Double-Barrel Shotgun: Probiotic Lactic Acid Bacteria with Antiviral Properties Modified to Serve as Vaccines
Abstract
:1. Introduction
2. Regulation of Gut Wall Permeability
3. Immune System Modulation and Activation by Probiotics
4. Production of Antiviral Substances and Direct Virus Interaction by Probiotics
5. Virus–Probiotic Viral Receptor Competition
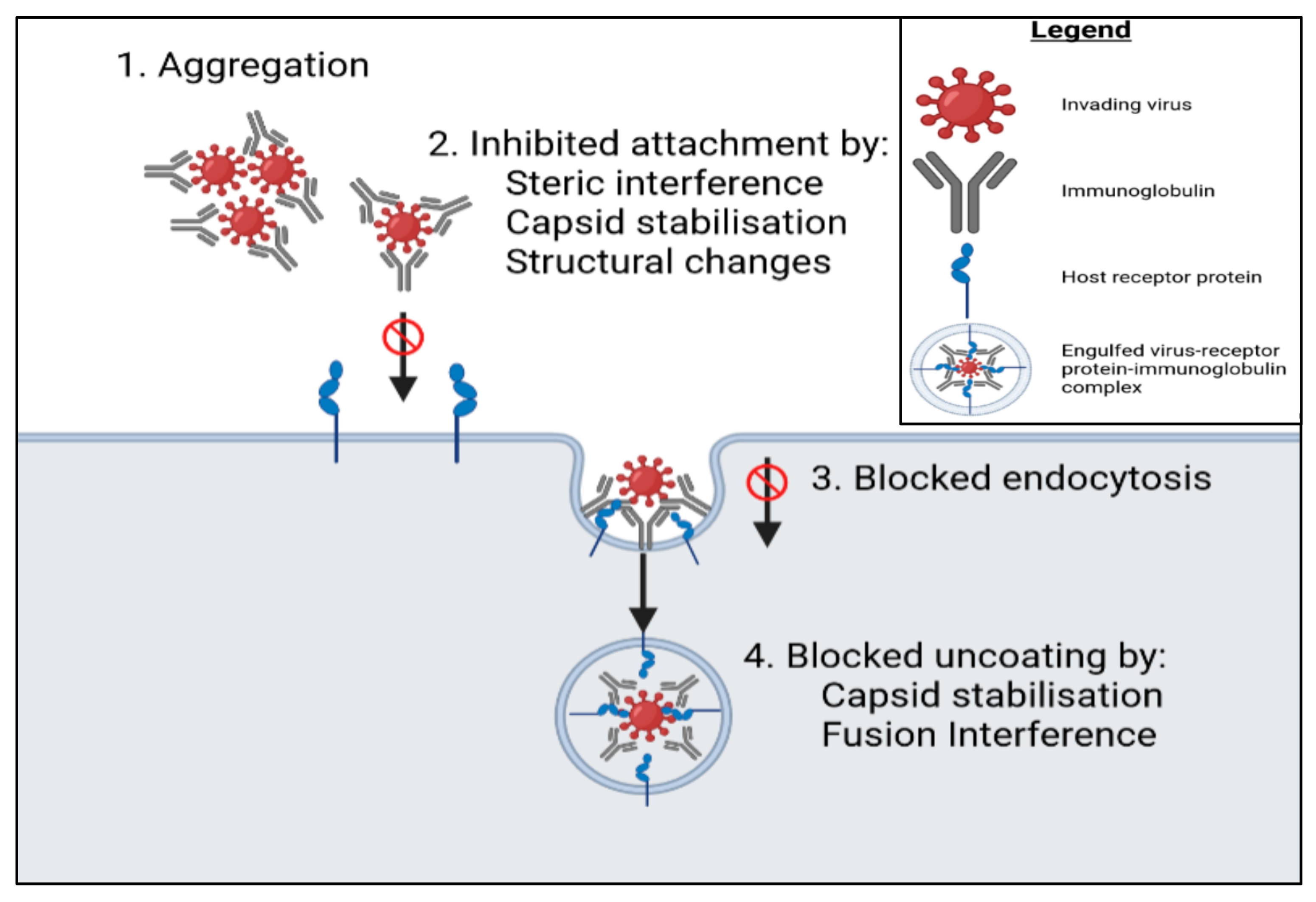
5.1. Binding of Proteins to Cell Membrane Receptors
5.2. Adhesion of Bacterial Cells to Virus Particles
5.3. Probiotic Biofilm Formation
5.4. Probiotic-Mediated Compositional Modulation of the Gut, Lung, and Respiratory Tract Microflora
6. SARS-CoV Probiotic-Based Vaccine and Secondary Symptom Treatment Possibilities
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. COVID-19 Vaccine Tracker and Landscape; WHO: Geneva, Switzerland, 2021; pp. 1–5. [Google Scholar]
- Anand, P.; Stahel, V.P. The safety of Covid-19 mRNA vaccines: A review. Patient Saf. Surg. 2021, 15, 1–9. [Google Scholar] [CrossRef]
- Food and Agriculture Organisation of the United Nations; World Health Organization. Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Hotel, A.; Cordoba, A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention 2001, 5, 1–34. [Google Scholar]
- Blottière, H.M.; De Vos, W.M.; Ehrlich, S.D.; Dore, J. Human intestinal metagenomics: State of the art and future. Curr. Opin. Microbiol. 2013, 16, 232–239. [Google Scholar] [CrossRef]
- Al Kassaa, I. Antiviral Probiotics: A New Concept in Medical Sciences. In New Insights on Antiviral Probiotics: From Research to Applications; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 1–46. ISBN 9783319496887. [Google Scholar]
- Chey, W.; Menees, S. The gut microbiome and irritable bowel syndrome. F1000Research 2018, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tran, D.Q.; Rhoads, J.M. Probiotics in Disease Prevention and Treatment. J. Clin. Pharmacol. 2018, 58, S164–S179. [Google Scholar] [CrossRef]
- Park, M.-K.; Ngo, V.; Moon, D.-W.; Jeong, E.-J.; Kim, M.-C.; Lee, Y.-N.; Jang, J.-H.; Oh, J.-S.; Kim, C.-H.; Kang, S.-M.; et al. Lactobacillus plantarum DK119 as a Probiotic Confers Protection against Influenza Virus by Modulating Innate Immunity. PLoS ONE 2013, 8, e75368. [Google Scholar] [CrossRef] [Green Version]
- Waki, N.; Yajima, N.; Suganuma, H.; Buddle, B.M.; Luo, D.; Heiser, A.; Zheng, T. Oral administration of Lactobacillus brevis KB290 to mice alleviates clinical symptoms following influenza virus infection. Lett. Appl. Microbiol. 2014, 58, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Yeo, J.-M.; Lee, H.-J.; Kim, J.-W.; Lee, J.-B.; Park, S.-Y.; Choi, I.-S.; Song, C.-S. Lactobacillus fermentum CJL-112 protects mice against influenza virus infection by activating T-helper 1 and eliciting a protective immune response. Int. Immunopharmacol. 2014, 18, 50–54. [Google Scholar] [CrossRef]
- Shibata, T.; Kanayama, M.; Haida, M.; Fujimoto, S.; Oroguchi, T.; Sata, K.; Mita, N.; Kutsuzawa, T.; Ikeuchi, M.; Kondo, M.; et al. Lactococcus lactis JCM5805 activates anti-viral immunity and reduces symptoms of common cold and influenza in healthy adults in a randomized controlled trial. J. Funct. Foods 2016, 24, 492–500. [Google Scholar] [CrossRef]
