Analysis of 56,348 Genomes Identifies the Relationship between Antibiotic and Metal Resistance and the Spread of Multidrug-Resistant Non-Typhoidal Salmonella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Salmonella enterica Genome Assembly Acquisition
2.2. Identification of Plasmid Replicons, Metal and Antibiotic Resistance Homologues
2.3. Co-Occurrence Identification
2.4. Phylogeny and S. enterica I,4,[5],12:i:- Analysis
3. Results
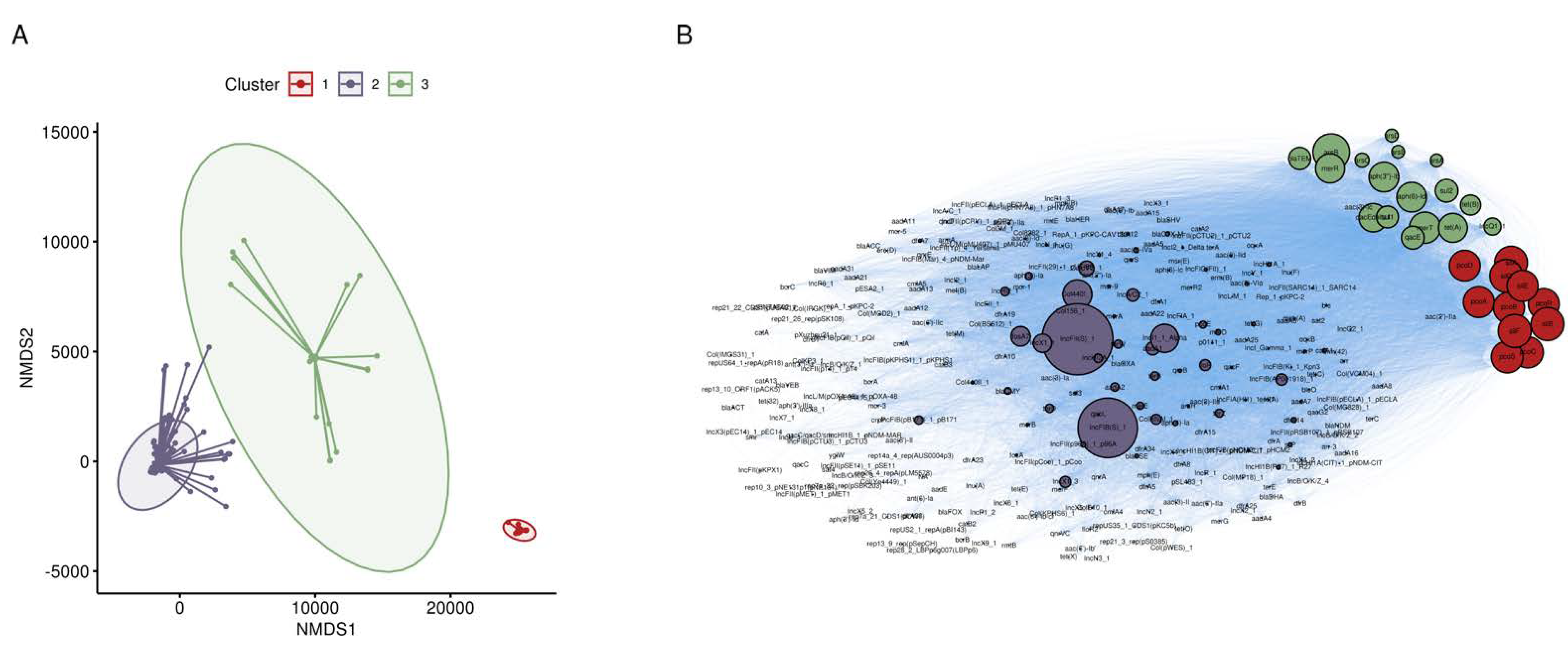
3.1. Broad Screen for Metal and Antibiotic Co-Occurrence in S. enterica
