Metatranscriptomic Analysis of Bacterial Communities on Laundered Textiles: A Pilot Case Study
Abstract
:1. Introduction
2. Materials and Methods
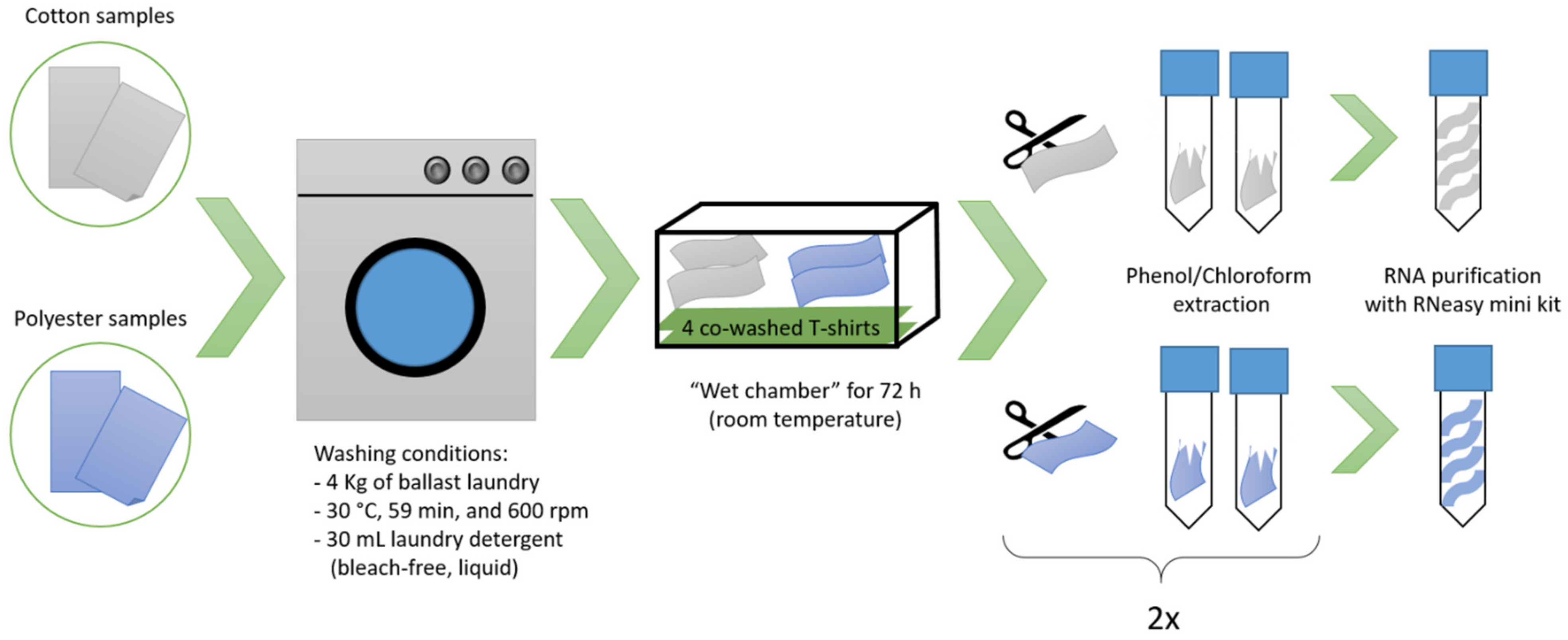
2.1. Sample Preparation
2.2. RNA Extraction and Sequencing
2.3. Sequence Data Analysis
3. Results and Discussion
3.1. Reads and de novo Transcriptome Assembly
3.2. Evaluation of the Different de novo Transcriptome Assemblies
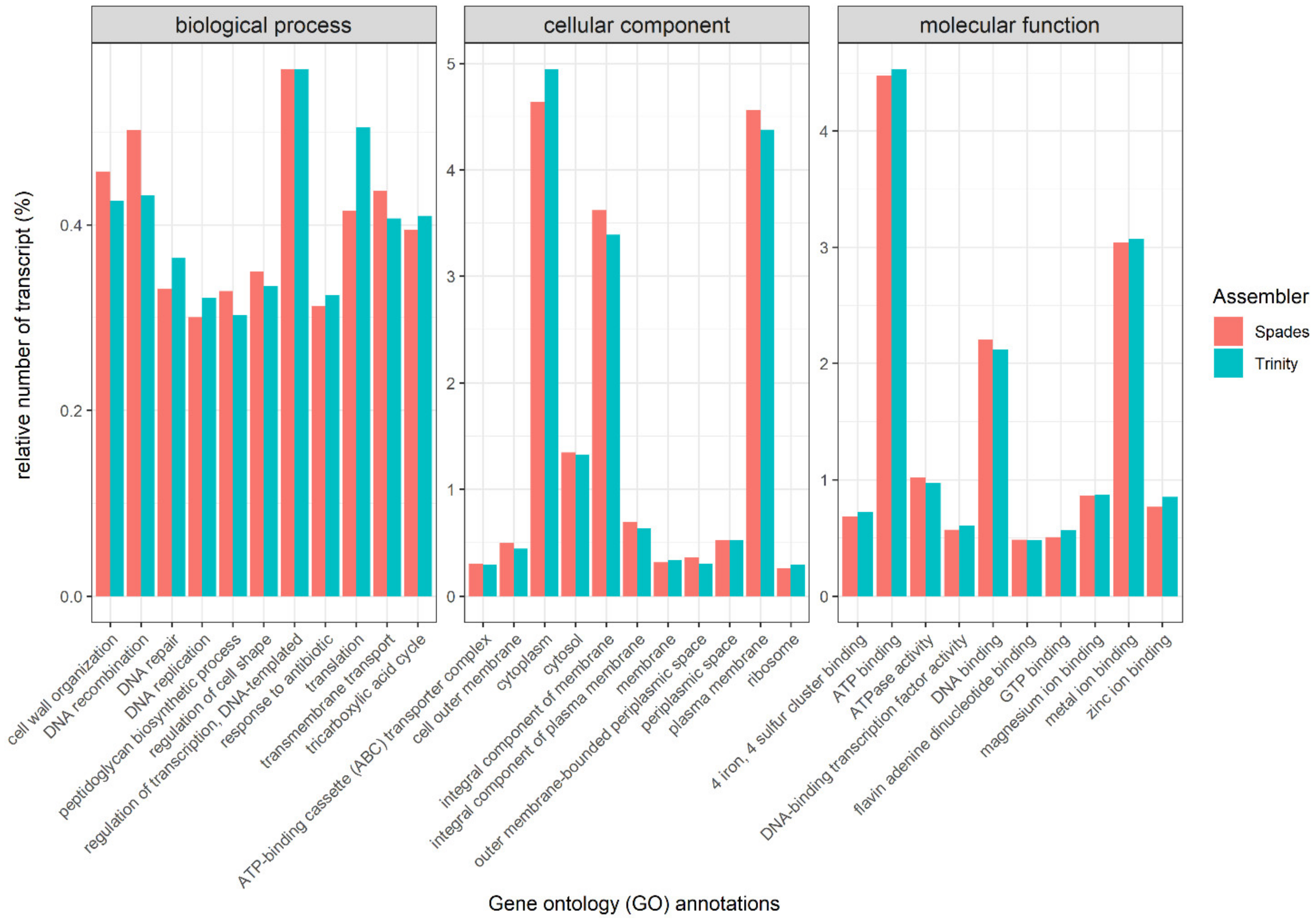
3.3. Transcript Annotation
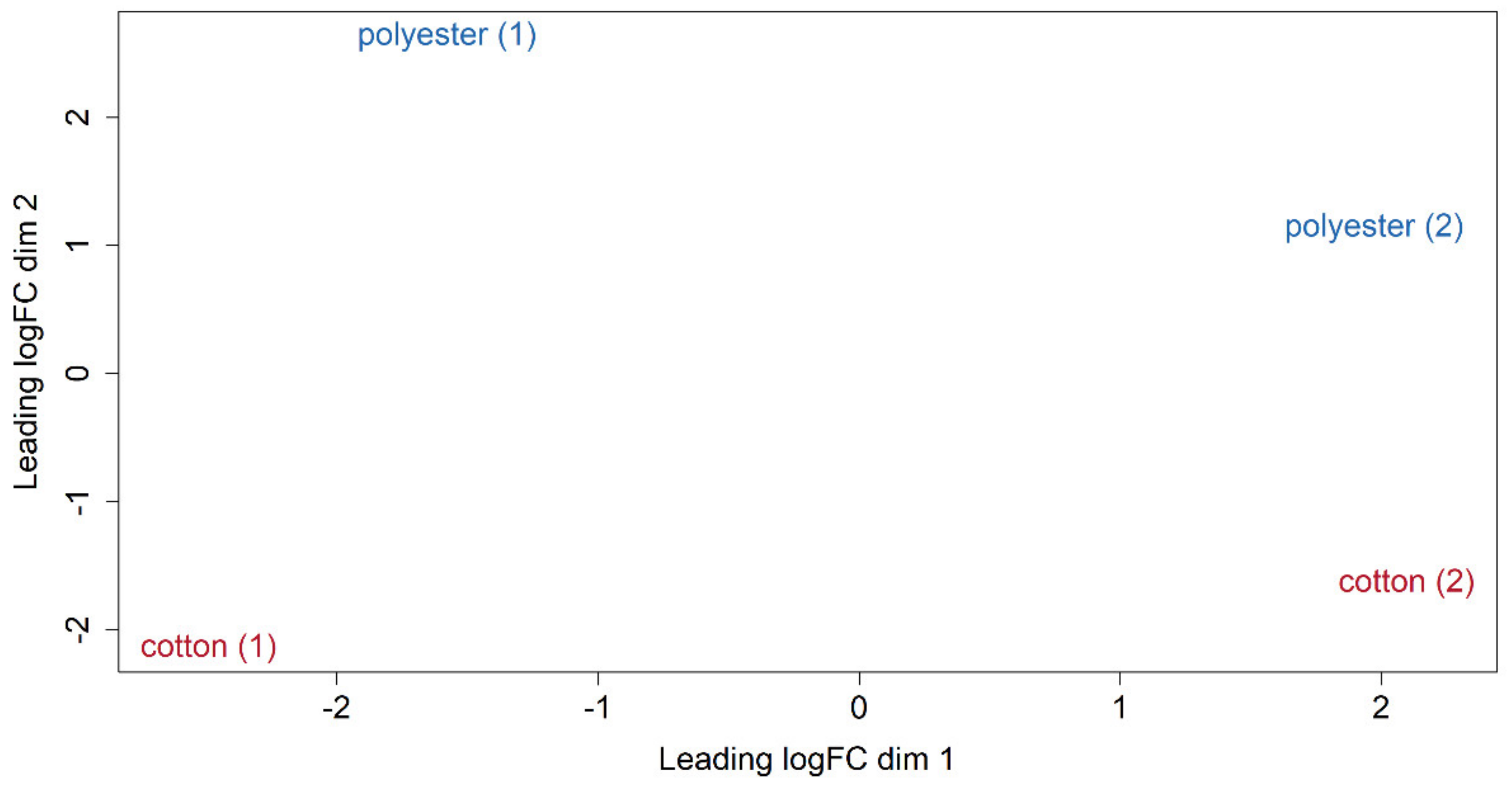
3.4. Differential Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Novak Babič, M.; Gostinčar, C.; Gunde-Cimerman, N. Microorganisms populating the water-related indoor biome. Appl. Microbiol. Biotechnol. 2020, 104, 6443–6462. [Google Scholar] [CrossRef]
- Munk, S.; Johansen, C.; Stahnke, L.H.; Adler-Nissen, J. Microbial survival and odor in laundry. J. Surfact. Deterg. 2001, 4, 385–394. [Google Scholar] [CrossRef]
- Nix, I.D.; Frontzek, A.; Bockmühl, D.P. Characterization of Microbial Communities in Household Washing Machines. Tenside. Surfactants Deterg. 2015, 52, 432–440. [Google Scholar] [CrossRef]
- Egert, M. The BE-Microbiome-Communities with Relevance for Laundry and Home Care. SOFW J. 2017, 143, 44–48. [Google Scholar]
- Bloomfield, S.F.; Exner, M.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Krämer, I.; Merkens, W.; Oltmanns, P.; Rotter, M.; et al. Lesser-known or hidden reservoirs of infection and implications for adequate prevention strategies: Where to look and what to look for. GMS Hyg. Infect. Control 2015, 10, Doc04. [Google Scholar] [CrossRef]
- Gibson, L.L.; Rose, J.B.; Haas, C.N. Use of quantitative microbial risk assessment for evaluation of the benefits of laundry sanitation. Am. J. Infect. Control. 1999, 27, S34–S39. [Google Scholar] [CrossRef]
- Jacksch, S.; Kaiser, D.; Weis, S.; Weide, M.; Ratering, S.; Schnell, S.; Egert, M. Influence of Sampling Site and other Environmental Factors on the Bacterial Community Composition of Domestic Washing Machines. Microorganisms 2020, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Babič, M.N.; Zalar, P.; Ženko, B.; Schroers, H.-J.; Džeroski, S.; Gunde-Cimerman, N. Candida and Fusarium species known as opportunistic human pathogens from customer-accessible parts of residential washing machines. Fungal Biol. 2015, 119, 95–113. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, M.B.; Spijkerman, D.C.M.; Voor In ‘t Holt, A.F.; van der Laan, R.J.; Bode, L.G.M.; van Vianen, W.; Klaassen, C.H.W.; Vos, M.C.; Severin, J.A. An outbreak of ST307 extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in a rehabilitation center: An unusual source and route of transmission. Infect. Control Hosp. Epidemiol. 2020, 41, 31–36. [Google Scholar] [CrossRef]
- Callewaert, C.; van Nevel, S.; Kerckhof, F.M.; Granitsiotis, M.S.; Boon, N. Bacterial Exchange in Household Washing Machines. Front. Microbiol. 2015, 6, 1381. [Google Scholar] [CrossRef] [Green Version]
- Gattlen, J.; Amberg, C.; Zinn, M.; Mauclaire, L. Biofilms isolated from washing machines from three continents and their tolerance to a standard detergent. Biofouling 2010, 26, 873–882. [Google Scholar] [CrossRef]
- Honisch, M.; Stamminger, R.; Bockmühl, D.P. Impact of wash cycle time, temperature and detergent formulation on the hygiene effectiveness of domestic laundering. J. Appl. Microbiol. 2014, 117, 1787–1797. [Google Scholar] [CrossRef]
- Jacksch, S.; Zohra, H.; Weide, M.; Schnell, S.; Egert, M. Cultivation-Based Quantification and Identification of Bacteria at Two Hygienic Key Sides of Domestic Washing Machines. Microorganisms 2021, 9, 905. [Google Scholar] [CrossRef] [PubMed]
- Schmithausen, R.M.; Sib, E.; Exner, M.; Hack, S.; Rösing, C.; Ciorba, P.; Bierbaum, G.; Savin, M.; Bloomfield, S.F.; Kaase, M.; et al. The Washing Machine as a Reservoir for Transmission of Extended-Spectrum-Beta-Lactamase (CTX-M-15)-Producing Klebsiella oxytoca ST201 to Newborns. Appl. Environ. 2019, 85, e01435-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, H.; Mitani, A.; Niwano, Y.; Takeuchi, K.; Tanaka, A.; Yamaguchi, N.; Kawamura, Y.; Hitomi, J. Moraxella species are primarily responsible for generating malodor in laundry. Appl. Environ. Microbiol. 2012, 78, 3317–3324. [Google Scholar] [CrossRef] [Green Version]
- Stapleton, K.; Hill, K.; Day, K.; Perry, J.D.; Dean, J.R. The potential impact of washing machines on laundry malodour generation. Lett. Appl. Microbiol. 2013, 56, 299–306. [Google Scholar] [CrossRef]