- Tiihonen, K.; Ouwehand, A.C.; Rautonen, N. Human intestinal microbiota and healthy ageing. Ageing Res. Rev. 2010, 9, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Bolte, L.A.; Vich Vila, A.; Imhann, F.; Collij, V.; Gacesa, R.; Peters, V.; Wijmenga, C.; Kurilshikov, A.; Campmans-Kuijpers, M.J.E.; Fu, J.; et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021, 70, 1287–1298. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Świątecka, D.; Narbad, A.; Ridgway, P.K.; Kostyra, H. The study on the impact of glycated pea proteins on human intestinal bacteria. Int. J. Food Microbiol. 2011, 145, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.-Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Marco, M.L.; Pavan, S.; Kleerebezem, M. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 2006, 17, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T.; Botes, M. Probiotic lactic acid bacteria in the gastro-intestinal tract: Health benefits, safety and mode of action. Benef. Microbes 2010, 1, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Amara, A.A.; Shibl, A. Role of Probiotics in health improvement, infection control and disease treatment and management. Saudi Pharm. J. 2015, 23, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Sathyabama, S.; Vijayabharathi, R.; Bruntha Devi, P.; Ranjith Kumar, M.; Priyadarisini, V.B. Screening for probiotic properties of strains isolated from feces of various human groups. J. Microbiol. 2012, 50, 603–612. [Google Scholar] [CrossRef]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef]
- Bhat, A.A.; Uppada, S.; Achkar, I.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front. Physiol. 2019, 9, 1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohland, C.L.; Macnaughton, W.K. Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Liver Gastrointest. Physiol. 2010, 298, G807–G819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rani, R.A.; Ali, R.A.R.; Lee, Y.Y. Irritable bowel syndrome and inflammatory bowel disease overlap syndrome: Pieces of the puzzle are falling into place. Intest. Res. 2016, 14, 297–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Tajik, N.; Frech, M.; Schulz, O.; Schälter, F.; Lucas, S.; Azizov, V.; Dürholz, K.; Steffen, F.; Omata, Y.; Rings, A.; et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, J.; Wells, J.; Cani, P.D.; Garcia-Rodenas, C.L.; Macdonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.-J. Human Intestinal Barrier Function in Health and Disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef] [PubMed]
- Marquez, M.; Fernández Gutiérrez del Álamo, C.; Girón-González, J.A. Gut epithelial barrier dysfunction in human immunodeficiency virus-hepatitis C virus coinfected patients: Influence on innate and acquired immunity. World J. Gastroenterol. 2016, 22, 1433–1448. [Google Scholar] [CrossRef]
- Mao, X.; Gu, C.; Hu, H.; Tang, J.; Chen, D.; Yu, B.; He, J.; Yu, J.; Luo, J.; Tian, G. Dietary Lactobacillus rhamnosus GG Supplementation Improves the Mucosal Barrier Function in the Intestine of Weaned Piglets Challenged by Porcine Rotavirus. PLoS ONE 2016, 11, e0146312. [Google Scholar] [CrossRef]
- Álvarez-Olmos, M.I.; Oberhelman, R.A. Probiotic Agents and Infectious Diseases: A Modern Perspective on a Traditional Therapy. Clin. Infect. Dis. 2001, 32, 1567–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resta-Lenert, S.; Barrett, K.E. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 2003, 52, 988–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helbert, M. How Autoimmune Disease Develops. In Immunology for Medical Students; Elsevier Limited: Amsterdam, The Netherlands, 2017; Chapter 28; pp. 217–230. ISBN 978-0-7020-6801-0. [Google Scholar]