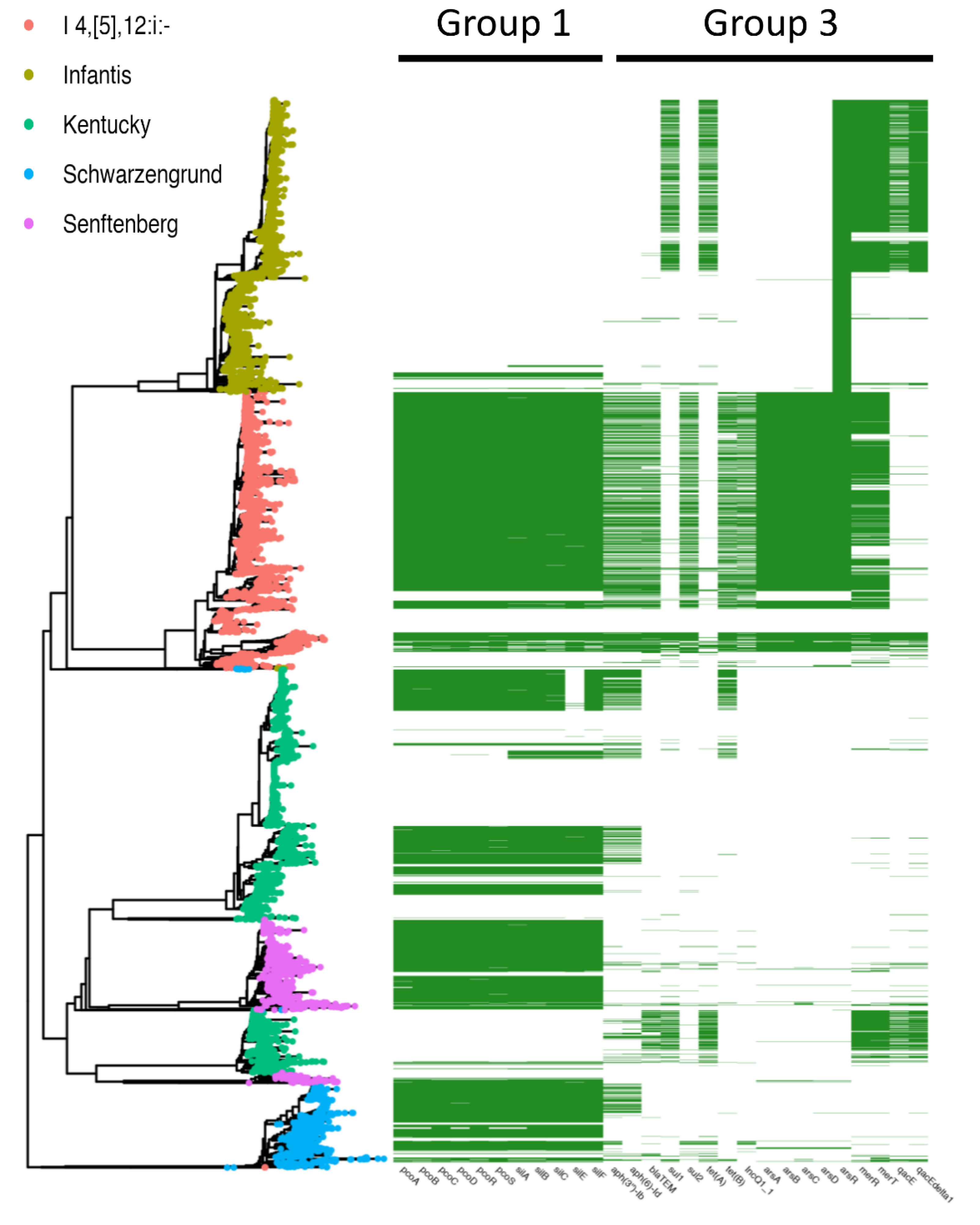
3.2. Co-Occurrence of Metal and Antibiotic Resistance
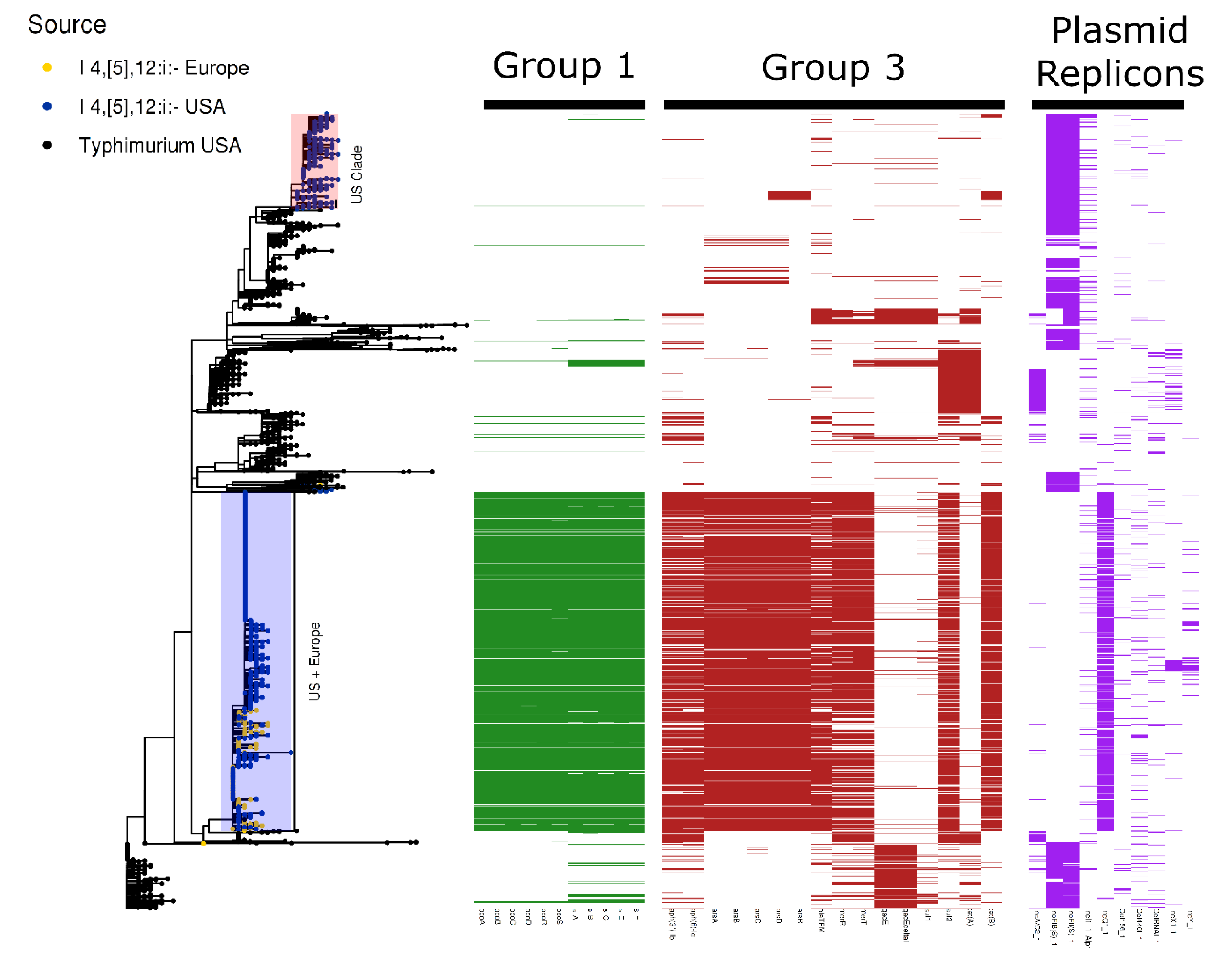
3.3. S. enterica I 4,[5],12:i:-
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 27 April 2020).
