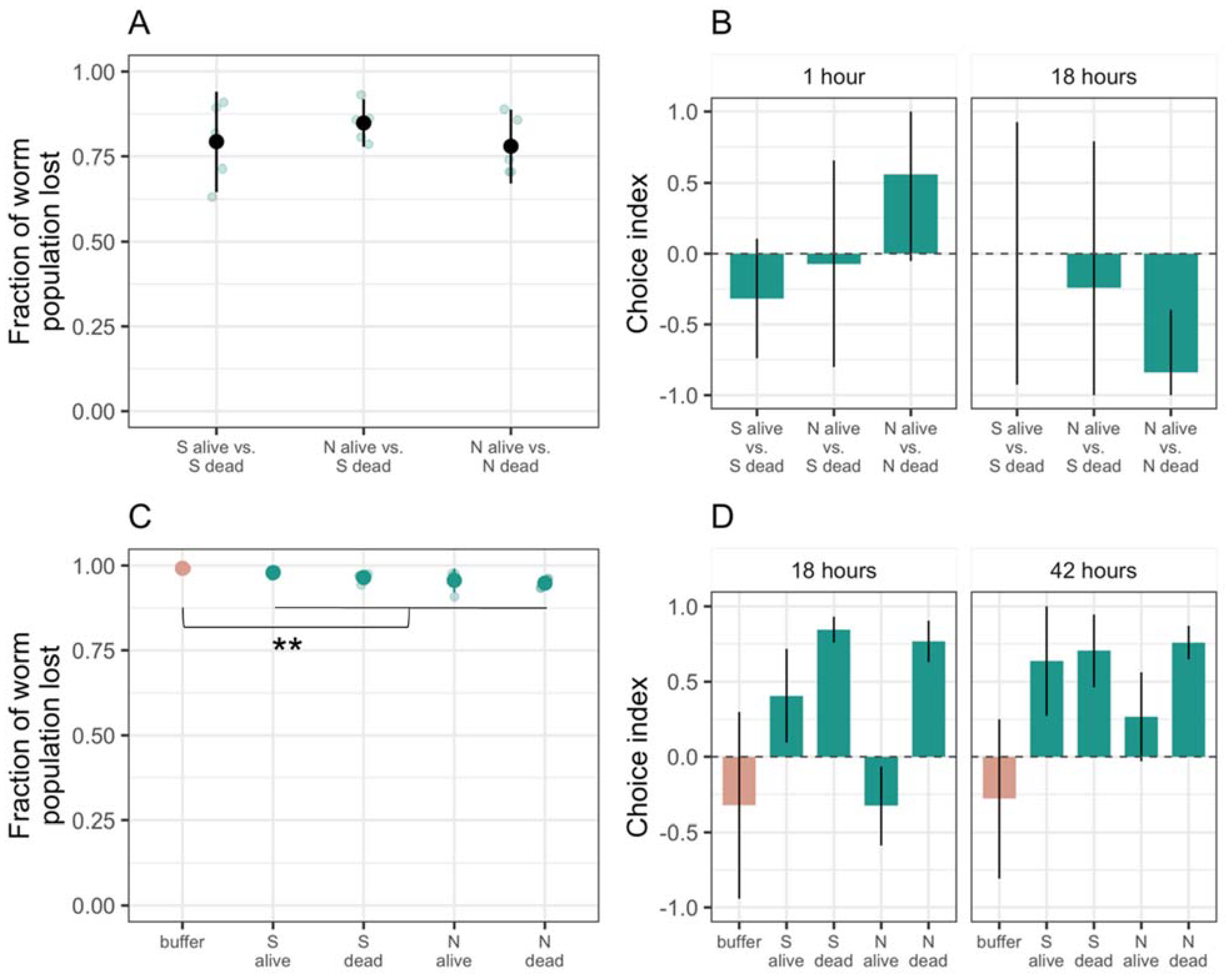
3.1. Only a Few C. elegans Worms Interact with M. xanthus Regardless of Whether It Is Alive or Dead
3.2. C. elegans Prefers Both Basal Prey Species over M. xanthus
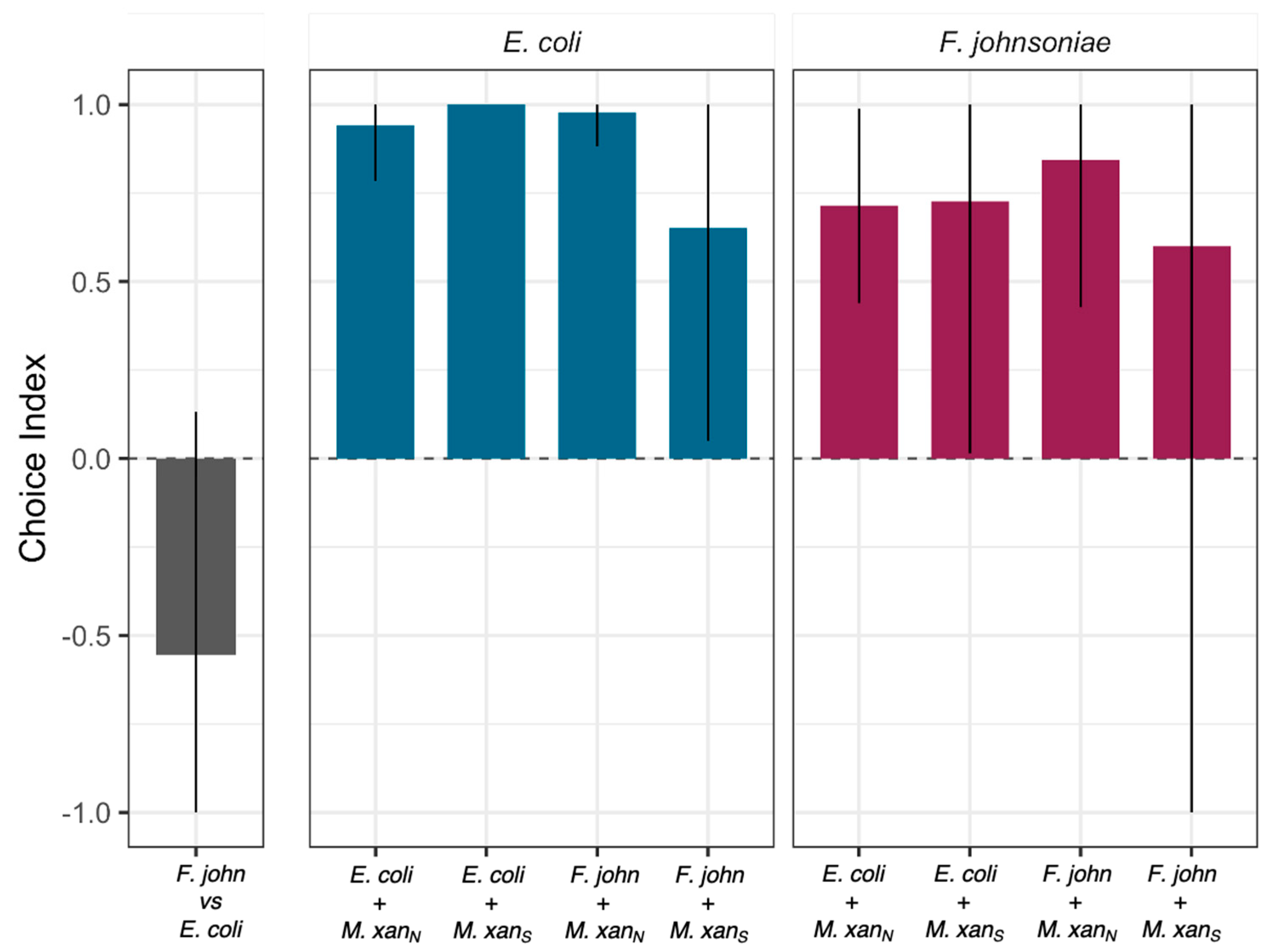
3.3. M. xanthus Responds Behaviorally to C. elegans in a Prey-Dependent Manner
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hairston, N.G.; Smith, F.E.; Slobodkin, L.B. Community Structure, Population Control, and Competition. Am. Nat. 1960, 94, 421–425. [Google Scholar] [CrossRef]
- Leibold, M.A.; Chase, J.M.; Shurin, J.B.; Downing, A.L. Species Turnover and the Regulation of Trophic Structure. Annu. Rev. Ecol. Syst. 1997, 28, 467–494. [Google Scholar] [CrossRef]
- Brodie, E.D.; Brodie, E.D. Predator-Prey Arms RacesAsymmetrical selection on predators and prey may be reduced when prey are dangerous. BioScience 1999, 49, 557–568. [Google Scholar] [CrossRef]
- Hass, C.C.; Valenzuela, D. Anti-predator benefits of group living in white-nosed coatis (Nasua narica). Behav. Ecol. Sociobiol. 2002, 51, 570–578. [Google Scholar] [CrossRef]
- Matz, C.; Webb, J.S.; Schupp, P.J.; Phang, S.Y.; Penesyan, A.; Egan, S.; Steinberg, P.; Kjelleberg, S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE 2008, 3, e2744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, A.R.E.; Mduma, S.; Brashares, J.S. Patterns of predation in a diverse predator–prey system. Nature 2003, 425, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Merilaita, S. Animal camouflage: Current issues and new perspectives. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 423–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarnita, C.E. The ecology and evolution of social behavior in microbes. J. Exp. Biol. 2017, 220, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnke, J.; Cohen, Y.; de Leeuw, M.; Kushmaro, A.; Jurkevitch, E.; Chatzinotas, A. Multiple micro-predators controlling bacterial communities in the environment. Curr. Opin. Biotechnol. 2014, 27, 185–190. [Google Scholar] [CrossRef]
- Fenchel, T.; Blackburn, H.; King, G.M.; Blackburn, T.H. Bacterial Biogeochemistry: The Ecophysiology of Mineral Cycling; Academic Press: Cambridge, MA, USA, 2012; ISBN 978-0-12-415836-8. [Google Scholar]