- Nainu, F.; Shiratsuchi, A.; Nakanishi, Y. Induction of Apoptosis and Subsequent Phagocytosis of Virus-Infected Cells as an Antiviral Mechanism. Front. Immunol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Koyama, S.; Ishii, K.J.; Coban, C.; Akira, S. Innate immune response to viral infection. Cytokine 2008, 43, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Murosaki, S.; Yamamoto, Y.; Yoshikai, Y.; Tsuru, T. Daily Intake of Heat-Killed Lactobacillus plantarum L-137 Augments Acquired Immunity in Healthy Adults. J. Nutr. 2006, 136, 3069–3073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimura, T.; Jounai, K.; Ohshio, K.; Tanaka, T.; Suwa, M.; Fujiwara, D. Immunomodulatory effect of Lactococcus lactis JCM5805 on human plasmacytoid dendritic cells. Clin. Immunol. 2013, 149, 509–518. [Google Scholar] [CrossRef]
- Laiño, J.E.; Villena, J.; Kanmani, P.; Kitazawa, H. Immunoregulatory Effects Triggered by Lactic Acid Bacteria Exopolysaccharides: New Insights into Molecular Interactions with Host Cells. Microorganisms 2016, 4, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonetti, F.R.; Islam, M.A.; Vizoso-Pinto, M.G.; Takahashi, H.; Kitazawa, H.; Villena, J. Nasal priming with immunobiotic lactobacilli improves the adaptive immune response against influenza virus. Int. Immunopharmacol. 2020, 78, 106115. [Google Scholar] [CrossRef] [PubMed]
- Taverniti, V.; Guglielmetti, S. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes Nutr. 2011, 6, 261–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response during SARS-CoV-2 Infection: Lessons from the Past. Front. Immunol. 2020, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.S.; Warrington, R.; Watson, W.; Kim, H.L. An introduction to immunology and immunopathology. Allergy Asthma Clin. Immunol. 2018, 14, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, M.A.; Hossain, N.; Kashem, M.A.; Shahid, M.A.; Alam, A. Immune response in COVID-19: A review. J. Infect. Public Health 2020, 13, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, L.B. The immune system. Essays Biochem. 2016, 60, 275–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiecki, M.; Colonna, M. The multifacted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015, 15, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Kanauchi, O.; Andoh, A.; AbuBakar, S.; Yamamoto, N. Probiotics and Paraprobiotics in Viral Infection: Clinical Application and Effects on the Innate and Acquired Immune Systems. Curr. Pharm. Des. 2018, 24, 710–717. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Ohshio, K.; Fujiwara, D. Lactococcus lactis subsplactis JCM 5805 activates natural killer cells via dendritic cells. Biosci. Biotechnol. Biochem. 2016, 80, 798–800. [Google Scholar] [CrossRef] [Green Version]
- Ekitazawa, H.; Evillena, J. Modulation of Respiratory TLR3-Anti-Viral Response by Probiotic Microorganisms: Lessons Learned from Lactobacillus rhamnosus CRL1505. Front. Immunol. 2014, 5, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Yasui, H.; Mike, A.; Ohwaki, M. Immunogenicity of Bifidobacterium breve and Change in Antibody Production in Peyer’s Patches After Oral Administration. J. Dairy Sci. 1989, 72, 30–35. [Google Scholar] [CrossRef]
- Greenspan, N.S.; Cavacini, L.A. Immunoglobulin Function. In Clinical Immunology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 223–233. [Google Scholar]