- Van Herreweghen, F.; Amberg, C.; Marques, R.; Callewaert, C. Biological and Chemical Processes that Lead to Textile Malodour Development. Microorganisms 2020, 8, 1709. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [Green Version]
- Cardoso-Silva, C.B.; Costa, E.A.; Mancini, M.C.; Balsalobre, T.W.; Canesin, L.E.; Pinto, L.R.; Carneiro, M.S.; Garcia, A.A.; de Souza, A.P.; Vicentini, R. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS ONE 2014, 9, e88462. [Google Scholar] [CrossRef] [Green Version]
- Bharti, R.; Grimm, D.G. Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [CrossRef] [Green Version]
- Ghurye, J.S.; Cepeda-Espinoza, V.; Pop, M. Metagenomic Assembly: Overview, Challenges and Applications. Yale J. Biol. Med. 2016, 89, 353–362. [Google Scholar]
- Wajid, B.; Serpedin, E. Do it yourself guide to genome assembly. Brief. Funct. Genom. 2016, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Koch, C.M.; Chiu, S.F.; Akbarpour, M.; Bharat, A.; Ridge, K.M.; Bartom, E.T.; Winter, D.R. A Beginner’s Guide to Analysis of RNA Sequencing Data. Am. J. Respir. Cell Mol. Biol. 2018, 59, 145–157. [Google Scholar] [CrossRef]
- Oshlack, A.; Robinson, M.D.; Young, M.D. From RNA-seq reads to differential expression results. Genome Biol. 2010, 11, 220. [Google Scholar] [CrossRef] [Green Version]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Schulz, M.H.; Zerbino, D.R.; Vingron, M.; Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 28, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yu, T.; Mu, Z.; Li, G. TransLiG: A de novo transcriptome assembler that uses line graph iteration. Genome Biol. 2019, 20, 81. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Lan, C.; Zhao, L.; Wang, J.; Chen, B.; Chen, Y.-P.P. Recent advances in sequence assembly: Principles and applications. Brief. Funct. Genom. 2017, 16, 361–378. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.; Bradnam, K.; St John, J.; Darling, A.; Lin, D.; Fass, J.; Yu, H.O.K.; Buffalo, V.; Zerbino, D.R.; Diekhans, M.; et al. Assemblathon 1: A competitive assessment of de novo short read assembly methods. Genome Res. 2011, 21, 2224–2241. [Google Scholar] [CrossRef] [Green Version]