- Griffiths, B.S. Microbial-feeding nematodes and protozoa in soil: Their effectson microbial activity and nitrogen mineralization in decomposition hotspots and the rhizosphere. Plant Soil 1994, 164, 25–33. [Google Scholar] [CrossRef]
- Kadouri, D.E.; To, K.; Shanks, R.M.Q.; Doi, Y. Predatory bacteria: A potential ally against multidrug-resistant Gram-negative pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef] [Green Version]
- Erken, M.; Lutz, C.; McDougald, D. The rise of pathogens: Predation as a factor driving the evolution of human pathogens in the environment. Microb. Ecol. 2013, 65, 860–868. [Google Scholar] [CrossRef] [Green Version]
- Davidov, Y.; Jurkevitch, E. Predation between prokaryotes and the origin of eukaryotes. BioEssays 2009, 31, 748–757. [Google Scholar] [CrossRef]
- Forterre, P. The Common Ancestor of Archaea and Eukarya Was Not an Archaeon. Archaea 2013, 2013, e372396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero, R.; Pedros-Alio, C.; Esteve, I.; Mas, J.; Chase, D.; Margulis, L. Predatory prokaryotes: Predation and primary consumption evolved in bacteria. Proc. Natl. Acad. Sci. USA 1986, 83, 2138–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Malley, M.A. The first eukaryote cell: An unfinished history of contestation. Stud. Hist. Philos. Biol. Biomed. Sci. 2010, 41, 212–224. [Google Scholar] [CrossRef]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Søgaard-Andersen, L.; van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Lyons, N.A.; Kolter, R. On the evolution of bacterial multicellularity. Curr. Opin. Microbiol. 2015, 24, 21–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gralka, M.; Szabo, R.; Stocker, R.; Cordero, O.X. Trophic Interactions and the Drivers of Microbial Community Assembly. Curr. Biol. 2020, 30, R1176–R1188. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.J.; Prakash, P.; Edwards, J.S. Advances in flux balance analysis. Curr. Opin. Biotechnol. 2003, 14, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Thiele, I.; Palsson, B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Edwards, J.S.; Covert, M.; Palsson, B. Metabolic modelling of microbes: The flux-balance approach. Environ. Microbiol. 2002, 4, 133–140. [Google Scholar] [CrossRef]
- Ram, Y.; Dellus-Gur, E.; Bibi, M.; Karkare, K.; Obolski, U.; Feldman, M.W.; Cooper, T.F.; Berman, J.; Hadany, L. Predicting microbial growth in a mixed culture from growth curve data. Proc. Natl. Acad. Sci. USA 2019, 116, 14698–14707. [Google Scholar] [CrossRef] [Green Version]