- Rhorer, J.; Ambrose, C.S.; Dickinson, S.; Hamilton, H.; Oleka, N.A.; Malinoski, F.J.; Wittes, J. Efficacy of live attenuated influenza vaccine in children: A meta-analysis of nine randomized clinical trials. Vaccine 2009, 27, 1101–1110. [Google Scholar] [CrossRef]
- Klasse, P.J. Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Adv. Biol. 2014, 2014, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Stieh, D.J.; King, D.F.; Klein, K.; Liu, P.; Shen, X.; Hwang, K.K.; Ferrari, G.; Montefiori, D.C.; Haynes, B.; Pitisuttithum, P.; et al. Aggregate complexes of HIV-1 induced by multimeric antibodies. Retrovirology 2014, 11, 1–16. [Google Scholar] [CrossRef]
- Wrighton, K.H. Blocking endocytosis gives therapeutic antibodies a boost. Nat. Rev. Drug Discov. 2020, 19, 237. [Google Scholar] [CrossRef]
- Yan, F.; Polk, D. Probiotics and immune health. Curr. Opin. Gastroenterol. 2011, 27, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Montero-Melendez, T. May Inflammation Be with You! Front. Young Minds 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Gabryszewski, S.J.; Bachar, O.; Dyer, K.D.; Percopo, C.M.; Killoran, K.E.; Domachowske, J.B.; Rosenberg, H.F. Lactobacillus-Mediated Priming of the Respiratory Mucosa Protects against Lethal Pneumovirus Infection. J. Immunol. 2011, 186, 1151–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botić, T.; Klingberg, T.D.; Weingartl, H.; Cencic, A. A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int. J. Food Microbiol. 2007, 115, 227–234. [Google Scholar] [CrossRef]
- Wang, Z.; Chai, W.; Burwinkel, M.; Twardziok, S.; Wrede, P.; Palissa, C.; Esch, B.; Schmidt, M.F.G. Inhibitory Influence of Enterococcus faecium on the Propagation of Swine Influenza A Virus In Vitro. PLoS ONE 2013, 8, e53043. [Google Scholar] [CrossRef] [Green Version]
- AL Kassaa, I.; Hamze, M.; Hober, D.; Chihib, N.-E.; Drider, D. Identification of Vaginal Lactobacilli with Potential Probiotic Properties Isolated from Women in North Lebanon. Microb. Ecol. 2014, 67, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Salminen, S.; Nybom, S.; Meriluoto, J.; Collado, M.C.; Vesterlund, S.; El-Nezami, H. Interaction of probiotics and pathogens—benefits to human health? Curr. Opin. Biotechnol. 2010, 21, 157–167. [Google Scholar] [CrossRef]
- Eschenbach, D.A.; Davick, P.R.; Williams, B.L.; Klebanoff, S.J.; Young-Smith, K.; Critchlow, C.M.; Holmes, K.K. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J. Clin. Microbiol. 1989, 27, 251–256. [Google Scholar] [CrossRef] [Green Version]
- AL Kassaa, I.; Hober, D.; Hamze, M.; Chihib, N.E.; Drider, D. Antiviral Potential of Lactic Acid Bacteria and Their Bacteriocins. Probiotics Antimicrob. Proteins 2014, 6, 177–185. [Google Scholar] [CrossRef]
- Zabihollahi, R.; Motevaseli, E.; Sadat, S.M.; Azizi-Saraji, A.R.; Asaadi-Dalaie, S.; Modarressi, M.H. Inhibition of HIV and HSV infection by vaginal lactobacilli in vitro and in vivo. DARU J. Pharm. Sci. 2012, 20, 53. [Google Scholar] [CrossRef] [Green Version]
- Seo, D.J.; Jung, D.; Jung, S.; Yeo, D.; Choi, C. Inhibitory effect of lactic acid bacteria isolated from kimchi against murine norovirus. Food Control 2020, 109, 106881. [Google Scholar] [CrossRef]
- Boomsma, B.; Bikker, E.; Lansdaal, E.; Stuut, P. L-Lactic Acid—A Safe Antimicrobial for Home- and Personal Care Formulations. SOFW 2015, 10, 141–145. [Google Scholar]
