Development and Comparison of a Panel of Modified CS17 Fimbrial Tip Adhesin Proteins as Components for an Adhesin-Based Vaccine against Enterotoxigenic Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. CS17 Vaccine Antigen Design by Allele Matching and Structural Modelling
2.2. Molecular Cloning and Protein Purification
2.3. Physiochemical Characterization of Proteins
2.4. Murine Immunizations
2.5. Immunological Sampling and Measurement of Immune Response
2.6. Statistical Analysis
3. Results
3.1. Antigen Allele Matching and in Silico Engineering
3.2. Physiochemical Characterization of Vaccine Antigens
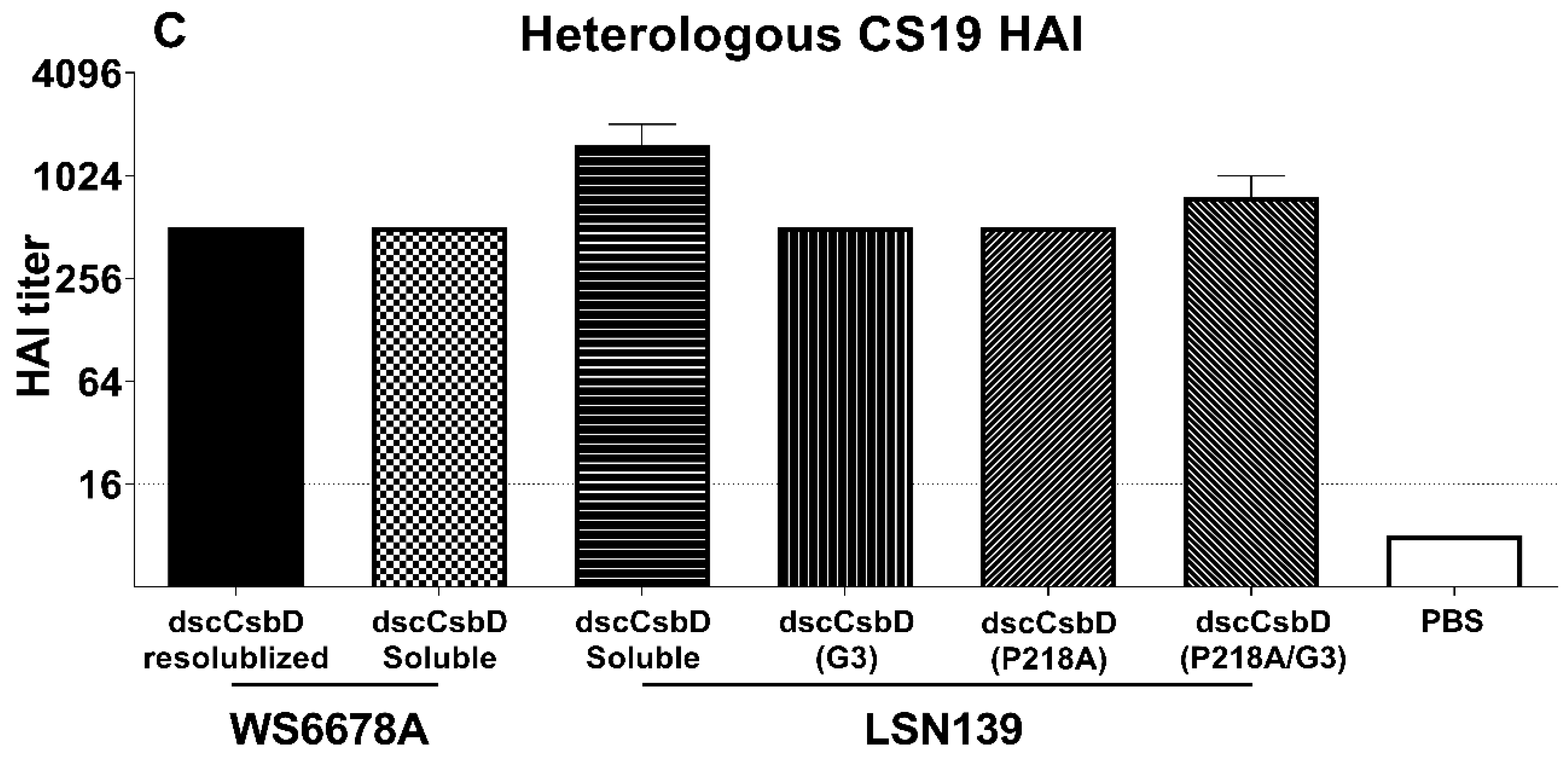
3.3. Immunogenicity and Functional Antibody Responses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Olson, S.; Hall, A.; Riddle, M.S.; Porter, C.K. Travelers’ diarrhea: Update on the incidence, etiology and risk in military and similar populations—1990–2005 versus 2005–2015, does a decade make a difference? Trop. Dis. Travel Med. Vaccines 2019, 5, 1. [Google Scholar] [CrossRef]
- Isidean, S.D.; Riddle, M.S.; Savarino, S.J.; Porter, C.K. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 2011, 29, 6167–6178. [Google Scholar] [CrossRef]
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G.; et al. Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: The Global Burden of Disease Study 1990-2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef] [Green Version]
- Begum, Y.A.; Talukder, K.A.; Azmi, I.J.; Shahnaij, M.; Sheikh, A.; Sharmin, S.; Svennerholm, A.-M.; Qadri, F. Resistance Pattern and Molecular Characterization of Enterotoxigenic Escherichia coli (ETEC) Strains Isolated in Bangladesh. PLoS ONE 2016, 11, e0157415. [Google Scholar] [CrossRef]
- Vidal, R.M.; Muhsen, K.; Tennant, S.M.; Svennerholm, A.M.; Sow, S.O.; Sur, D.; Zaidi, A.K.M.; Faruque, A.S.G.; Saha, D.; Adegbola, R.; et al. Colonization factors among enterotoxigenic Escherichia coli isolates from children with moderate-to-severe diarrhea and from matched controls in the Global Enteric Multicenter Study (GEMS). PLoS Negl. Trop. Dis. 2019, 13, e0007037. [Google Scholar] [CrossRef] [Green Version]
- Anantha, R.P.; McVeigh, A.L.; Lee, L.H.; Agnew, M.K.; Cassels, F.J.; Scott, D.A.; Whittam, T.S.; Savarino, S.J. Evolutionary and functional relationships of colonization factor antigen i and other class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect. Immun. 2004, 72, 7190–7201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, D.G.; Satterwhite, T.K.; Evans, D.J., Jr.; DuPont, H.L. Differences in serological responses and excretion patterns of volunteers challenged with enterotoxigenic Escherichia coli with and without the colonization factor antigen. Infect. Immun. 1978, 19, 883–888. [Google Scholar] [CrossRef] [Green Version]