- Johnke, J.; Baron, M.; de Leeuw, M.; Kushmaro, A.; Jurkevitch, E.; Harms, H.; Chatzinotas, A. A Generalist Protist Predator Enables Coexistence in Multitrophic Predator-Prey Systems Containing a Phage and the Bacterial Predator Bdellovibrio. Front. Ecol. Evol. 2017, 5. [Google Scholar] [CrossRef] [Green Version]
- Miele, V.; Guill, C.; Ramos-Jiliberto, R.; Kéfi, S. Non-trophic interactions strengthen the diversity—functioning relationship in an ecological bioenergetic network model. PLoS Comput. Biol. 2019, 15, e1007269. [Google Scholar] [CrossRef] [Green Version]
- McClean, D.; Friman, V.-P.; Finn, A.; Salzberg, L.I.; Donohue, I. Coping with multiple enemies: Pairwise interactions do not predict evolutionary change in complex multitrophic communities. Oikos 2019, 128, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- Johnke, J.; Boenigk, J.; Harms, H.; Chatzinotas, A. Killing the killer: Predation between protists and predatory bacteria. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Athar, R.; Zheng, G.; Williams, H.N. Prey bacteria shape the community structure of their predators. ISME J. 2011, 5, 1314–1322. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, A.; Cherrier, J.; Williams, H.N. Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc. Natl. Acad. Sci. USA 2009, 106, 4301–4306. [Google Scholar] [CrossRef] [Green Version]
- Griffin, J.N.; Byrnes, J.E.K.; Cardinale, B.J. Effects of predator richness on prey suppression: A meta-analysis. Ecology 2013, 94, 2180–2187. [Google Scholar] [CrossRef]
- Kandel, P.P.; Pasternak, Z.; van Rijn, J.; Nahum, O.; Jurkevitch, E. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol. Ecol. 2014, 89, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Pérez, J.; Moraleda-Muñoz, A.; Marcos-Torres, F.J.; Muñoz-Dorado, J. Bacterial predation: 75 years and counting! Environ. Microbiol. 2016, 18, 766–779. [Google Scholar] [CrossRef]
- Berleman, J.E.; Scott, J.; Chumley, T.; Kirby, J.R. Predataxis behavior in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 2008, 105, 17127–17132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, A.D.; MacLean, R.C.; Hillesland, K.L.; Velicer, G.J. Comparative analysis of Myxococcus predation on soil bacteria. Appl. Environ. Microbiol. 2010, 76, 6920–6927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, R.R.; Vasse, M.; Wielgoss, S.; Sun, L.; Yu, Y.-T.N.; Velicer, G.J. Bacterial predator-prey coevolution accelerates genome evolution and selects on virulence-associated prey defences. Nat. Commun. 2019, 10, 4301. [Google Scholar] [CrossRef] [Green Version]