- Dicks, L.M.T.; Dreyer, L.; Smith, C.; Van Staden, A.D. A Review: The Fate of Bacteriocins in the Human Gastro-Intestinal Tract: Do They Cross the Gut–Blood Barrier? Front. Microbiol. 2018, 9, 2297. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, H.; Saeed, S.; Ahmed, S.; Rasool, S.A. Coliphage Hsa as a Model for Antiviral studies/spectrum by Some Indigenous Bacteriocin like Inhibitory Substances (BLIS). Pak. J. Pharm. Sci. 2006, 19, 182–185. [Google Scholar] [CrossRef]
- Saeed, S.; Rasool, S.A.; Ahmad, S.; Zaidi, S.Z.; Rehmani, S. Antiviral Activity of Staphylococcin 188: A Purified Bacteriocin like Inhibitory Substance Isolated from Staphylococcus aureus AB188. Res. J. Microbiol. 2007, 2, 796–806. [Google Scholar] [CrossRef] [Green Version]
- Todorov, S.D.; Wachsman, M.; Tomé, E.; Dousset, X.; Destro, M.T.; Dicks, L.M.T.; de Melo Franco, B.D.G.; Vaz-Velho, M.; Drider, D. Characterisation of an antiviral pediocin-like bacteriocin produced by Enterococcus faecium. Food Microbiol. 2010, 27, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Wachsman, M.B.; Castilla, V.; De Ruiz Holgado, A.P.; De Torres, R.A.; Sesma, F.; Coto, C.E. Enterocin CRL35 inhibits late stages of HSV-1 and HSV-2 replication in vitro. Antivir. Res. 2003, 58, 17–24. [Google Scholar] [CrossRef]
- Lehtoranta, L.; Pitkäranta, A.; Korpela, R. Probiotics in respiratory virus infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1289–1302. [Google Scholar] [CrossRef] [PubMed]
- Salas-Cárdenas, S.P.; Olaya-Galán, N.N.; Fernández, K.; Velez, F.; Guerrero, C.A.; Guitiérrez, M.F. Decreased rotavirus infection of MA104 cells via probiotic extract binding to Hsc70 and ß3 integrin receptors. Univ. Sci. 2018, 23, 219–239. [Google Scholar] [CrossRef]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; García, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef]
- Marionneau, S.; Cailleau-Thomas, A.; Rocher, J.; Le Moullac-Vaidye, B.; Ruvoën, N.; Clément, M.; Le Pendu, J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 2001, 83, 565–573. [Google Scholar] [CrossRef]
- Heggelund, J.E.; Varrot, A.; Imberty, A.; Krengel, U. Histo-blood group antigens as mediators of infections. Curr. Opin. Struct. Biol. 2017, 44, 190–200. [Google Scholar] [CrossRef] [Green Version]
- Haga, K.; Ettayebi, K.; Tenge, V.R.; Karandikar, U.C.; Lewis, M.A.; Lin, S.-C.; Neill, F.H.; Ayyar, B.V.; Zeng, X.-L.; Larson, G.; et al. Genetic Manipulation of Human Intestinal Enteroids Demonstrates the Necessity of a Functional Fucosyltransferase 2 Gene for Secretor-Dependent Human Norovirus Infection. mBio 2020, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Aspinall, G.O.; Monteiro, M.A. Lipopolysaccharides ofHelicobacter pyloriStrains P466 and MO19: Structures of the O Antigen and Core Oligosaccharide Regions. Biochemistry 1996, 35, 2498–2504. [Google Scholar] [CrossRef]
- Sindhu, K.N.C.; Sowmyanarayanan, T.V.; Paul, A.; Babji, S.; Ajjampur, S.S.R.; Priyadarshini, S.; Sarkar, R.; Balasubramanian, K.A.; Wanke, C.A.; Ward, H.D.; et al. Immune Response and Intestinal Permeability in Children with Acute Gastroenteritis Treated with Lactobacillus rhamnosus GG: A Randomized, Double-Blind, Placebo-Controlled Trial. Clin. Infect. Dis. 2014, 58, 1107–1115. [Google Scholar] [CrossRef]