- Freedman, D.J.; Tacket, C.O.; Delehanty, A.; Maneval, D.R.; Nataro, J.; Crabb, J.H. Milk immunoglobulin with specific activity against purified colonization factor antigens can protect against oral challenge with enterotoxigenic Escherichia coli. J. Infect. Dis. 1998, 177, 662–667. [Google Scholar] [CrossRef] [Green Version]
- Sakellaris, H.; Munson, G.P.; Scott, J.R. A conserved residue in the tip proteins of CS1 and CFA/I pili of enterotoxigenic Escherichia coli that is essential for adherence. Proc. Natl. Acad. Sci. USA 1999, 96, 12828–12832. [Google Scholar] [CrossRef] [Green Version]
- Poole, S.T.; McVeigh, A.L.; Anantha, R.P.; Lee, L.H.; Akay, Y.M.; Pontzer, E.A.; Scott, D.A.; Bullitt, E.; Savarino, S.J. Donor strand complementation governs intersubunit interaction of fimbriae of the alternate chaperone pathway. Mol. Microbiol. 2007, 63, 1372–1384. [Google Scholar] [CrossRef]
- Baker, K.K.; Levine, M.M.; Morison, J.; Phillips, A.; Barry, E.M. CfaE tip mutations in enterotoxigenic Escherichia coli CFA/I fimbriae define critical human intestinal binding sites. Cell Microbiol. 2009, 11, 742–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luiz, W.B.; Rodrigues, J.F.; Crabb, J.H.; Savarino, S.J.; Ferreira, L.C. Maternal vaccination with a fimbrial tip adhesin and passive protection of neonatal mice against lethal human enterotoxigenic Escherichia coli challenge. Infect. Immun. 2015, 83, 4555–4564. [Google Scholar] [CrossRef] [Green Version]
- Rollenhagen, J.E.; Jones, F.; Hall, E.; Maves, R.; Nunez, G.; Espinoza, N.; O’Dowd, A.; Prouty, M.G.; Savarino, S.J. Establishment, Validation, and Application of a New World Primate Model of Enterotoxigenic Escherichia coli Disease for Vaccine Development. Infect. Immun. 2019, 87, e00634-18. [Google Scholar] [CrossRef] [Green Version]
- Giuntini, S.; Stoppato, M.; Sedic, M.; Ejemel, M.; Pondish, J.R.; Wisheart, D.; Schiller, Z.A.; Thomas, W.D., Jr.; Barry, E.M.; Cavacini, L.A.; et al. Identification and Characterization of Human Monoclonal Antibodies for Immunoprophylaxis against Enterotoxigenic Escherichia coli Infection. Infect. Immun. 2018, 86, e00355-18. [Google Scholar] [CrossRef] [Green Version]
- Stoppato, M.; Gaspar, C.; Regeimbal, J.; Nunez, R.G.; Giuntini, S.; Schiller, Z.A.; Gawron, M.A.; Pondish, J.R.; Martin, J.C., 3rd; Schneider, M.I.; et al. Oral administration of an anti-CfaE secretory IgA antibody protects against Enterotoxigenic Escherichia coli diarrheal disease in a nonhuman primate model. Vaccine 2020, 38, 2333–2339. [Google Scholar] [CrossRef] [Green Version]
