Subfractional Spectrum of Serum Lipoproteins and Gut Microbiota Composition in Healthy Individuals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Recruitment of Study Participants
2.2. Sample Collection and Gut Microbiota Analysis
2.3. Lipoproteins Subfractional Spectrum Analysis
2.4. Intima-Media Thickness Assessment
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Gut Microbiota Composition
3.2. Association between the Gut Microbiota Composition and Subfractional Spectrum of Apo B-Containing Lipoproteins
3.3. Association between Gut Microbiota Diversity and Metabolic Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the Management of Dyslipidaemias: Lipid Modification to Reduce Cardiovascular Risk. Atherosclerosis 2019, 290. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-J.; Zhang, Y.; Li, S.; Cui, C.-J.; Zhu, C.-G.; Guo, Y.-L.; Wu, N.-Q.; Xu, R.-X.; Liu, G.; Dong, Q.; et al. Large HDL Subfraction But Not HDL-C Is Closely Linked with Risk Factors, Coronary Severity and Outcomes in a Cohort of Nontreated Patients With Stable Coronary Artery Disease: A Prospective Observational Study. Medicine 2016, 95. [Google Scholar] [CrossRef]
- Hernáez, Á.; Soria-Florido, M.T.; Schröder, H.; Ros, E.; Pintó, X.; Estruch, R.; Salas-Salvadó, J.; Corella, D.; Arós, F.; Serra-Majem, L.; et al. Role of HDL Function and LDL Atherogenicity on Cardiovascular Risk: A Comprehensive Examination. PLoS ONE 2019, 14, e0218533. [Google Scholar] [CrossRef]
- Le Roy, T.; Lécuyer, E.; Chassaing, B.; Rhimi, M.; Lhomme, M.; Boudebbouze, S.; Ichou, F.; Haro Barceló, J.; Huby, T.; Guerin, M.; et al. The Intestinal Microbiota Regulates Host Cholesterol Homeostasis. BMC Biol. 2019, 17, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kang, J.; Choi, S.; Park, H.; Hwang, E.; Kang, Y.; Kim, A.; Holzapfel, W.; Ji, Y. Cholesterol-Lowering Effect of Lactobacillus Rhamnosus BFE5264 and Its Influence on the Gut Microbiome and Propionate Level in a Murine Model. PLoS ONE 2018, 13, e0203150. [Google Scholar] [CrossRef] [Green Version]
- Antharam, V.C.; McEwen, D.C.; Garrett, T.J.; Dossey, A.T.; Li, E.C.; Kozlov, A.N.; Mesbah, Z.; Wang, G.P. An Integrated Metabolomic and Microbiome Analysis Identified Specific Gut Microbiota Associated with Fecal Cholesterol and Coprostanol in Clostridium Difficile Infection. PLoS ONE 2016, 11, e0148824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriaa, A.; Bourgin, M.; Potiron, A.; Mkaouar, H.; Jablaoui, A.; Gérard, P.; Maguin, E.; Rhimi, M. Microbial Impact on Cholesterol and Bile Acid Metabolism: Current Status and Future Prospects. J. Lipid Res. 2019, 60, 323. [Google Scholar] [CrossRef] [Green Version]