- Bradnam, K.R.; Fass, J.N.; Alexandrov, A.; Baranay, P.; Bechner, M.; Birol, I.; Boisvert, S.; Chapman, J.A.; Chapuis, G.; Chikhi, R.; et al. Assemblathon 2: Evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2013, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Falk, N.A. Surfactants as Antimicrobials: A Brief Overview of Microbial Interfacial Chemistry and Surfactant Antimicrobial Activity. J Surfactants Deterg. 2019, 22, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Schages, J.; Stamminger, R.; Bockmühl, D.P. A New Method to Evaluate the Antimicrobial Efficacy of Domestic Laundry Detergents. J Surfactants Deterg. 2020, 23, 629–639. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Booijink, C.C.; Klaassens, E.S.; Heilig, H.G.; Kleerebezem, M.; Smidt, H.; de Vos, W.M. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 2006, 1, 954–959. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore. Babraham Institute. 2012. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 28 April 2021).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [Green Version]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC. Babraham Institute. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 28 April 2021).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Florea, L. Rcorrector: Efficient and accurate error correction for Illumina RNA-seq reads. Gigascience 2015, 4, 48. [Google Scholar] [CrossRef] [Green Version]
- Macmanes, M.D.; Eisen, M.B. Improving transcriptome assembly through error correction of high-throughput sequence reads. PeerJ 2013, 1, e113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Santillán, D.D.; Machain-Williams, C.; Hernández-Montes, G.; Ortega, J. De Novo Transcriptome Assembly and Functional Annotation in Five Species of Bats. Sci. Rep. 2019, 9, 6222. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, A.; Ichihashi, Y.; Farhi, M.; Zumstein, K.; Townsley, B.; David-Schwartz, R.; Sinha, N.R. De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol. 2014, 166, 1186–1199. [Google Scholar] [CrossRef] [Green Version]
- Carradec, Q.; Pelletier, E.; Da Silva, C.; Alberti, A.; Seeleuthner, Y.; Blanc-Mathieu, R.; Lima-Mendez, G.; Rocha, F.; Tirichine, L.; Labadie, K.; et al. A global ocean atlas of eukaryotic genes. Nat. Commun. 2018, 9, 373. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Suvorov, V.; Prjibelski, A.D. rnaQUAST: A quality assessment tool for de novo transcriptome assemblies. Bioinformatics 2016, 32, 2210–2212. [Google Scholar] [CrossRef] [Green Version]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Blast. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B. TransDecoder. Available online: https://github.com/TransDecoder/TransDecoder.wiki.git (accessed on 28 April 2021).