- Savarino, S.J.; McKenzie, R.; Tribble, D.R.; Porter, C.K.; O’Dowd, A.; Cantrell, J.A.; Sincock, S.A.; Poole, S.T.; DeNearing, B.; Woods, C.M.; et al. Prophylactic Efficacy of Hyperimmune Bovine Colostral Antiadhesin Antibodies Against Enterotoxigenic Escherichia coli Diarrhea: A Randomized, Double-Blind, Placebo-Controlled, Phase 1 Trial. J. Infect. Dis. 2017, 216, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Savarino, S.J.; McKenzie, R.; Tribble, D.R.; Porter, C.K.; O’Dowd, A.; Sincock, S.A.; Poole, S.T.; DeNearing, B.; Woods, C.M.; Kim, H.; et al. Hyperimmune Bovine Colostral Anti-CS17 Antibodies Protect Against Enterotoxigenic Escherichia coli Diarrhea in a Randomized, Doubled-Blind, Placebo-Controlled Human Infection Model. J. Infect. Dis. 2019, 220, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Tchesnokova, V.; McVeigh, A.; Kisiela, D.I.; Dori, K.; Navarro, A.; Sokurenko, E.V.; Savarino, S.J. Adaptive evolution of class 5 fimbrial genes in enterotoxigenic Escherichia coli and its functional consequences. J. Biol. Chem. 2012, 287, 6150–6158. [Google Scholar] [CrossRef] [Green Version]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2006, 5, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobling, M.G.; Poole, S.T.; Rasulova-Lewis, F.; O’Dowd, A.; McVeigh, A.L.; Balakrishnan, A.; Sincock, S.A.; Prouty, M.G.; Holmes, R.K.; Savarino, S.J. Biochemical and immunological characterization of an ETEC CFA/I adhesin cholera toxin B subunit chimera. PLoS ONE 2020, 15, e0230138. [Google Scholar] [CrossRef] [PubMed]
- Koelle, K.; Rasmussen, D.A. Influenza: Prediction is worth a shot. Nature 2014, 507, 47–48. [Google Scholar] [CrossRef]
- Fletcher, L.D.; Bernfield, L.; Barniak, V.; Farley, J.E.; Howell, A.; Knauf, M.; Ooi, P.; Smith, R.P.; Weise, P.; Wetherell, M. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect. Immun. 2004, 72, 2088–2100. [Google Scholar] [CrossRef] [Green Version]
- Volkin, D.B.; Middaugh, C.R. Vaccines as physically and chemically well-defined pharmaceutical dosage forms. Expert Rev. Vaccines 2010, 9, 689–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, K.; Hilliard, G.M.; Hamilton, D.J.; Luo, J.; Ostmann, M.M.; Fleckenstein, J.M. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 2009, 457, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Luo, Q.; Vickers, T.J.; Sheikh, A.; Lewis, W.G.; Fleckenstein, J.M. EatA, an immunogenic protective antigen of enterotoxigenic Escherichia coli, degrades intestinal mucin. Infect. Immun. 2014, 82, 500–508. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Esser, L.; Interlandi, G.; Kisiela, D.I.; Tchesnokova, V.; Thomas, W.E.; Sokurenko, E.; Xia, D.; Savarino, S.J. Tight conformational coupling between the domains of the enterotoxigenic Escherichia coli fimbrial adhesin CfaE regulates binding state transition. J. Biol. Chem. 2013, 288, 9993–10001. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Shahabudin, S.; Farid, S.; Lee, L.H.; McVeigh, A.L.; Maciel, M., Jr.; Poole, S.T.; Broadman, M.; Prouty, M.G.; Savarino, S.J. Cross-Reactivity, Epitope Mapping, and Potency of Monoclonal Antibodies to Class 5 Fimbrial Tip Adhesins of Enterotoxigenic Escherichia coli. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Li, Y.F.; Poole, S.; Nishio, K.; Jang, K.; Rasulova, F.; McVeigh, A.; Savarino, S.J.; Xia, D.; Bullitt, E. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2009, 106, 10793–10798. [Google Scholar] [CrossRef] [Green Version]
- Walker, R.I.; Wierzba, T.F.; Mani, S.; Bourgeois, A.L. Vaccines against Shigella and enterotoxigenic Escherichia coli: A summary of the 2016 VASE Conference. Vaccine 2017, 35, 6775–6782. [Google Scholar] [CrossRef] [PubMed]
- Barry, E.; Cassels, F.; Riddle, M.; Walker, R.; Wierzba, T. Vaccines Against Shigella and Enterotoxigenic Escherichia coli: A summary of the 2018 VASE Conference. Vaccine 2019, 37, 4768–4774. [Google Scholar] [CrossRef] [PubMed]



| Antigens * | Purity (%) | Endotoxin (EU/mg) | Solubility ¶ (mg/mL) | SE-HPLC (% AUC) | Secondary Structure † | Melting Temperature (°C) † |
|---|---|---|---|---|---|---|
| dscCsbDWS6788 (resolubilized) | 97 | 231 | NA | NA | β-sheets | NA |
| dscCsbDWS6788 (soluble) | 100 | 15 | ≥8.0 | 95 | β-sheets | 71 |
| dscCsbDLSN139 | 100 | 22 | ≥8.0 | 98 | β-sheets | 71 |
| dscCsbDLSN139(P218A) | 100 | 279 | ≥8.0 | 89 | β-sheets | 70 |
| dscCsbDLSN139(G3) | 100 | 19 | ≥8.0 | 97 | β-sheets | 73 |
| dscCsbDLSN139(P218A/G3) | 100 | 261 | ≥8.0 | 88 | β-sheets | 73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Maciel, M., Jr.; O’Dowd, A.; Poole, S.T.; Rollenhagen, J.E.; Etobayeva, I.V.; Savarino, S.J. Development and Comparison of a Panel of Modified CS17 Fimbrial Tip Adhesin Proteins as Components for an Adhesin-Based Vaccine against Enterotoxigenic Escherichia coli. Microorganisms 2021, 9, 1646. https://doi.org/10.3390/microorganisms9081646
Liu Y, Maciel M Jr., O’Dowd A, Poole ST, Rollenhagen JE, Etobayeva IV, Savarino SJ. Development and Comparison of a Panel of Modified CS17 Fimbrial Tip Adhesin Proteins as Components for an Adhesin-Based Vaccine against Enterotoxigenic Escherichia coli. Microorganisms. 2021; 9(8):1646. https://doi.org/10.3390/microorganisms9081646
Chicago/Turabian StyleLiu, Yang, Milton Maciel, Jr., Aisling O’Dowd, Steven T. Poole, Julianne E. Rollenhagen, Irina V. Etobayeva, and Stephen J. Savarino. 2021. "Development and Comparison of a Panel of Modified CS17 Fimbrial Tip Adhesin Proteins as Components for an Adhesin-Based Vaccine against Enterotoxigenic Escherichia coli" Microorganisms 9, no. 8: 1646. https://doi.org/10.3390/microorganisms9081646