- Petters, S.; Groß, V.; Söllinger, A.; Pichler, M.; Reinhard, A.; Bengtsson, M.M.; Urich, T. The soil microbial food web revisited: Predatory myxobacteria as keystone taxa? ISME J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Berleman, J.E.; Kirby, J.R. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 2009, 33, 942–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livingstone, P.G.; Millard, A.D.; Swain, M.T.; Whitworth, D.E. Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding. Microb. Genom. 2018, 4, e000152. [Google Scholar] [CrossRef] [Green Version]
- Pham, V.D.; Shebelut, C.W.; Diodati, M.E.; Bull, C.T.; Singer, M. Mutations affecting predation ability of the soil bacterium Myxococcus xanthus. Microbiol. Read. Engl. 2005, 151, 1865–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berleman, J.E.; Chumley, T.; Cheung, P.; Kirby, J.R. Rippling Is a Predatory Behavior in Myxococcus xanthus. J. Bacteriol. 2006, 188, 5888–5895. [Google Scholar] [CrossRef] [Green Version]
- Hillesland, K.L.; Lenski, R.E.; Velicer, G.J. Ecological variables affecting predatory success in Myxococcus xanthus. Microb. Ecol. 2007, 53, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Soares, H.; Velicer, G.J. Decomposing predation: Testing for parameters that correlate with predatory performance by a social bacterium. Microb. Ecol. 2013, 65, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Hillesland, K.L.; Velicer, G.J.; Lenski, R.E. Experimental evolution of a microbial predator’s ability to find prey. Proc. R. Soc. B Biol. Sci. 2009, 276, 459–467. [Google Scholar] [CrossRef] [Green Version]
- Rønn, R.; McCaig, A.E.; Griffiths, B.S.; Prosser, J.I. Impact of Protozoan Grazing on Bacterial Community Structure in Soil Microcosms. Appl. Environ. Microbiol. 2002, 68, 6094–6105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prugh, L.R.; Stoner, C.J.; Epps, C.W.; Bean, W.T.; Ripple, W.J.; Laliberte, A.S.; Brashares, J.S. The Rise of the Mesopredator. BioScience 2009, 59, 779–791. [Google Scholar] [CrossRef]