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. HMMER. HMMER. 2020. Available online: http://hmmer.org/ (accessed on 28 April 2021).
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.-H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Trinotate. Available online: https://github.com/Trinotate/Trinotate.github.io/wiki (accessed on 28 April 2020).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [Green Version]
- Stubben, C. trinotateR. 2016. Available online: https://github.com/cstubben/trinotateR (accessed on 28 April 2021).
- The R Project for Statistical Computing. 2020; Available online: https://www.R-project.org/ (accessed on 28 April 2021).
- RStudio Team. R Studio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016; Available online: http://www.rstudio.com/ (accessed on 28 April 2021).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hölzer, M.; Marz, M. De novo transcriptome assembly: A comprehensive cross-species comparison of short-read RNA-Seq assemblers. Gigascience 2019, 8, giz039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Carruthers, M.; Yurchenko, A.A.; Augley, J.J.; Adams, C.E.; Herzyk, P.; Elmer, K.R. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genom. 2018, 19, 32. [Google Scholar] [CrossRef]
- Riesgo, A.; Andrade, S.C.S.; Sharma, P.P.; Novo, M.; Pérez-Porro, A.R.; Vahtera, V.; González, V.L.; Kawauchi, G.Y.; Giribet, G. Comparative description of ten transcriptomes of newly sequenced invertebrates and efficiency estimation of genomic sampling in non-model taxa. Front. Zool. 2012, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [Green Version]
- Geniza, M.; Jaiswal, P. Tools for building de novo transcriptome assembly. Curr. Plant Biol. 2017, 11–12, 41–45. [Google Scholar] [CrossRef]