- Jespersen, L.; Tarnow, I.; Eskesen, D.; Morberg, C.M.; Michelsen, B.; Bugel, S.; Dragsted, L.O.; Rijkers, G.T.; Calder, P.C. Effect of Lactobacillus paracasei subsp. paracasei, L. casei 431 on immune response to influenza vaccination and upper respiratory tract infections in healthy adult volunteers: A randomized, double-blind, placebo-controlled, parallel-group study. Am. J. Clin. Nutr. 2015, 101, 1188–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holscher, H.D.; Czerkies, L.A.; Tappenden, K.; Cekola, P.; Litov, R.; Benbow, M.; Santema, S.; Alexander, D.D.; Perez, V.; Sun, S.; et al. Bifidobacterium lactisBb12 Enhances Intestinal Antibody Response in Formula-Fed Infants: A randomized, double-blind, controlled trial. J. Parenter. Enter. Nutr. 2012, 36, 106S–117S. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Wang, L.; Xiong, Y.; Wang, Z.; Qiu, Y.; Wen, X.; Jiang, Z.; Yang, X.; Ma, X. Lactobacillus reuteri LR1 Improved Expression of Genes of Tight Junction Proteins via the MLCK Pathway in IPEC-1 Cells during Infection with Enterotoxigenic Escherichia coli K88. Mediat. Inflamm. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, R.; McBain, A.J.; O’Neill, C.A. Strain-Dependent Augmentation of Tight-Junction Barrier Function in Human Primary Epidermal Keratinocytes by Lactobacillus and Bifidobacterium Lysates. Appl. Environ. Microbiol. 2013, 79, 4887–4894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastromarino, P.; Cacciotti, F.; Masci, A.; Mosca, L. Antiviral activity of Lactobacillus brevis towards herpes simplex virus type 2: Role of cell wall associated components. Anaerobe 2011, 17, 334–336. [Google Scholar] [CrossRef]
- Maragkoudakis, P.A.; Chingwaru, P.W.; Gradisnik, L.; Tsakalidou, E.; Cencic, A. Lactic acid bacteria efficiently protect human and animal intestinal epithelial and immune cells from enteric virus infection. Int. J. Food Microbiol. 2010, 141, S91–S97. [Google Scholar] [CrossRef] [PubMed]
- Colbère-Garapin, F.; Martin-Latil, S.; Blondel, B.; Mousson, L.; Pelletier, I.; Autret, A.; François, A.; Niborski, V.; Grompone, G.; Catonnet, G.; et al. Prevention and treatment of enteric viral infections: Possible benefits of probiotic bacteria. Microbes Infect. 2007, 9, 1623–1631. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Kothari, D.; Patel, S.; Kim, S.-K. Probiotic supplements might not be universally-effective and safe: A review. Biomed. Pharmacother. 2019, 111, 537–547. [Google Scholar] [CrossRef]
- ISAPP Board of Directors. How Some Probiotic Scientists Are Working to Address COVID-19; ISAPP: Sacramento, CA, USA, 2020. [Google Scholar]
- LWrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Marelli, B.; Pérez, A.R.; Banchio, C.; De Mendoza, D.; Magni, C. Oral immunization with live Lactococcus lactis expressing rotavirus VP8* subunit induces specific immune response in mice. J. Virol. Methods 2011, 175, 28–37. [Google Scholar] [CrossRef]
- Akour, A. Probiotics and COVID-19: Is there any link? Lett. Appl. Microbiol. 2020, 71, 229–234. [Google Scholar] [CrossRef]
- Kaur, G.; Lungarella, G.; Rahman, I. SARS-CoV-2 COVID-19 susceptibility and lung inflammatory storm by smoking and vaping. J. Inflamm. 2020, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N-and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. bio-Rxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Dimopoulou Agri, V.; Gibson, G.R.; Reid, G.; Giannoni, E. Using Probiotics to Flatten the Curve of Coronavirus Disease COVID-2019 Pandemic. Front. Public Health 2020, 8, 186. [Google Scholar] [CrossRef] [PubMed]



| Immune System Component: | Examples: | Function: |