- Prete, R.; Long, S.L.; Gallardo, A.L.; Gahan, C.G.; Corsetti, A.; Joyce, S.A. Beneficial Bile Acid Metabolism from Lactobacillus Plantarum of Food Origin. Sci. Rep. 2020, 10, 1165. [Google Scholar] [CrossRef] [Green Version]
- Kashtanova, D.A.; Tkacheva, O.N.; Doudinskaya, E.N.; Strazhesko, I.D.; Kotovskaya, Y.V.; Popenko, A.S.; Tyakht, A.V.; Alexeev, D.G. Gut Microbiota in Patients with Different Metabolic Statuses: Moscow Study. Microorganisms 2018, 6, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efimova, D.; Tyakht, A.; Popenko, A.; Vasilyev, A.; Altukhov, I.; Dovidchenko, N.; Odintsova, V.; Klimenko, N.; Loshkarev, R.; Pashkova, M.; et al. Knomics-Biota—A System for Exploratory Analysis of Human Gut Microbiota Data. BioData Min. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera-Pinto, J.; Egozcue, J.J.; Pawlowsky-Glahn, V.; Paredes, R.; Noguera-Julian, M.; Calle, M.L. Balances: A New Perspective for Microbiome Analysis. MSystems 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palarea-Albaladejo, J.; Martín-Fernández, J.A. zCompositions—R Package for Multivariate Imputation of Left-Censored Data under a Compositional Approach. Chemom. Intell. Lab. Syst. 2015, 143, 85–96. [Google Scholar] [CrossRef]
- Tyakht, A.V.; Kostryukova, E.S.; Popenko, A.S.; Belenikin, M.S.; Pavlenko, A.V.; Larin, A.K.; Karpova, I.Y.; Selezneva, O.V.; Semashko, T.A.; Ospanova, E.A.; et al. Human Gut Microbiota Community Structures in Urban and Rural Populations in Russia. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Giuffrè, M.; Campigotto, M.; Campisciano, G.; Comar, M.; Crocè, L.S. A Story of Liver and Gut Microbes: How Does the Intestinal Flora Affect Liver Disease? A Review of the Literature. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G889–G906. [Google Scholar] [CrossRef]
- Xu, Z.; McClure, S.; Appel, L. Dietary Cholesterol Intake and Sources among U.S Adults: Results from National Health and Nutrition Examination Surveys (NHANES), 2001–2014. Nutrients 2018, 10, 771. [Google Scholar] [CrossRef] [Green Version]
- Annema, W.; von Eckardstein, A. High-Density Lipoproteins. Multifunctional but Vulnerable Protections from Atherosclerosis. Circ. J. 2013, 77, 2432–2448. [Google Scholar] [CrossRef] [Green Version]
- Feingold, K.R. Introduction to Lipids and Lipoproteins; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., de Herder, W.W., Dhatariya, K., Dungan, K., Grossman, A., Hershman, J.M., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2021. [Google Scholar] [PubMed]