- Sahraeian, S.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Schein, J.; Chiu, R.; Corbett, R.; Field, M.; Jackman, S.D.; Mungall, K.; Lee, S.; Okada, H.M.; Qian, J.Q.; et al. De novo assembly and analysis of RNA-seq data. Nat. Methods 2010, 7, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.Y.; Wang, Y.; Kong, Y.M.; Luo, D.; Li, X.; Hao, P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: A comparative study. BMC Bioinform. 2011, 12 (Suppl. 14), S2. [Google Scholar] [CrossRef] [Green Version]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [Green Version]
- Pal, B.; Chen, Y.; Vaillant, F.; Capaldo, B.D.; Joyce, R.; Song, X.; Bryant, V.L.; Penington, J.S.; Di Stefano, L.; Tubau Ribera, N.; et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021, 40, e107333. [Google Scholar] [CrossRef]
- Denawaka, C.J.; Fowlis, I.A.; Dean, J.R. Source, impact and removal of malodour from soiled clothing. J. Chromatogr. A 2016, 1438, 216–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frickey, T.; Lupas, A.N. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004, 146, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef] [Green Version]
- Postma, P.W.; Lengeler, J.W.; Jacobson, G.R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 1993, 57, 543–594. [Google Scholar] [CrossRef]
- St Martin, E.J.; Wittenberger, C.L. Regulation and function of sucrose 6-phosphate hydrolase in Streptococcus mutans. Infect. Immun. 1979, 26, 487–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callewaert, C.; de Maeseneire, E.; Kerckhof, F.-M.; Verliefde, A.; van de Wiele, T.; Boon, N. Microbial odor profile of polyester and cotton clothes after a fitness session. Appl. Environ. Microbiol. 2014, 80, 6611–6619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Blaxter, M.L. Comparing de novo assemblers for 454 transcriptome data. BMC Genom. 2010, 11, 571. [Google Scholar] [CrossRef] [Green Version]
- Moreton, J.; Dunham, S.P.; Emes, R.D. A consensus approach to vertebrate de novo transcriptome assembly from RNA-seq data: Assembly of the duck (Anas platyrhynchos) transcriptome. Front. Genet. 2014, 5, 190. [Google Scholar] [CrossRef] [Green Version]
- Scholz, M.; Lo, C.-C.; Chain, P.S.G. Improved assemblies using a source-agnostic pipeline for MetaGenomic Assembly by Merging (MeGAMerge) of contigs. Sci. Rep. 2014, 4, 6480. [Google Scholar] [CrossRef] [Green Version]
- Wences, A.H.; Schatz, M.C. Metassembler: Merging and optimizing de novo genome assemblies. Genome Biol. 2015, 16, 207. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