- Singh, A.K.; Bhunia, A.K. Animal-Use Antibiotics Induce Cross-Resistance in Bacterial Pathogens to Human Therapeutic Antibiotics. Curr. Microbiol. 2019, 76, 1112–1117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Espinosa, C.D.; Abelilla, J.J.; Casas, G.A.; Lagos, L.V.; Lee, S.A.; Kwon, W.B.; Mathai, J.K.; Navarro, D.M.; Jaworski, N.W.; et al. Non-antibiotic feed additives in diets for pigs: A review. Anim. Nutr. 2018, 4, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, C.B.; Woodworth, J.C.; DeRouchey, J.M.; Tokach, M.D.; Goodband, R.D.; Dritz, S.S.; Wu, F.; Usry, J.L. Effects of increasing copper from tri-basic copper chloride or a copper-methionine chelate on growth performance of nursery pigs1,2. Transl. Anim. Sci. 2018, 3, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Lucas, I.A.M.; Livingstone, R.M.; McDonald, I. Copper sulphate as a growth stimulant for pigs: Effect of level and purity. Anim. Sci. 1961, 3, 111–119. [Google Scholar] [CrossRef]
- Slade, R.; Kyriazakis, I.; Carroll, S.; Reynolds, F.; Wellock, I.; Broom, L.; Miller, H. Effect of rearing environment and dietary zinc oxide on the response of group-housed weaned pigs to enterotoxigenic Escherichia coli O149 challenge. Animal 2011, 5, 1170–1178. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Dong, Z.; Li, G.; Wang, J.; Li, Y.; Wan, D.; Yang, H.; Yin, Y. Effect of Dietary Copper on Intestinal Microbiota and Antimicrobial Resistance Profiles of Escherichia coli in Weaned Piglets. Front. Microbiol. 2019, 10, 2808. [Google Scholar] [CrossRef]
- Hasman, H.; Aarestrup, F.M. tcrB, a Gene Conferring Transferable Copper Resistance in Enterococcus faecium: Occurrence, Transferability, and Linkage to Macrolide and Glycopeptide Resistance. Antimicrob. Agents Chemother. 2002, 46, 1410. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Dickinson, A.; Power, A.; Hansen, M.; Brandt, K.; Piliposian, G.; Appleby, P.; O’Neill, P.; Jones, R.; Sierocinski, P.; Koskella, B.; et al. Heavy metal pollution and co-selection for antibiotic resistance: A microbial palaeontology approach. Environ. Int. 2019, 132, 105117. [Google Scholar] [CrossRef]
- Branchu, P.; Charity, O.; Bawn, M.; Thilliez, G.; Dallman, T.J.; Petrovska, L.; Kingsley, R.A. SGI-4 in Monophasic Salmonella Typhimurium ST34 Is a Novel ICE That Enhances Resistance to Copper. Front. Microbiol. 2019, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Mastrorilli, E.; Pietrucci, D.; Barco, L.; Ammendola, S.; Petrin, S.; Longo, A.; Mantovani, C.; Battistoni, A.; Ricci, A.; Desideri, A.; et al. A Comparative Genomic Analysis Provides Novel Insights Into the Ecological Success of the Monophasic Salmonella Serovar 4,[5],12:i:-. Front. Microbiol. 2018, 9, 715. [Google Scholar] [CrossRef] [PubMed]
- Dyall-Smith, M.L.; Liu, Y.; Billman-Jacobe, H. Genome Sequence of an Australian Monophasic Salmonella enterica subsp. enterica Typhimurium Isolate (TW-Stm6) Carrying a Large Plasmid with Multiple Antimicrobial Resistance Genes. Genome Announc. 2017, 5, e00793-17. [Google Scholar] [CrossRef] [PubMed]
- Petrovska, L.; Mather, A.E.; AbuOun, M.; Branchu, P.; Harris, S.R.; Connor, T.; Hopkins, K.L.; Underwood, A.; Lettini, A.A.; Page, A.; et al. Microevolution of Monophasic Salmonella Typhimurium during Epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. J. 2016, 22, 617. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.J.; Nash, J.H.E.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate. 2018. Available online: https://github.com/tseemann/abricate (accessed on 11 October 2019).
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In SilicoDetection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Suzuki, S.; Kakuta, M.; Ishida, T.; Akiyama, Y. Faster sequence homology searches by clustering subsequences. Bioinformatics 2015, 31, 1183–1190. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Inter. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Bateman, A.; Durbin, R. QuickTree: Building huge Neighbour-Joining trees of protein sequences. Bioinformatics 2002, 18, 1546–1547. [Google Scholar] [CrossRef]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads. 2015. Available online: https://github.com/tseemann/snippy (accessed on 15 April 2020).
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- European Food Safety, Authority and European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar]
- CDC. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet 2015 Surveillance Report (Final Data). 2017. Available online: https://www.cdc.gov/foodnet/reports/index.html (accessed on 27 March 2020).