|---|---|---|
| Pathogen-associated molecular patterns (PAMPs) | DNA, RNA, surface glycoproteins | Act as ligands belonging to the virus in the form of conserved sequences or structures. Recognition molecules of the host identify the ligands and triggers the appropriate immune response. |
| Damage-associated molecular patterns (DAMPs) | Molecules such as ATP, DNA, hyaluronan fragments, and the chromatin-associated leaderless secreted protein HMGB1 secreted from damaged host cells | Unlike PAMPs, DAMPs originate from virus-infected host cells. DAMPs are specific in sequence and structure. |
| Pattern recognition sequences (PRRs) | Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) | PRRs are sensors mainly encoded by cells of the innate immune system (e.g., DCs, macrophages, neutrophils, etc.) that detect PAMPs and DAMPs. This initiates the release of cytokines and leads to an antigen-specific systemic immune response. |
| Cytokines | Interferons (IFNs) such as type 1 IFN-α/β and IFN-γ, interleukins IL-1, 2, 6, 8, 10, 15, and 18, tumour necrosis factor (TNF), cytokines TNF-α, and chemokines such as CXCL-8 | Cytokines are small signalling proteins, peptides, or glycoproteins that play an integral part in inflammation and immunity regulation. Type-1 IFNs are the only cytokines solely associated with viral immunity, as opposed to also being involved in bacterial immunity. |
| Macrophages | Employ phagocytosis to “digest” viral particles. While doing so, macrophages release cytokines of their own upon PAMP recognition by their PRRs. | |
| Granulocytes | Neutrophils, eosinophils, and basophils | Neutrophils are the most prominent white blood cells in the human body. These phagocytotic cells have a short half-life, followed by apoptosis. A major chemoattractant of neutrophils are chemokines, particularly CXCL-8. |
| Dendritic Cells | Nasal, epidermal, intestinal, pulmonary, tracheal, etc. Dendritic cells are external cells that come into contact with the external environment | Initiate both innate and system immune responses. They too are phagocytotic cells and can thus engulf viral particles. Furthermore, once infected by a virus, DCs initiate a T-cell response by displaying viral antigens on the type-I major histocompatibility complex (MHC). If the DC has only phagocytotically engulfed the virus, it can display the associated antigens on a type-II MHC which will elicit a systemic immune response. DCs can also produce cytokines such as type-I IFNs when their TLRs bind to viral particles such as RNA. |
| T-cells | Cytotoxic T-cells (TC cells) such as CD8 and T8, and T-helper cells (TH cells) | TH cells initiate B cells to produce antibodies and in turn are extremely important in the systemic immune response, whilst TC cells, such as CD8-T cells in particular, form pores in infected cells after which cytotoxins are released by the cells killing the infected cell as well as any viruses inside it. |
| Natural killer (NK) cells | NK cells chemotoxically kill infected cells and their infiltrating viruses. However, NK- and CD8- T cells recognise these infected cells differently. | |
| Peripheral blood mononuclear cells | Any blood cell with a circular nucleus such as T cells, B cells, and NK cells | Engulf and phagocytose viruses whilst releasing pro-inflammatory cytokines |
| Mechanism of Action: | Probiotic Strain: | Studied Virus: | Test Patients /Tissue: | Result: | Reference: |
|---|---|---|---|---|---|