| Assembly | Gene | Uniprot Entry/ Blast Accession # | Percent Identity | E-Value | Ontology | Name | Genus | log FC | log CPM | FDR | Regulation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trinity | TRINITY_DN12211_c0_g1 | WP_082183191.1 | 100.00 | 2.0 × 10−08 | Energy production and conversion | FAD-binding oxidoreductase | Rhizobium | −9.871 | 5.431 | 0.003 | down |
| TRINITY_DN12794_c0_g1 | WP_126090172.1 | 73.08 | 2.5 × 10−02 | Transcription | LysR family transcriptional regulator | none available | −9.988 | 5.545 | 0.003 | down | |
| TRINITY_DN16017_c0_g1 | KIV68812.1 | 100.00 | 2.0 × 10−36 | Carbohydrate transport and metabolism | Sucrose−6-phosphate hydrolase | Rhizobium | −9.852 | 5.412 | 0.003 | down | |
| TRINITY_DN16123_c0_g1 | IHFA_CHRVO | 41.54 | 1.3 × 10−10 | Transcription | Integration host factor subunit alpha | none available | −9.635 | 5.202 | 0.008 | down | |
| TRINITY_DN19317_c0_g1 | GLO22_ECOLI | 51.90 | 4.3 × 10−15 | Inorganic ion transport and metabolism | Hydroxyacylglutathione hydrolase GloC | Rhizobium | −9.756 | 5.319 | 0.010 | down | |
| TRINITY_DN6555_c0_g1 | WP_164056586.1 | 100.00 | 2.0 × 10−17 | Replication, recombination and repair | AAA family ATPase | Rhizobium | −9.705 | 5.270 | 0.005 | down | |
| TRINITY_DN8425_c0_g1 | WP_142779495.1 | 100.00 | 1.0 × 10−18 | Cell cycle control, cell division, chromosome partitioning | ParA family protein | Rhizobium | −9.602 | 5.170 | 0.008 | down | |
| TRINITY_DN12673_c0_g1 | WP_042878669.1 | 97.53 | 2.0 × 10−25 | Amino acid transport and metabolism, Carbohydrate transport and metabolism | DMT family transporter | Aeromonas | 9.611 | 5.184 | 0.008 | up | |
| TRINITY_DN14275_c0_g1 | WP_174060752.1 | 100.00 | 5.0 × 10−23 | Amino acid transport and metabolism, Inorganic ion transport and metabolism | ABC transporter permease | Rhizobium | 9.590 | 5.163 | 0.010 | up | |
| TRINITY_DN14310_c0_g1 | WP_124801776.1 | 100.00 | 6.0 × 10−11 | Inorganic ion transport and metabolism | cation:proton antiporter | Epilithonimonas | 9.577 | 5.150 | 0.008 | up | |
| TRINITY_DN16876_c0_g1 | YDDG_ECOLI | 66.67 | 6.9 × 10−08 | Amino acid transport and metabolism, Carbohydrate transport and metabolism | Aromatic amino acid exporter YddG | Acinetobacter | 9.751 | 5.318 | 0.003 | up | |