- Larsen, J.M. The Immune Response to Prevotella Bacteria in Chronic Inflammatory Disease. Immunology 2017, 151. [Google Scholar] [CrossRef] [Green Version]
- Iljazovic, A.; Roy, U.; Gálvez, E.J.C.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C.; et al. Perturbation of the Gut Microbiome by Prevotella Spp. Enhances Host Susceptibility to Mucosal Inflammation. Mucosal Immunol. 2020, 14, 113–124. [Google Scholar] [CrossRef]
- Hu, H.-J.; Park, S.-G.; Jang, H.B.; Choi, M.-G.; Park, K.-H.; Kang, J.H.; Park, S.I.; Lee, H.-J.; Cho, S.-H. Obesity Alters the Microbial Community Profile in Korean Adolescents. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut Microbiota Dysbiosis Contributes to the Development of Hypertension. Microbiome 2017, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K.; et al. Disentangling the Effects of Type 2 Diabetes and Metformin on the Human Gut Microbiota. Nature 2015, 528, 262. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Ma, L.; Ma, Y.; Zhang, F.; Zhao, C.; Nie, Y. Insights into the Role of Gut Microbiota in Obesity: Pathogenesis, Mechanisms, and Therapeutic Perspectives. Protein Cell 2018, 9, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the Gut Microbiota in Nutrition and Health. BMJ 2018, 361. [Google Scholar] [CrossRef] [Green Version]

| Total Cohort (n = 304) | Males (n = 104) | Females (n = 200) | p-Value | FDR | Young (n = 139) | Age Risk Group (n = 165) | p-Value | FDR | |
|---|---|---|---|---|---|---|---|---|---|
| Age, years | 51.5 ± 13.26 | 48.1 ± 12.03 | 53.27 ± 13.55 | 0.0008 | 0.0014 | 40.45 ± 8.55 | 60.82 ± 8.52 | - | - |
| BMI, kg/m2 | 27.36 ± 5.14 | 28.59 ± 4.22 | 26.72 ± 5.46 | 0.0011 | 0.0017 | 26.5 ± 5.55 | 28.08 ± 4.67 | 0.0086 | 0.0118 |
| SBP, mmHg | 125.33 ± 16.42 | 129.52 ± 14.61 | 123.14 ± 16.91 | 0.0007 | 0.0014 | 120.09 ± 15.26 | 129.74 ± 16.11 | <0.00001 | <0.00001 |
| DBP, mmHg | 78.16 ± 10.26 | 79.88 ± 10.16 | 77.26 ± 10.22 | 0.0342 | 0.0376 | 77.24 ± 10.39 | 78.93 ± 10.12 | 0.1555 | 0.1711 |
| Smokers, n | 58 (19%) | 30 (29%) | 28 (14%) | 0.0171 | 0.0210 | 27 (19%) | 31 (19%) | 1.0000 | 1.0000 |
| TC, mmol/L | 5.65 ± 1.14 | 5.61 ± 1.02 | 5.68 ± 1.21 | 0.5958 | 0.5958 | 5.4 ± 0.97 | 5.86 ± 1.24 | 0.0003 | 0.0005 |
| TG, mmol/L | 1.28 ± 0.85 | 1.6 ± 1.07 | 1.12 ± 0.65 | <0.00001 | 0.0001 | 1.09 ± 0.68 | 1.45 ± 0.94 | 0.0001 | 0.0002 |
| HDL-C, mmol/L | 1.22 ± 0.31 | 1.03 ± 0.27 | 1.32 ± 0.29 | <0.00001 | <0.00001 | 1.25 ± 0.31 | 1.19 ± 0.32 | 0.1078 | 0.1318 |