| Immunoregulation | L. delbrueckii ssp. bulgaricus OLL1073R-1 | Common cold symptoms | Elderly people | A significant increase in NK cell cytotoxicity, resulting in a reduced risk of ailing from cold symptoms. | [82] |
| L. paracasei ssp. paracasei, L. casei 431 | Influenza | Healthy adults with influenza vaccination | A significant increase in influenza-specific IgG, IgG1, and IgG3 in plasma and IgA in saliva. | [83] | |
| B. animalis (Bb12) | Polio and rotavirus | Healthy 6-week-old infants | A significant increase in polio and rotavirus-specific IgA antibodies. | [84] | |
| L. lactis JCM5805 | Common cold | Healthy adults | Activation of pDCs amongst peripheral blood mononuclear cells (PBMCs) and as such, a significant reduction in morbidity attributed to the common cold. | [38] | |
| Influenza | Healthy adults | A significant increase in IFN-α mRNA in PBMCs, meaning a significant decrease in the number of days ailing from influenza symptoms such as sore throats and coughs. | [12] | ||
| Tight junction maintenance and functional improvement | L. reuteri LR1 | Intestinal porcine epithelial cells | MLCK-dependent dephosphorylation of TJ subunit proteins such as ZO-1 and occluding resulting in a decreased pathogen flooding of the lamina propria | [85] | |
| B. longum and LGG lysates | Normal human epidermal keratinocytes | A lysate-induced increase in claudin 1 levels in keratinocytes correlating with the decreased pathogen flooding of the lamina propria. | [86] | ||
| Direct virus inactivation by probiotics/probiotic compounds | Lactobacillus brevis | Herpes simplex virus 2 | Vero cells | Cell wall interaction with virus envelope resulting in reduced viral replication | [87] |
| Herpes simplex virus 2 | Vero cells | No proteinaceous heat resistant proteins isolated from L. brevis extract interacted with HSV2 envelopes. | [87] | ||
| L. plantarum PCA236 | RV and transmissible gastroenteritis virus (TGSV) | Human and animal intestinal and macrophage cell lines | Reactive oxygen species (ROS), nitric oxide (NO-), and H2O2 interaction with RV and TGSV virion envelopes. | [88] | |
| Epithelial cell virus receptor interference | L. casei DN114 001 and Bacteroides thetaiotaomicron | RV | Human epithelial cells | Soluble compound production binding to viral receptors, resulting in glycosylation and thus structural isomerisation of the receptor, making it unable to attach and identify RV virions. | [89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dicks, L.M.T.; Grobbelaar, M.J. Double-Barrel Shotgun: Probiotic Lactic Acid Bacteria with Antiviral Properties Modified to Serve as Vaccines. Microorganisms 2021, 9, 1565. https://doi.org/10.3390/microorganisms9081565
Dicks LMT, Grobbelaar MJ. Double-Barrel Shotgun: Probiotic Lactic Acid Bacteria with Antiviral Properties Modified to Serve as Vaccines. Microorganisms. 2021; 9(8):1565. https://doi.org/10.3390/microorganisms9081565
Chicago/Turabian StyleDicks, Leon M. T., and Matthew J. Grobbelaar. 2021. "Double-Barrel Shotgun: Probiotic Lactic Acid Bacteria with Antiviral Properties Modified to Serve as Vaccines" Microorganisms 9, no. 8: 1565. https://doi.org/10.3390/microorganisms9081565
APA StyleDicks, L. M. T., & Grobbelaar, M. J. (2021). Double-Barrel Shotgun: Probiotic Lactic Acid Bacteria with Antiviral Properties Modified to Serve as Vaccines. Microorganisms, 9(8), 1565. https://doi.org/10.3390/microorganisms9081565