| TRINITY_DN16965_c0_g1 | STY97430.1 | 95.16 | 1.0 × 10−33 | Coenzyme transport and metabolism | Dihydroneopterin aldolase | Moraxella | 9.920 | 5.481 | 0.003 | up | |
| TRINITY_DN19448_c0_g1 | WP_204155761.1 | 82.05 | 4.0 × 10−15 | Cell motility, Intracellular trafficking, secretion, and vesicular transport | prepilin-type N-terminal cleavage/methylation domain-containing protein | Moraxella | 9.832 | 5.396 | 0.003 | up | |
| TRINITY_DN19763_c0_g1 | Y2604_PSEAE | 69.09 | 6.5 × 10−21 | Cell wall/membrane/envelope biogenesis | Uncharacterized protein PA2604 | Pseudomonas | 9.985 | 5.544 | 0.004 | up | |
| TRINITY_DN24104_c0_g1 | PTSBC_SALTM | 79.22 | 7.3 × 10−33 | Carbohydrate transport and metabolism | PTS system sucrose-specific EIIBC component | Aeromonas | 10.140 | 5.695 | 0.005 | up | |
| TRINITY_DN9443_c0_g1 | WP_074855682.1 | 100.00 | 4.0 × 10−18 | Inorganic ion transport and metabolism | ArsC family reductase | Pseudomonas | 9.803 | 5.368 | 0.003 | up | |
| Spades | NODE_11013_length_508_cov_2.086614_g10506 | YOXD_BACSU | 46.39 | 1.7 × 10−35 | Function unknown | Uncharacterized oxidoreductase YoxD | Epilithonimonas | 9.511 | 5.043 | 0.000 | up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacksch, S.; König, C.; Kaiser, D.; Weide, M.; Ratering, S.; Schnell, S.; Egert, M. Metatranscriptomic Analysis of Bacterial Communities on Laundered Textiles: A Pilot Case Study. Microorganisms 2021, 9, 1591. https://doi.org/10.3390/microorganisms9081591
Jacksch S, König C, Kaiser D, Weide M, Ratering S, Schnell S, Egert M. Metatranscriptomic Analysis of Bacterial Communities on Laundered Textiles: A Pilot Case Study. Microorganisms. 2021; 9(8):1591. https://doi.org/10.3390/microorganisms9081591
Chicago/Turabian StyleJacksch, Susanne, Christoph König, Dominik Kaiser, Mirko Weide, Stefan Ratering, Sylvia Schnell, and Markus Egert. 2021. "Metatranscriptomic Analysis of Bacterial Communities on Laundered Textiles: A Pilot Case Study" Microorganisms 9, no. 8: 1591. https://doi.org/10.3390/microorganisms9081591
APA StyleJacksch, S., König, C., Kaiser, D., Weide, M., Ratering, S., Schnell, S., & Egert, M. (2021). Metatranscriptomic Analysis of Bacterial Communities on Laundered Textiles: A Pilot Case Study. Microorganisms, 9(8), 1591. https://doi.org/10.3390/microorganisms9081591