- Dahl, J.L.; Ulrich, C.H.; Kroft, T.L. Role of phase variation in the resistance of Myxococcus xanthus fruiting bodies to Caenorhabditis elegans predation. J. Bacteriol. 2011, 193, 5081–5089. [Google Scholar] [CrossRef] [Green Version]
- Steffan, S.A.; Chikaraishi, Y.; Currie, C.R.; Horn, H.; Gaines-Day, H.R.; Pauli, J.N.; Zalapa, J.E.; Ohkouchi, N. Microbes are trophic analogs of animals. Proc. Natl. Acad. Sci. USA 2015, 112, 15119–15124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heithaus, M.R. Habitat selection by predators and prey in communities with asymmetrical intraguild predation. Oikos 2001, 92, 542–554. [Google Scholar] [CrossRef]
- Ritchie, E.G.; Johnson, C.N. Predator interactions, mesopredator release and biodiversity conservation. Ecol. Lett. 2009, 12, 982–998. [Google Scholar] [CrossRef]
- Hodgkin, J.; Kaiser, D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Two gene systems control movement. Mol. Genet. Genom. MGG 1979, 171, 177–191. [Google Scholar] [CrossRef]
- Velicer, G.J.; Raddatz, G.; Keller, H.; Deiss, S.; Lanz, C.; Dinkelacker, I.; Schuster, S.C. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. USA 2006, 103, 8107–8112. [Google Scholar] [CrossRef] [Green Version]
- Velicer, G.J.; Yu, Y.N. Evolution of novel cooperative swarming in the bacterium Myxococcus xanthus. Nature 2003, 425, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Stiernagle, T. Maintenance of C. elegans. WormBook 2006. [Google Scholar] [CrossRef] [Green Version]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Hagen, D.C.; Bretscher, A.P.; Kaiser, D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 1978, 64, 284–296. [Google Scholar] [CrossRef]
- Pires-daSilva, A. Pristionchus pacificus protocols. WormBook 2013, 1–20. [Google Scholar] [CrossRef]
- Kauffman, A.; Parsons, L.; Stein, G.; Wills, A.; Kaletsky, R.; Murphy, C. C. elegans Positive Butanone Learning, Short-term, and Long-term Associative Memory Assays. JoVE J. Vis. Exp. 2011, e2490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bretscher, A.P.; Kaiser, D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 1978, 133, 763–768. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- RStudio|Open Source & Professional Software for Data Science Teams. Available online: https://rstudio.com/ (accessed on 10 December 2020).