- Branchu, P.; Bawn, M.; Kingsley, R.A. Genome Variation and Molecular Epidemiology of Salmonella enterica Serovar Typhimurium Pathovariants. Infect. Immun. 2018, 86, 00079-18. [Google Scholar] [CrossRef] [PubMed]
- Falgenhauer, L.; Ghosh, H.; Guerra, B.; Yao, Y.; Fritzenwanker, M.; Fischer, J.; Helmuth, R.; Imirzalioglu, C.; Chakraborty, T. Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Veter- Microbiol. 2017, 200, 114–117. [Google Scholar] [CrossRef]
- Nair, D.; Gupta, N.; Kabra, S.; Ahuja, R.B.; Prakash, S. Salmonella senftenberg: A new pathogen in the burns ward. Burns 1999, 25, 723–727. [Google Scholar] [CrossRef]
- Scott, A.; Vadalasetty, K.P.; Łukasiewicz, M.; Jaworski, S.; Wierzbicki, M.; Chwalibog, A.; Sawosz, E. Effect of different levels of copper nanoparticles and copper sulphate on performance, metabolism and blood biochemical profiles in broiler chicken. J. Anim. Physiol. Anim. Nutr. 2017, 102, e364–e373. [Google Scholar] [CrossRef]
- Swiatkiewicz, S.; Arczewska-Włosek, A.; Józefiak, D. The efficacy of organic minerals in poultry nutrition: Review and implications of recent studies. World’s Poult. Sci. J. 2014, 70, 475–486. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Hendriksen, R.S.; Lockett, J.; Gay, K.; Teates, K.; McDermott, P.F.; White, D.G.; Hasman, H.; Sørensen, G.; Bangtrakulnonth, A.; et al. International Spread of Multidrug-resistantSalmonellaSchwarzengrund in Food Products. Emerg. Infect. Dis. 2007, 13, 726–731. [Google Scholar] [CrossRef]
- Yang, S.; Deng, W.; Liu, S.; Yu, X.; Mustafa, G.R.; Chen, S.; He, L.; Ao, X.; Yang, Y.; Zhou, K.; et al. Presence of heavy metal resistance genes in Escherichia coli and Salmonella isolates and analysis of resistance gene structure in E. coli E308. J. Glob. Antimicrob. Resist. 2020, 21, 420–426. [Google Scholar] [CrossRef]
- Wilson, A.; Fox, E.; Fegan, N.; Kurtböke, D. Ípek Comparative Genomics and Phenotypic Investigations Into Antibiotic, Heavy Metal, and Disinfectant Susceptibilities of Salmonella enterica Strains Isolated in Australia. Front. Microbiol. 2019, 10, 1620. [Google Scholar] [CrossRef]
- Mourao, J.; Marçal, S.; Ramos, P.; Campos, J.; Machado, J.; Peixe, L.; Noais, C.; Antunes, P. Tolerance to multiple metal stressors in emerging non-typhoidal MDR Salmonella serotypes: A relevant role for copper in anaerobic conditions. J. Antimicrob. Chemother. 2016, 71, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Billman-Jacobe, H.; Liu, Y.; Haites, R.; Weaver, T.; Robinson, L.; Marenda, M.; Dyall-Smith, M. pSTM6-275, a Conjugative IncHI2 Plasmid of Salmonella enterica That Confers Antibiotic and Heavy-Metal Resistance under Changing Physiological Conditions. Antimicrob. Agents Chemother. 2018, 62, e02357-17. [Google Scholar] [CrossRef] [PubMed]
- Arai, N.; Sekizuka, T.; Tamamura, Y.; Kusumoto, M.; Hinenoya, A.; Yamasaki, S.; Iwata, T.; Watanabe-Yanai, A.; Kuroda, M.; Akiba, M. Salmonella Genomic Island 3 Is an Integrative and Conjugative Element and Contributes to Copper and Arsenic Tolerance of Salmonella enterica. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Elnekave, E.; Hong, S.; Mather, A.E.; Boxrud, D.; Taylor, A.J.; Lappi, V.; Johnson, T.; Vannucci, F.; Davies, P.; Hedberg, C.; et al. Salmonella enterica Serotype 4,[5],12:i:- in Swine in the United States Midwest: An Emerging Multidrug-Resistant Clade. Clin. Infect. Dis. 2018, 66, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, D.E.; Tietze, E. Comparative Biology of IncQ and IncQ-Like Plasmids. Microbiol. Mol. Biol. Rev. 2001, 65, 481–496. [Google Scholar] [CrossRef]