| AIP | −0.07 ± 0.72 | 0.31 ± 0.72 | −0.27 ± 0.64 | <0.00001 | <0.00001 | −0.27 ± 0.71 | 0.09 ± 0.7 | <0.00001 | <0.00001 |
| Fasting plasma glucose, mmol/L | 5.75 ± 1.4 | 6.1 ± 1.61 | 5.56 ± 1.24 | 0.0036 | 0.0049 | 5.3 ± 0.94 | 6.12 ± 1.6 | <0.00001 | <0.00001 |
| WC, cm | 89.54 ± 15.3 | 98.67 ± 12.46 | 84.84 ± 14.5 | <0.00001 | <0.00001 | 85.17 ± 15.01 | 93.21 ± 14.59 | <0.00001 | <0.00001 |
| Cholesterol, mg/dL | Total Cohort (n = 304) | Males (n = 104) | Females (n = 200) | p-Value | FDR | Young (n = 139) | Age Risk Group (n = 165) | p-Value | FDR |
|---|---|---|---|---|---|---|---|---|---|
| VLDL | 15.49 ± 4.44 | 17.14 ± 4.10 | 14.68 ± 4.39 | 0.0002 | 0.0012 | 14.87 ± 4.29 | 16.36 ± 4.53 | 0.0223 | 0.0580 |
| IDL-C | 23.08 ± 8.32 | 23.49 ± 7.66 | 22.88 ± 8.65 | 0.6225 | 0.6744 | 22.38 ± 8.17 | 24.08 ± 8.48 | 0.1662 | 0.2700 |
| IDL-B | 18.92 ± 5.88 | 18.89 ± 4.34 | 18.93 ± 6.53 | 0.9644 | 0.9644 | 17.39 ± 4.86 | 21.08 ± 6.52 | <0.00001 | 0.0002 |
| IDL-A | 21.34 ± 8.09 | 20.38 ± 8.47 | 21.82 ± 7.89 | 0.2569 | 0.4564 | 20.04 ± 7.77 | 23.18 ± 8.22 | 0.0084 | 0.0273 |
| LDL-1 | 39.77 ± 13.7 | 38.08 ± 12.68 | 40.61 ± 14.15 | 0.2109 | 0.4564 | 38 ± 11.84 | 42.27 ± 15.71 | 0.0417 | 0.0902 |
| LDL-2 | 18.8 ± 12.27 | 21.17 ± 11.99 | 17.63 ± 12.28 | 0.0573 | 0.2484 | 16.2 ± 11.23 | 22.48 ± 12.79 | 0.0006 | 0.0024 |
| LDL-3 | 3.65 ± 5.00 | 4.61 ± 5.82 | 3.17 ± 4.49 | 0.0853 | 0.2773 | 3.19 ± 4.88 | 4.3 ± 5.12 | 0.1310 | 0.2434 |
| LDL-4 | 0.61 ± 2.15 | 0.86 ± 3.29 | 0.49 ± 1.24 | 0.3862 | 0.4564 | 0.58 ± 2.58 | 0.65 ± 1.33 | 0.8172 | 0.8172 |
| LDL-5 | 0.08 ± 0.88 | 0.20 ± 1.50 | 0.02 ± 0.18 | 0.3234 | 0.4564 | 0.12 ± 1.14 | 0.01 ± 0.11 | 0.3055 | 0.3776 |
| LDL-6 | 0.01 ± 0.14 | 0.03 ± 0.25 | 0.00 ± 0.00 | 0.3211 | 0.4564 | 0.02 ± 0.19 | 0 ± 0 | 0.3195 | 0.3776 |
| LDL-7 | 0.21 ± 2.40 | 0.03 ± 0.25 | 0.30 ± 2.93 | 0.2984 | 0.4564 | 0.02 ± 0.19 | 0.49 ± 3.71 | 0.2618 | 0.3776 |
| Total LDL | 127.23 ± 34.74 | 130.20 ± 27.56 | 125.75 ± 37.84 | 0.3551 | 0.4564 | 118.26 ± 29.49 | 140.08 ± 37.72 | <0.00001 | 0.0002 |
| HDL | 56.16 ± 15.3 | 47.83 ± 13.21 | 60.29 ± 14.61 | <0.00001 | <0.00001 | 56.94 ± 14.19 | 55.05 ± 16.78 | 0.4135 | 0.4480 |
| Cholesterol, mg/dL | Linear Regression Coefficient | p-Value | FDR Adjusted p-Value |
|---|---|---|---|
| VLDL | 0.0136 | <0.00001 | 0.0001 |
| IDL-C | 0.0035 | 0.0324 | 0.0789 |
| IDL-B | 0.0080 | 0.0004 | 0.0025 |
| IDL-A | 0.0016 | 0.3564 | 0.4633 |
| LDL-1 | −0.0011 | 0.2864 | 0.4137 |
| LDL-2 | 0.0025 | 0.0237 | 0.0769 |