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 10 December 2020).
- Lenth, R.V.; Buerkner, P.; Herve, M.; Love, J.; Riebl, H.; Singmann, H. Emmeans: Estimated Marginal Means, aka Least-Squares Means. 2020. Available online: https://cran.r-project.org/web/packages/emmeans/index.html (accessed on 10 December 2020).
- Moore, R.S.; Kaletsky, R.; Murphy, C.T. Piwi/PRG-1 Argonaute and TGF-β Mediate Transgenerational Learned Pathogenic Avoidance. Cell 2019, 177, 1827–1841.e12. [Google Scholar] [CrossRef]
- Brook, L.A.; Johnson, C.N.; Ritchie, E.G. Effects of predator control on behaviour of an apex predator and indirect consequences for mesopredator suppression. J. Appl. Ecol. 2012, 49, 1278–1286. [Google Scholar] [CrossRef]
- Crooks, K.R.; Soulé, M.E. Mesopredator release and avifaunal extinctions in a fragmented system. Nature 1999, 400, 563–566. [Google Scholar] [CrossRef]
- Wikenros, C.; Ståhlberg, S.; Sand, H. Feeding under high risk of intraguild predation: Vigilance patterns of two medium-sized generalist predators. J. Mammal. 2014, 95, 862–870. [Google Scholar] [CrossRef] [Green Version]
- Félix, M.-A.; Duveau, F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Shtonda, B.B.; Avery, L. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 2006, 209, 89–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meisel, J.D.; Kim, D.H. Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol. 2014, 35, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Keane, R.; Berleman, J. The predatory life cycle of Myxococcus xanthus. Microbiol. Read. Engl. 2016, 162, 1–11. [Google Scholar] [CrossRef]
- Berleman, J.E.; Allen, S.; Danielewicz, M.A.; Remis, J.P.; Gorur, A.; Cunha, J.; Hadi, M.Z.; Zusman, D.R.; Northen, T.R.; Witkowska, H.E.; et al. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front. Microbiol. 2014, 5, 474. [Google Scholar] [CrossRef]
- Thiery, S.; Kaimer, C. The Predation Strategy of Myxococcus xanthus. Front. Microbiol. 2020, 11, 2. [Google Scholar] [CrossRef]
- Findlay, B.L. The Chemical Ecology of Predatory Soil Bacteria. ACS Chem. Biol. 2016, 11, 1502–1510. [Google Scholar] [CrossRef]
- Pradel, E.; Zhang, Y.; Pujol, N.; Matsuyama, T.; Bargmann, C.I.; Ewbank, J.J. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2007, 104, 2295–2300. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.T.; Alvarez, I.V.; Saïdi, F.; Guiseppi, A.; Vinogradov, E.; Sharma, G.; Espinosa, L.; Morrone, C.; Brasseur, G.; Guillemot, J.-F.; et al. Modulation of bacterial multicellularity via spatio-specific polysaccharide secretion. PLoS Biol. 2020, 18, e3000728. [Google Scholar] [CrossRef] [PubMed]
- Stasulli, N.M.; Shank, E.A. Profiling the metabolic signals involved in chemical communication between microbes using imaging mass spectrometry. FEMS Microbiol. Rev. 2016, 40, 807–813. [Google Scholar] [CrossRef] [Green Version]
- Wikenros, C.; Jarnemo, A.; Frisén, M.; Kuijper, D.P.J.; Schmidt, K. Mesopredator behavioral response to olfactory signals of an apex predator. J. Ethol. 2017, 35, 161–168. [Google Scholar] [CrossRef] [Green Version]
- Hauzy, C.; Hulot, F.D.; Gins, A.; Loreau, M. Intra- and interspecific density-dependent dispersal in an aquatic prey–predator system. J. Anim. Ecol. 2007, 76, 552–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shores, C.R.; Dellinger, J.A.; Newkirk, E.S.; Kachel, S.M.; Wirsing, A.J. Mesopredators change temporal activity in response to a recolonizing apex predator. Behav. Ecol. 2019, 30, 1324–1335. [Google Scholar] [CrossRef]