- Yau, S.; Liu, X.; Djordjevic, S.P.; Hall, R.M. RSF1010-Like Plasmids in Australian Salmonella enterica Serovar Typhimurium and Origin of Their sul2-strA-strB Antibiotic Resistance Gene Cluster. Microb. Drug Resist. 2010, 16, 249–252. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Willem, V.S.; Van Schaik, W.; Schürch, A.C. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb. Genom. 2017, 3, e000128. [Google Scholar] [CrossRef] [PubMed]

| Serotype | Number of Genomes | Median Length (Mb) | Date Range |
|---|---|---|---|
| Enteritidis | 11325 | 4.69 | 1950–2019 |
| Typhimurium | 8561 | 4.91 | 1958–2019 |
| Kentucky | 4131 | 4.92 | 1972–2019 |
| Infantis | 3866 | 4.94 | 1971–2019 |
| I 4,[5],12:i:- | 3657 | 4.95 | 1985–2019 |
| Newport | 3646 | 4.76 | 1975–2019 |
| Typhi | 2736 | 4.74 | 1958–2019 |
| Heidelberg | 2454 | 4.89 | 1979–2019 |
| Montevideo | 1841 | 4.65 | 1997–2019 |
| Agona | 1718 | 4.82 | 1952–2019 |
| Muenchen | 1641 | 4.79 | 1987–2019 |
| Saintpaul | 1570 | 4.79 | 1974–2019 |
| Anatum | 1560 | 4.73 | 1993–2019 |
| Senftenberg | 1329 | 4.81 | 2001–2019 |
| Schwarzengrund | 1130 | 4.81 | 2000–2019 |
| Mbandaka | 1083 | 4.75 | 2000–2019 |
| Braenderup | 1044 | 4.69 | 1999–2019 |
| Hadar | 1038 | 4.74 | 1988–2019 |
| Derby | 1023 | 4.87 | 1986–2019 |
| Javiana | 995 | 4.61 | 1995–2019 |
| Cluster | Gene | Class | Subclass |
|---|---|---|---|
| 1 | pcoA | Copper | Copper |
| pcoB | Copper | Copper | |
| pcoC | Copper | Copper | |
| pcoD | Copper | Copper | |
| pcoR | Copper | Copper | |
| pcoS | Copper | Copper | |
| silA | Silver | Silver | |
| silB | Silver | Silver | |
| silC | Silver | Silver | |
| silE | Silver | Silver | |
| silF | Silver | Silver | |
| 3 | aph(3’’)-Ib | Aminoglycoside | Streptomycin |
| aph(6)-Id | Aminoglycoside | Streptomycin | |
| arsA | Arsenic | Arsenite | |
| arsB | Arsenic | Arsenite | |
| arsC | Arsenic | Arsenate | |
| arsD | Arsenic | Arsenite | |
| arsR | Arsenic | Arsenite | |
| blaTEM | Beta-Lactam | Beta-Lactam | |
| IncQ1_1 | Plasmid Replicon | NA | |
| merR | Mercury | Mercury | |
| merT | Mercury | Mercury | |
| qacE | Quaternary Ammonium | Quaternary Ammonium | |
| qacEdelta1 | Quaternary Ammonium | Quaternary Ammonium | |
| sul1 | Sulfonamide | Sulfonamide | |
| sul2 | Sulfonamide | Sulfonamide | |
| tet(A) | Tetracycline | Tetracycline | |
| tet(B) | Tetracycline | Tetracycline |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenske, G.J.; Scaria, J. Analysis of 56,348 Genomes Identifies the Relationship between Antibiotic and Metal Resistance and the Spread of Multidrug-Resistant Non-Typhoidal Salmonella. Microorganisms 2021, 9, 1468. https://doi.org/10.3390/microorganisms9071468
Fenske GJ, Scaria J. Analysis of 56,348 Genomes Identifies the Relationship between Antibiotic and Metal Resistance and the Spread of Multidrug-Resistant Non-Typhoidal Salmonella. Microorganisms. 2021; 9(7):1468. https://doi.org/10.3390/microorganisms9071468
Chicago/Turabian StyleFenske, Gavin J., and Joy Scaria. 2021. "Analysis of 56,348 Genomes Identifies the Relationship between Antibiotic and Metal Resistance and the Spread of Multidrug-Resistant Non-Typhoidal Salmonella" Microorganisms 9, no. 7: 1468. https://doi.org/10.3390/microorganisms9071468
APA StyleFenske, G. J., & Scaria, J. (2021). Analysis of 56,348 Genomes Identifies the Relationship between Antibiotic and Metal Resistance and the Spread of Multidrug-Resistant Non-Typhoidal Salmonella. Microorganisms, 9(7), 1468. https://doi.org/10.3390/microorganisms9071468