| LDL-3 | 0.0056 | 0.0364 | 0.0789 |
| LDL-4 | 0.0089 | 0.1543 | 0.2508 |
| LDL-5 | −0.0022 | 0.8844 | 0.8844 |
| LDL-6 | −0.0282 | 0.7638 | 0.8275 |
| LDL-7 | 0.0098 | 0.0801 | 0.1487 |
| Total LDL | 0.0009 | 0.0173 | 0.0751 |
| HDL | −0.0004 | 0.6894 | 0.8147 |
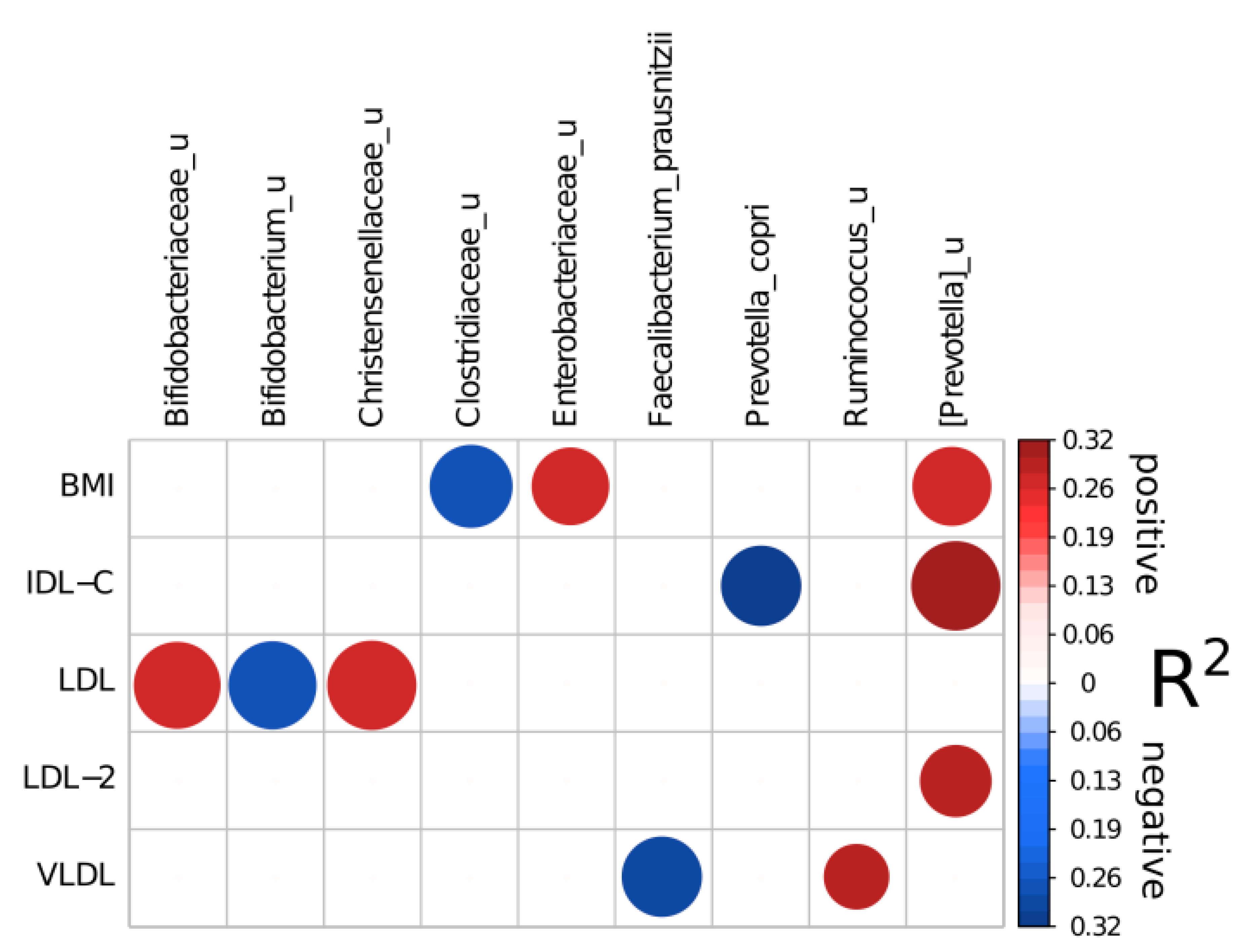
| Factor | Full Balance | Adjusted R2 | Reproducible Taxon | Direction of Association | Percent of Times Included in a Balance |
|---|---|---|---|---|---|
| BMI | [Prevotella]_u | Enterobacteriaceae_u)/Clostridiaceae_u | 0.26515 | [Prevotella]_u | + | 72.5 |
| Enterobacteriaceae_u | + | 70 | |||
| Clostridiaceae_u | - | 80 | |||
| LDL | Bifidobacteriaceae_u|Christensenellaceae_u/Bifidobacterium_u | 0.26381 | Bifidobacteriaceae_u | + | 87.5 |
| Christensenellaceae_u | + | 92.5 | |||
| Bifidobacterium_u | - | 90 | |||
| VLDL | Ruminococcus_u/Faecalibacterium_prausnitzii | 0.29299 | Ruminococcus_u | + | 50 |
| Faecalibacterium_prausnitzii | - | 75 | |||
| IDL-C | [Prevotella]_u/Prevotella_copri | 0.32018 | [Prevotella]_u | + | 92.5 |
| Prevotella_copri | - | 75 | |||
| LDL-2 | [Prevotella]_u/[Ruminococcus]_gnavus | 0.28927 | [Prevotella]_u | + | 60 |
| p-Value | Linear Model Coefficient | FDR | Alpha Diversity Metric | |
|---|---|---|---|---|
| BMI | 0.0017 | −17.5031 | 0.0250 | Chao1 |
| Abdominal obesity | 0.0083 | −194.5565 | 0.0622 | Chao1 |
| BMI | 0.0029 | −0.0344 | 0.0147 | Shannon |
| Abdominal obesity | 0.0018 | −0.3968 | 0.0138 | Shannon |
| AIP | 0.0015 | −0.3065 | 0.0138 | Shannon |
| HDL-C | 0.0050 | 0.6334 | 0.0187 | Shannon |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashtanova, D.A.; Klimenko, N.S.; Tkacheva, O.N.; Strazhesko, I.D.; Metelskaya, V.A.; Gomyranova, N.V.; Boytsov, S.A. Subfractional Spectrum of Serum Lipoproteins and Gut Microbiota Composition in Healthy Individuals. Microorganisms 2021, 9, 1461. https://doi.org/10.3390/microorganisms9071461
Kashtanova DA, Klimenko NS, Tkacheva ON, Strazhesko ID, Metelskaya VA, Gomyranova NV, Boytsov SA. Subfractional Spectrum of Serum Lipoproteins and Gut Microbiota Composition in Healthy Individuals. Microorganisms. 2021; 9(7):1461. https://doi.org/10.3390/microorganisms9071461
Chicago/Turabian StyleKashtanova, Daria A., Natalia S. Klimenko, Olga N. Tkacheva, Irina D. Strazhesko, Victoria A. Metelskaya, Natalia V. Gomyranova, and Sergey A. Boytsov. 2021. "Subfractional Spectrum of Serum Lipoproteins and Gut Microbiota Composition in Healthy Individuals" Microorganisms 9, no. 7: 1461. https://doi.org/10.3390/microorganisms9071461