- Creel, S.; Christianson, D.; Liley, S.; Winnie, J.A. Predation Risk Affects Reproductive Physiology and Demography of Elk. Science 2007, 315, 960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creel, S.; Christianson, D. Relationships between direct predation and risk effects. Trends Ecol. Evol. 2008, 23, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Ripple, W.J.; Beschta, R.L. Wolves and the Ecology of Fear: Can Predation Risk Structure Ecosystems? BioScience 2004, 54, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Elmhagen, B.; Rushton, S.P. Trophic control of mesopredators in terrestrial ecosystems: Top-down or bottom-up? Ecol. Lett. 2007, 10, 197–206. [Google Scholar] [CrossRef]
- Lynam, C.P.; Llope, M.; Möllmann, C.; Helaouët, P.; Bayliss-Brown, G.A.; Stenseth, N.C. Interaction between top-down and bottom-up control in marine food webs. Proc. Natl. Acad. Sci. USA 2017, 114, 1952–1957. [Google Scholar] [CrossRef] [Green Version]
- Whalen, M.A.; Duffy, J.E.; Grace, J.B. Temporal shifts in top-down vs. bottom-up control of epiphytic algae in a seagrass ecosystem. Ecology 2013, 94, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Elmhagen, B.; Ludwig, G.; Rushton, S.P.; Helle, P.; Lindén, H. Top predators, mesopredators and their prey: Interference ecosystems along bioclimatic productivity gradients. J. Anim. Ecol. 2010, 79, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Ellner, S.P.; Becks, L. Rapid prey evolution and the dynamics of two-predator food webs. Theor. Ecol. 2010, 4, 133–152. [Google Scholar] [CrossRef]
- Werner, E.E.; Peacor, S.D. A Review of Trait-Mediated Indirect Interactions in Ecological Communities. Ecology 2003, 84, 1083–1100. [Google Scholar] [CrossRef] [Green Version]
- Hiltunen, T.; Jones, L.E.; Ellner, S.P.; Hairston, N.G. Temporal dynamics of a simple community with intraguild predation: An experimental test. Ecology 2013, 94, 773–779. [Google Scholar] [CrossRef] [Green Version]
- Hiltunen, T.; Ayan, G.B.; Becks, L. Environmental fluctuations restrict eco-evolutionary dynamics in predator–prey system. Proc. R. Soc. B Biol. Sci. 2015, 282, 20150013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaitala, V.; Hiltunen, T.; Becks, L.; Scheuerl, T. Co-evolution as an important component explaining microbial predator-prey interaction. J. Theor. Biol. 2020, 486, 110095. [Google Scholar] [CrossRef] [PubMed]
- Scheuerl, T.; Cairns, J.; Becks, L.; Hiltunen, T. Predator coevolution and prey trait variability determine species coexistence. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190245. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayrhofer, N.; Velicer, G.J.; Schaal, K.A.; Vasse, M. Behavioral Interactions between Bacterivorous Nematodes and Predatory Bacteria in a Synthetic Community. Microorganisms 2021, 9, 1362. https://doi.org/10.3390/microorganisms9071362
Mayrhofer N, Velicer GJ, Schaal KA, Vasse M. Behavioral Interactions between Bacterivorous Nematodes and Predatory Bacteria in a Synthetic Community. Microorganisms. 2021; 9(7):1362. https://doi.org/10.3390/microorganisms9071362
Chicago/Turabian StyleMayrhofer, Nicola, Gregory J. Velicer, Kaitlin A. Schaal, and Marie Vasse. 2021. "Behavioral Interactions between Bacterivorous Nematodes and Predatory Bacteria in a Synthetic Community" Microorganisms 9, no. 7: 1362. https://doi.org/10.3390/microorganisms9071362
APA StyleMayrhofer, N., Velicer, G. J., Schaal, K. A., & Vasse, M. (2021). Behavioral Interactions between Bacterivorous Nematodes and Predatory Bacteria in a Synthetic Community. Microorganisms, 9(7), 1362. https://doi.org/10.3390/microorganisms9071362





